Abstract
l-nucleoside analogs represent an important class of small molecules for treating both viral infections and cancers. These pro-drugs achieve pharmacological activity only after enzyme-catalyzed conversion to their tri-phosphorylated forms. Herein, we report the crystal structures of human deoxycytidine kinase (dCK) in complex with the l-nucleosides (−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC)—an approved anti-human immunodeficiency virus (HIV) agent—and troxacitabine (TRO)—an experimental anti-neoplastic agent. The first step in activating these agents is catalyzed by dCK. Our studies reveal how dCK, which normally catalyzes phosphorylation of the natural d-nucleosides, can efficiently phosphorylate substrates with non-physiologic chirality. The capability of dCK to phosphorylate both d- and l-nucleosides and nucleoside analogs derives from structural properties of both the enzyme and the substrates themselves. First, the nucleoside-binding site tolerates substrates with different chiral configurations by maintaining virtually all of the protein-ligand interactions responsible for productive substrate positioning. Second, the pseudo-symmetry of nucleosides and nucleoside analogs in combination with their conformational flexibility allows the l- and d-enantiomeric forms to adopt similar shapes when bound to the enzyme. This is the first analysis of the structural basis for activation of l-nucleoside analogs, providing further impetus for discovery and clinical development of new agents in this molecular class.
INTRODUCTION
Chirality is a hallmark of biological systems: proteins are composed of l-amino acids and DNA and RNA are composed of β-d-nucleotides. It has, therefore, been widely accepted that enzymes that must correctly bind ligands and catalyze chemistry on their respective substrates, will invariably show high chiral selectivity. Quite unexpectedly, however, it was demonstrated that it is the l-component of the racemic mixture (±)-BCH-189 [(±)-β-2′,3′-dideoxy-3′-thiacytidine], which possesses significantly better anti-human immunodeficiency virus (HIV) properties than its d-enantiomeric counterpart (1,2). Since this discovery, two non-physiologic l-nucleoside analogs [(−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC), the l-enantiomer of BCH-189, and its 5-fluorinated analog FTC] have been approved for treatment of human acquired immune deficiency syndrome (AIDS). An additional six d-nucleoside analogs have been approved as AIDS therapeutics, and it is customary for patients to receive both l- and d-enantiomer anti-HIV agents simultaneously. The pharmacological potential of l-nucleoside analogs has been recently reviewed (3–5). The dioxolane analog of 3TC, troxacitabine [TRO, (−)-l-β-2′,3′-dideoxy-3′-oxacytidine; Figure 1A] is currently in a Phase II/III clinical trial for third-line treatment of acute myelogenous leukemia (AML), a blood cancer. The l-nucleoside analogs 3TC and FTC are preferred as anti-virals over their d-analogs [(+)-BCH-189 and (+)-FTC, respectively], primarily because these HIV reverse transcriptase inhibitors do not affect cellular DNA polymerases (4), resulting in minimal toxicity. In contrast, TRO, the first l-nucleoside analog to be used as a cytotoxic anti-cancer agent (6), which differs from 3TC by only one atom (S versus O in the five membered ring, Figure 1A), has potent anti-viral activity yet displays substantial cellular toxicity due to inhibition of DNA polymerases α, β and γ (7). Unlike the d-nucleoside analog anti-cancer agent cytarabine (AraC), TRO is not a substrate for the drug-modifying enzyme cytidine deaminase (6) that confers cytarabine resistance in AML (8).
Figure 1.
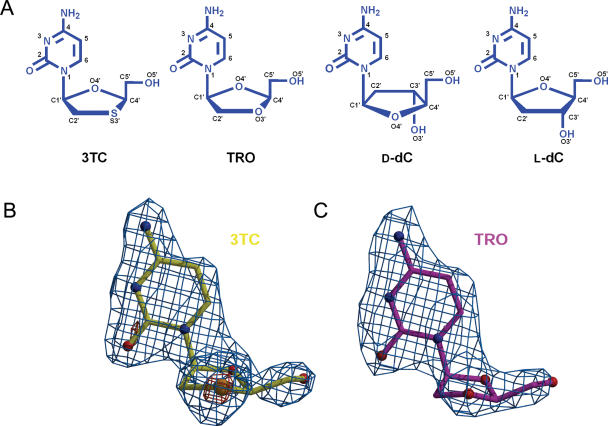
(A) Schematic of the d- and l-nucleosides (all in β-form): 3TC, (−)-l-2′,3′-dideoxy-3′-thiacytidine (Lamivudine, Epivir®); TRO, (−)-l-2′,3′-dideoxy-3′-oxacytidine (Troxacitabine, Troxatyl™); d-dC, d-2′-deoxycytidine, and l-dC, l-2′-deoxycytidine. Experimental electron density map for the l-nucleoside analogs: (B) 3TC is shown in yellow and (C) TRO in magenta. The difference map (|Fobs| − |Fcalc|) is contoured at 3σ (light blue) and 8σ (orange). Note the strong signal given by the sulfur atom in 3TC even at the high sigma cut-off. Figures were generated using Bobscript (26) and Molscript (27), and rendered with Raster3D (28).
Metabolic conversion of 3TC and TRO to pharmacologically active drugs requires serial phosphorylation to their triphosphorylated forms. The essential first step in this process is catalyzed by deoxycytidine kinase (dCK) (6,9–11), which together with the two mitochondrial enzymes thymidine kinase (TK2) and deoxyguanosine kinase (dGK), constitute a family of three closely related non-enantioselective enzymes responsible for nucleoside phosphorylation (12,13). Lack of selectivity pertains to both substrates and enantiomers. dCK phosphorylates dC, dA and dG. TK2 phosphorylates both dC and deoxythymidine (dT), and dGK phosphorylates both dA and dG. The overlapping substrate specificity of these nucleoside kinases reflects their differing sub-cellular localization: dCK is cytosolic, whereas TK2 and dGK are mitochondrial. All three enzymes are capable of phosphorylating both l- and d-enantiomers (14–16).
The question that we set out to answer is what structural features endow certain enzymes, such as dCK, the ability to accept substrates with non-physiological chirality? Answering this question is significant not only for our understanding of enzyme specificity, but also has medicinal ramifications as more l-nucleoside analogs enter clinical trials. To address this issue, we solved the crystal structures of human dCK in complex with the two clinically relevant l-nucleoside analogs, 3TC and TRO. This is the first report of an enzyme in complex with an l-nucleoside.
MATERIALS AND METHODS
Protein crystal preparation
Initial attempts to crystallize wild-type dCK with 3TC and TRO were hampered by reproducibility. Modification of all four surface-exposed cysteine residues yielded a significantly better behaved crystallization candidate, thereby making this study feasible. The dCK variant containing the C9S/C45S/C59S/C146S mutations, designated C4S-dCK, was created from the wild-type dCK gene, using QuickChange® (Stratagene, Inc.). The protein was produced in Escherichia coli and purified to homogeneity (E. Sabini, S. Hazra, M. Konrad and A. Lavie, manuscript in preparation) In short, E.coli BL21(DE3) harboring the pET14b expression plasmids were grown in 2YT media at 37°C, induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and harvested by centrifugation after 4 h. After lysis by sonication and ultracentrifugation, the supernatant was loaded onto a HisTrap HP Ni Sepharose column (GE Healthcare) and washed with buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl and 20 mM imidazole. The enzyme was eluted by the same washing buffer containing 200 mM imidazole. After protein concentration by centrifugation, the protein was injected onto a S-200 gel filtration column (GE Healthcare) equilibrated with 20 mM HEPES (pH 7.5), 200 mM sodium citrate and 2 mM EDTA. The fractions containing the enzyme were concentrated to ∼20 mg/ml and stored at −80°C.
Ternary complexes were formed by mixing each nucleoside analog (3TC, obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS; TRO, provided by SGX Pharmaceuticals, Inc.) with adenosine diphosphate (ADP) (final concentrations 5 mM each) and enzyme (10–20 mg/ml) in 5 mM MgCl2. Crystals were obtained under identical conditions as for wild-type dCK (17) via hanging drop vapor diffusion against a reservoir containing 0.95–1.5 M sodium citrate and 100 mM HEPES (pH 7.5).
Data collection and processing
X-ray data were obtained under standard cryogenic conditions at the Advanced Photon Source using SERCAT beamline BM-22. Diffraction data were indexed, scaled and merged using XDS and XSCALE (18) (see Table 1 for data collection statistics).
Table 1.
Data collection and refinement statistics
| Human C4S-dCK | ||
|---|---|---|
| 3TC-ADP | TRO-ADP | |
| PDB ID | 2NOA | 2NO9 |
| Beamline | SERCAT BM-22 | SERCAT BM-22 |
| Wavelength (Å) | 1.0 | 1.0 |
| Temperature (K) | 100 | 100 |
| Resolution (Å) | 1.8 | 2.15 |
| (1.8–1.9) | (2.15–2.20) | |
| Reflections | ||
| Observed | 156 829 | 156 130 |
| Unique | 50 033 | 30 536 |
| Completeness (%) | 95.3 (74.8) | 99.7 (100.0) |
| Rsym(%) | 5.9 (43.5) | 9.9 (51.1) |
| I/σ(I) | 12.0 (2.3) | 10.9 (4.4) |
| Space group | C2221 | C2221 |
| Unit cell (Å) | ||
| a | 52.6 | 52.9 |
| b | 134.2 | 134.5 |
| c | 154.9 | 155.3 |
| Molecules per a.u. | 2 | 2 |
| Refinement statistics | ||
| Rcryst (%) | 19.2 | 19.5 |
| Rfree (%) | 23.3 | 25.5 |
| Resolution range (Å) | 30–1.8 | 30–2.15 |
| Number of atoms | ||
| Protein | 2014 (chain A) | 1978 (A) |
| 1842 (chain B) | 1872 (B) | |
| Nucleoside | 15 × 2 | 15 × 2 |
| ADP | 27 × 2 | 27 × 2 |
| Water | 197 | 79 |
| R.m.s. deviation | ||
| Bond length (Å) | 0.015 | 0.016 |
| Bond angles (°) | 1.665 | 1.718 |
| Average B-factors (Å2) | ||
| Protein | 26 (A) | 36 (A) |
| 28 (B) | 38 (B) | |
| Main chain | 25 (A) | 35 (A) |
| 27 (B) | 37 (B) | |
| Side chain | 27 (A) | 37 (A) |
| 29 (B) | 39 (B) | |
| ADP | 19 (A) | 29 (A) |
| 21 (B) | 28 (B) | |
| Nucleoside | 20 (A) | 28 (A) |
| 24 (B) | 32 (B) | |
| Waters | 32 | 38 |
Structure determination and refinement
Ternary complex structures were determined via molecular replacement [MOLREP (19)] using the structure of wild-type dCK-d-dC/ADP (17) [PDB Code: 1P60 (20)] as the search model. Refinement was carried out using REFMAC (21) and simulated annealing maps were calculated using CNS (22) (see Table 1 for refinement statistics).
Steady-state kinetic assay
A spectroscopic enzyme-coupled assay was used to determine enzyme activity as described previously (23). This assay couples the production of ADP or UDP by dCK to the phosphorylation of phosphoenolpyruvate (PEP) by pyruvate kinase, followed by oxidation of NADH by lactate dehydrogenase. The decrease in NADH absorption is proportional to the ADP/UDP produced by dCK. The extinction coefficient of NADH limits the precise measurement of Km values below ∼3 μM. For this reason, substrates displaying a Km value below 3 μM are reported in Table 2 to this higher limit. The measurement of kcat values is unaffected by this technical limitation of the assay. Experiments were conducted at 37°C in a kinetic buffer containing 50 mM Tris–HCl (pH 7.5), 100 mM KCl and 5 mM MgCl2 with dCK: 0.35 μM, adenosine triphosphate (ATP)-Mg: 1 mM and uridine triphosphate (UTP)-Mg: 1 mM. Nucleoside/nucleoside analog concentrations were varied between 5 and 100 μM.
Table 2.
Steady state kinetic data of WT and C4S-dCKa
| Enzyme-substrate | Km (ATP) μM | Km (UTP) μM | kcat (ATP) s−1 | kcat (UTP) s−1 | kcat/Km (ATP) s−1M−1 | kcat/Km (UTP) s−1M−1 |
|---|---|---|---|---|---|---|
| WT-TRO | 13.2 ± 0.8 | 10.6 ± 1.3 | 0.095 ± 0.002 | 0.179 ± 0.006 | 7.2 × 103 | 16.9 × 103 |
| C4S-TRO | 56.8 ± 3.0 | 57.8 ± 7.9 | 0.192 ± 0.004 | 0.524 ± 0.030 | 3.4 × 103 | 9.1 × 103 |
| WT-3TC | 3.4 ± 1.0 | 8.0 ± 1.0 | 0.030 ± 0.001 | 0.102 ± 0.001 | 8.8 × 103 | 12.8 × 103 |
| C4S-3TC | 8.4 ± 1.4 | 14.9 ± 1.3 | 0.028 ± 0.002 | 0.072 ± 0.002 | 3.3 × 103 | 4.8 × 103 |
| WT-l-dC | <3 | <3 | 0.012 ± 0.001 | 0.010 ± 0.001 | >4 × 103 | >3 × 103 |
| C4S-l-dC | <3 | <3 | 0.036 ± 0.001 | 0.042 ± 0.001 | >12 × 103 | >14 × 103 |
| WT-d-dC | <3 | <3 | 0.033 ± 0.001 | 0.049 ± 0.001 | >11 × 103 | >16 × 103 |
| C4S-d-Dc | 3.0 ± 0.2 | <3 | 0.173 ± 0.002 | 0.217 ± 0.005 | 57.7 × 103 | >72 × 103 |
aValues shown are the averages of at least two experiments and SDs are shown.
Validation of choice of crystallization protein
To verify that the Cys→Ser mutations in C4S-dCK did not perturb the structure, ternary complex crystals of the C4S variant were obtained in the presence of d-dC/ADP (PDB ID 2NO1) and gemcitabine/ADP (PDB ID 2NO0) and compared to structures previously determined with wild-type enzyme (17). These control complexes crystallized in the same space group as the l-nucleoside complexes, and diffracted to 1.9 (d-dC/ADP complex) and 1.8 Å (gemcitabine/ADP) resolution (E. Sabini, S. Hazra, M. Konrad and A. Lavie, unpublished data). No significant structural differences were noted between corresponding polypeptide chain structures [root mean square deviation (r.m.s.d.) between 241 α carbon atomic positions: wild-type dCK-d-dC/ADP versus C4S-dCK-d-dC/ADP = 0.16 Å; dCK-gemcitabine/ADP versus C4S-dCK-gemcitabine/ADP = 0.60 Å] or substrate positions. Finally, no significant differences in catalytic efficiency were observed between wild-type and C4S-dCK for two distinct phosphate donors and multiple acceptors (Table 2). Together, these control experiments validate the use of the C4S mutant for structure determination. When referring to structures of dCK herein, we make no distinction between wild type and the C4S mutant, unless otherwise stated.
RESULTS AND DISCUSSION
The ternary complex structures of dCK-3TC/ADP and dCK-TRO/ADP were determined at 1.8 and 2.15 Å resolution, respectively. Excellent difference electron density maps were observed for the l-nucleoside analogs (Figure 1B and C). After several rounds of model building and refinement, in which the nucleotide ADP and the respective l-nucleoside analogs were modeled into the density, and addition of water molecules, the final crystallographic R-factors (Rfrees) converged to 19.2% (23.3%) and 19.5% (25.5%) for the 3TC and TRO complexes, respectively (Table 1).
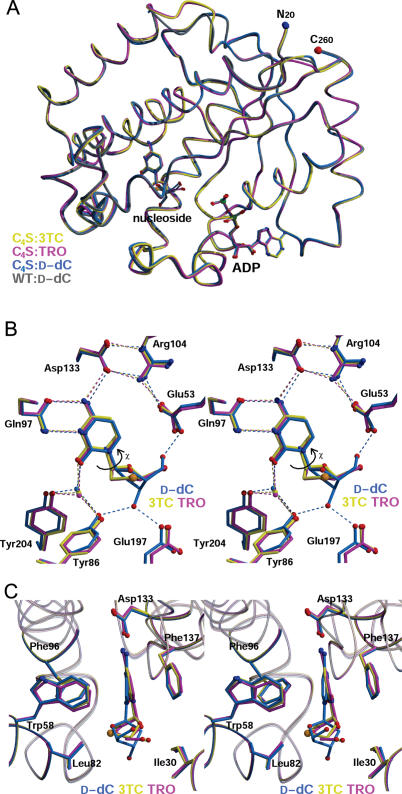
When compared to the ternary complex structures of dCK-d-dC/ADP and dCK-l-dC/ADP (E. Sabini, S. Hazra, M. Konrad and A. Lavie, unpublished data), binding of these l-nucleoside analogs does not affect the structure of the polypeptide chain r.m.s.d. for 239 common α-carbon atoms among pairs of ternary complex structures = 0.18–0.30 Å] nor the positions of the ADP molecules (Figure 2A). In contrast, the mode of l-nucleoside/l-nucleoside analog binding differs from that of d-nucleosides (Figure 2B and C).
Figure 2.
Structures of dCK-3TC/TRO complexes. (A) Shown are four superimposed structures of human dCK: the C4S variant in complex with 3TC/ADP (yellow), TRO/ADP (magenta) and d-dC/ADP (blue); the WT in complex with d-dC/ADP (gray; PDB ID 1P5Z). The structures in complex with d-dC/ADP of the dCK C4S variant and the WT dCK are basically identical, apart from one loop involved in crystal contact. This validates the use of the C4S mutant as a representative of WT dCK. Binding of l-nucleosides also does not elicit major structural perturbations. Non-carbon atom color coding: oxygen-red, nitrogen-blue, phosphorous-green and sulfur-orange. The N- and C-terminal residues visible in the experimental electron density map are labeled N20 and C260, respectively. (B) Stereo representation of the active site showing hydrogen bonding interactions (some water molecules omitted, for clarity). Curved arrow signifies the χ torsion angle around the glycosidic bond. (C) Stereo representation of the active site in an orientation rotated ∼90° relative to (B) depicting the cytosine base in the aromatic slot with Leu82 and Ile30 sandwiching the sugar. Structures have been deposited in the PDB with accession ID 2NOA for C4S-3TC/ADP, 2NO9 for C4S-TRO/ADP and 2NO1 for C4S-d-dC/ADP.
The active site environment surrounding each l-nucleoside analog is shown in Figure 3A, accompanied by similar views for dCK-d-dC/ADP and dCK-l-dC/ADP (Figure 3B). d-dC is the mirror image of l-dC (chirality being determined by the location of the CH2OH group at the C4′ chiral centre or ribose 4′ position, Figures 1A and 3B). 3TC and TRO closely resemble one another, and only differ from l-dC at the 3′ position (Figure 1A). All three l-nucleosides bind similarly to dCK. Detailed comparison of the protein–ligand interactions, presented schematically in Figure 4, reveals the structural basis for non-enantioselective phosphorylation by dCK. For both 3TC and TRO, the enzyme satisfies the same requirements for substrate recognition/catalysis that govern phosphorylation of its primary physiologic substrate, d-dC. First, hydrogen bonding interactions between the protein and the ligand permit the protein to detect the presence of a hydrogen bond donor (NH2) and two hydrogen bond acceptors (N; O) characteristic of the cytosine base (Figure 2B). Second, the five-membered ring resembling the ribose is positioned so as to place the site of phosphorylation (5′-OH) in the appropriate location for phosphoryl transfer from ATP.
Figure 3.
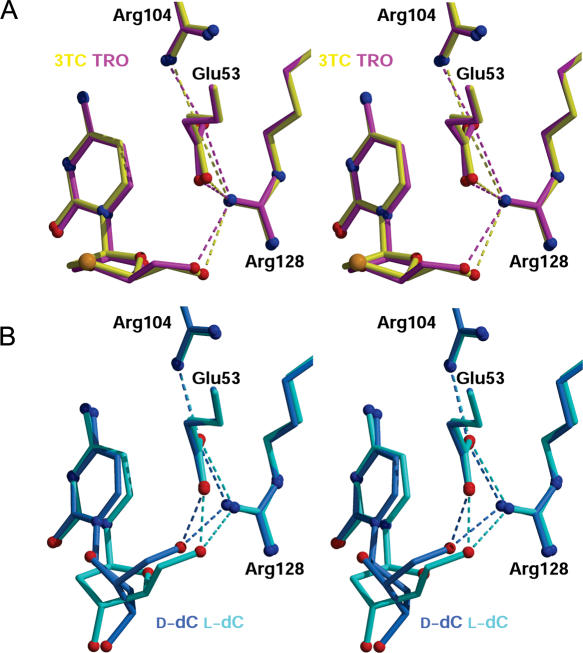
Interactions activating the 5′-OH of the nucleosides for nucleophilic attack. (A) Stereoview depicting the interactions made by the 5′-OH groups of 3TC (yellow) and TRO (magenta). (B) Stereoview depicting the analogous interactions for d-dC (blue) and l-dC (cyan).
Figure 4.
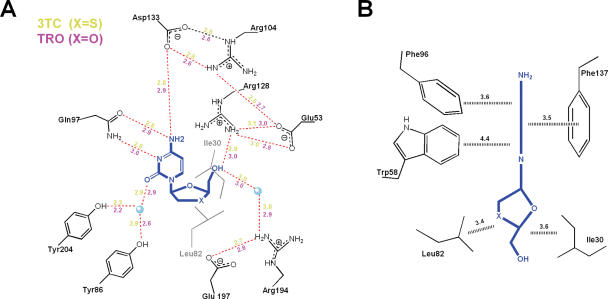
Enzyme-substrate interactions. Schematic representation of the (A) polar and (B) weakly polar and hydrophobic interactions made by 3TC and TRO with the residues in the dCK active site. Distances (Å) are color-coded in (A) with 3TC-yellow and TRO-magenta, while in (B) they are in black and correspond to the average closest contacts for both structures. Water molecules are represented as cyan balls.
Despite the difference in chirality between the physiologic substrate d-dC and the l-nucleoside analogs 3TC/TRO, the location of the cytosine base and its interactions with the enzyme are comparable. In all three ternary complex structures depicted in Figure 2B, Asp133 forms hydrogen bonds with the cytosine amino group. However, it is Gln97 that serves as the primary cytosine recognition element contributing two hydrogen bonds between its side chain and the components of the base that normally engage in Watson–Crick type hydrogen bonding with guanosine (Gln97 O→NH2; Gln97 NH2→N; Figures 2B and 4A). The fourth hydrogen bonding interaction between the protein and the base appears as a water bridge (Tyr204/Tyr86→H2O→O; Figures 2B and 4A). Analogous interactions with comparable hydrogen bond geometries occur between dCK and the cytosine bases of d-dC and l-dC (data not shown). The base itself sits deep in an aromatic slot, which is lined by the side chains of Phe137, Phe96 and Trp58 (all conserved among dCK, dGK and TK2). The aromatic cytosine base is pinioned between Phe137 and Phe96/Trp58. On one face, Phe137 engages in π–π stacking against the cytosine base (interplanar distance ∼3.5 Å). On the opposite face, Phe96 and Trp58 both engage in edge-to-face interactions that place electropositive aromatic hydrogen atoms close to the electronegative π-electron cloud of the cytosine base (Figures 2C and 4B). These aromatic–aromatic interactions exploit enthalpically favorable quadrupole–quadrupole interactions characteristic of aromatic amino acid side chains in proteins as also seen in crystalline benzene (24). The important systematic difference in the position of the cytosine bases of 3TC and TRO versus that of d-dC is a 10° tilt (Figure 2C). We believe that this difference is a general feature of l-enantiomers, as the same base tilt is reproduced in the structure of l-dC bound to dCK (E. Sabini, S. Hazra, M. Konrad and A. Lavie, unpublished data).
In contrast to the numerous interactions between the cytosine base and the aromatic slot, the five membered rings of 3TC and TRO make only limited enzyme contacts (Figures 3A and 4). Critical for catalysis is correct positioning of the 5′-OH, which receives the transferred phosphate group from ATP. In the l-nucleoside analog structures depicted in Figure 3A, the 5′-OH group is hydrogen bonded to the side chain of Arg128. Consequently in each case, the O5′-hydroxyl is close to a side chain oxygen atom of Glu53 (3.1–3.6 Å), the general base that activates 5′-OH group attack of the γ-phosphate of ATP (see Figures 2B and 3). To achieve this critical interaction, concomitant with base tilting, the five-membered rings of 3TC and TRO rotate ∼30° around the glycosidic bond (χ, marked with an arrow in Figure 2B) relative to the dCK-bound conformation of d-dC. A comparable χ rotation observed in the l-dC structure (E. Sabini, S. Hazra, M. Konrad and A. Lavie, unpublished data) provides additional evidence that our structural findings represent the way in which l-nucleosides and l-nucleoside analogs bind to dCK.
The four structures depicted in Figure 3 document that positioning of the 5′-OH group in the active site of dCK is independent of chirality. This remarkable property derives from the internal symmetry and conformational flexibility (i.e. rotation about the glycosidic torsion angle χ) of nucleosides and nucleoside analogs. Once the cytosine base is docked into the aromatic slot, the paucity of protein–ligand interactions involving the five-membered ring and the conformational properties of the nucleoside permit appropriate positioning of the 5′-OH group. Within the aromatic slot, possible glycosidic bond torsion angles are limited by the hydrophobic residues Ile30 and Leu82 (Figures 2C and 4). An additional degree of freedom dictating the precise location of the hydroxyl is the pucker of the five-membered ring. For 3TC and TRO, the ring pucker is equivalent to C3′ endo, whereas for l-dC and d-dC the sugar pucker is C3′ exo. This difference can be attributed in part to the absence of a 3′-OH group on 3TC and on TRO, both of which can act as DNA chain terminators. Finally, a torsion angle rotation about the C4′ and C5′ bond, from gauche in the d-dC structure to trans in the complex structures of 3TC, TRO and l-dC, direct the 5′-OH group towards Glu53. Although these features of 3TC and TRO conformational flexibility bring the 5′-OH group near to Glu53 (3.1–3.6 Å; Figure 3A), they do not stabilize the very close, so-called catalytic, interactions seen with l-dC and d-dC (2.4–2.5 Å; Figure 3B). However, the fact that dCK shows similar catalytic efficiency towards 3TC/TRO and l-dC (Table 2) suggests that the slightly sub-optimal position observed for the 5′-OH group in the 3TC/TRO structures does not impede catalysis.
In conclusion, the structures reported herein document that dCK phosphorylation of l-nucleosides and l-nucleoside analogs is due to both the nature of the enzyme active site and the nature of the substrates. We suggest that lack of enantiomeric selectivity may reflect the central role dCK plays in nucleotide salvage, i.e. phosphorylating both the pyrimidine dC and the purines dA and dG within the cytosolic compartment. The by-product of such promiscuity appears to be non-enantioselectivity. Similar arguments can be invoked to explain why the related enzymes dGK, which phosphorylates dA and dG, and TK2, which phosphorylates dT and dC, are not enantioselective. In contrast, human TK1, which is restricted to d-dT phosphorylation, is exquisitely enantioselective (25). We suggest that enzymes that evolved to catalyze more than one chemically distinct nucleoside substrate acquired less restrictive active sites, which in turn endowed such enzymes with hitherto unexpected catalytic activities for substrates with non-physiologic chirality.
Acknowledgments
The authors thank the SERCAT staff for help in data collection, and Dr Chung K. (David) Chu for a troxacitabine sample and helpful discussions. The authors acknowledge Dr Gilles Gosselin for providing a sample of l-dC. This work was supported by the NIH (E.S., S.H. and A.L.) and the Max Planck Society (M.K.). 3TC was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Funding to pay the Open Access publication charges for this article was provided by NIH grant RO1 CA95687.
Conflict of interest statement. One of the authors (S.K.B.) is an employee of SGX Pharmaceuticals, which is testing the compound troxacitabine as a therapeutic agent.
REFERENCES
- 1.Schinazi R.F., Chu C.K., Peck A., McMillan A., Mathis R., Cannon D., Jeong L.S., Beach J.W., Choi W.B., Yeola S., et al. Activities of the four optical isomers of 2′,3′-dideoxy-3′-thiacytidine (BCH-189) against human immunodeficiency virus type 1 in human lymphocytes. Antimicrob. Agents Chemother. 1992;36:672–676. doi: 10.1128/aac.36.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coates J.A., Cammack N., Jenkinson H.J., Mutton I.M., Pearson B.A., Storer R., Cameron J.M., Penn C.R. The separated enantiomers of 2′-deoxy-3′-thiacytidine (BCH 189) both inhibit human immunodeficiency virus replication in vitro Antimicrob. Agents Chemother. 1992;36:202–205. doi: 10.1128/aac.36.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Focher F., Spadari S., Maga G. Antivirals at the mirror: the lack of stereospecificity of some viral and human enzymes offers novel opportunities in antiviral drug development. Curr. Drug Targets Infect. Disord. 2003;3:41–53. doi: 10.2174/1568005033342163. [DOI] [PubMed] [Google Scholar]
- 4.Gumina G., Chong Y., Choo H., Song G.-Y., Chu C. l-Nucleosides: antiviral activity and molecular mechanism. Curr. Top Med. Chem. 2002;2:1065–1086. doi: 10.2174/1568026023393138. [DOI] [PubMed] [Google Scholar]
- 5.Maury G. The enantioselectivity of enzymes involved in current antiviral therapy using nucleoside analogues: a new strategy? Antivir. Chem. Chemother. 2000;11:165–189. doi: 10.1177/095632020001100301. [DOI] [PubMed] [Google Scholar]
- 6.Grove K.L., Guo X., Liu S.H., Gao Z., Chu C.K., Cheng Y.C. Anticancer activity of beta-l-dioxolane-cytidine, a novel nucleoside analogue with the unnatural L configuration. Cancer Res. 1995;55:3008–3011. [PubMed] [Google Scholar]
- 7.Kukhanova M., Liu S.H., Mozzherin D., Lin T.S., Chu C.K., Cheng Y.C. l- and d-enantiomers of 2′,3′-dideoxycytidine 5′-triphosphate analogs as substrates for human DNA polymerases. Implications for the mechanism of toxicity. J. Biol. Chem. 1995;270:23055–23059. doi: 10.1074/jbc.270.39.23055. [DOI] [PubMed] [Google Scholar]
- 8.Galmarini C.M., Mackey J.R., Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 9.Chang C.N., Skalski V., Zhou J.H., Cheng Y.C. Biochemical pharmacology of (+)- and (−)-2′,3′-dideoxy-3′-thiacytidine as anti-hepatitis B virus agents. J. Biol. Chem. 1992;267:22414–22420. [PubMed] [Google Scholar]
- 10.Shewach D.S., Liotta D.C., Schinazi R.F. Affinity of the antiviral enantiomers of oxathiolane cytosine nucleosides for human 2′-deoxycytidine kinase. Biochem. Pharmacol. 1993;45:1540–1543. doi: 10.1016/0006-2952(93)90058-5. [DOI] [PubMed] [Google Scholar]
- 11.Grove K.L., Cheng Y.C. Uptake and metabolism of the new anticancer compound beta-L-(−)-dioxolane-cytidine in human prostate carcinoma DU-145 cells. Cancer Res. 1996;56:4187–4191. [PubMed] [Google Scholar]
- 12.Eriksson S., Munch-Petersen B., Johansson K., Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell. Mol. Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Chattopadhyaya J., Eriksson S. The enantioselectivity of the cellular deoxynucleoside kinases. Nucleosides Nucleotides. 1999;18:807–810. doi: 10.1080/15257779908041567. [DOI] [PubMed] [Google Scholar]
- 14.Gaubert G., Gosselin G., Boudou V., Imbach J.L., Eriksson S., Maury G. Low enantioselectivities of human deoxycytidine kinase and human deoxyguanosine kinase with respect to 2′-deoxyadenosine, 2′-deoxyguanosine and their analogs. Biochimie. 1999;81:1041–1047. doi: 10.1016/s0300-9084(99)00331-4. [DOI] [PubMed] [Google Scholar]
- 15.Verri A., Focher F., Priori G., Gosselin G., Imbach J.L., Capobianco M., Garbesi A., Spadari S. Lack of enantiospecificity of human 2′-deoxycytidine kinase: relevance for the activation of beta-L-deoxycytidine analogs as antineoplastic and antiviral agents. Mol. Pharmacol. 1997;51:132–138. doi: 10.1124/mol.51.1.132. [DOI] [PubMed] [Google Scholar]
- 16.Verri A., Priori G., Spadari S., Tondelli L., Focher F. Relaxed enantioselectivity of human mitochondrial thymidine kinase and chemotherapeutic uses of L-nucleoside analogues. Biochem. J. 1997;328:317–320. doi: 10.1042/bj3280317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabini E., Ort S., Monnerjahn C., Konrad M., Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nature Struct. Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 19.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 20.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A.T., Adams P.D., Clore G.M., Delano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewskli J., Nilges N., Read R.J., et al. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Agarwal K.C., Miech R.P., Parks R.E., Jr . Guanylate kinases from human erythrocytes, hog brain, and rat liver. In: Carter C.W., Sweet R.M., editors. Methods Enzymol. Vol. 51. 1978. pp. 483–490. [DOI] [PubMed] [Google Scholar]
- 24.Burley S.K., Petsko G.A. Weakly polar interactions in proteins. Adv. Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Choudhury D., Chattopadhyaya J., Eriksson S. Stereoisomeric selectivity of human deoxyribonucleoside kinases. Biochemistry. 1999;38:16993–16999. doi: 10.1021/bi9908843. [DOI] [PubMed] [Google Scholar]
- 26.Esnouf R. An extensively modified version of molscript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 28.Merrit E.A., Murphy M.E.P. Raster3D Version 2.0—a program for photorealistic molecular graphics. Acta Cryst. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]