Abstract
Abf1 and Rap1 are general regulatory factors (GRFs) that contribute to transcriptional activation of a large number of genes, as well as to replication, silencing and telomere structure in yeast. In spite of their widespread roles in transcription, the scope of their functional targets genome-wide has not been previously determined. Here, we use microarrays to examine the contribution of these essential GRFs to transcription genome-wide, by using ts mutants that dissociate from their binding sites at 37°C. We then combine this data with published ChIP-chip studies and motif analysis to identify probable direct targets for Abf1 and Rap1. We also identify a substantial number of genes likely to bind Rap1 or Abf1, but not affected by loss of GRF binding. Interestingly, the results strongly suggest that Rap1 can contribute to gene activation from farther upstream than can Abf1. Also, consistent with previous work, more genes that bind Abf1 are unaffected by loss of binding than those that bind Rap1. Finally, we show for several such genes that the Abf1 C-terminal region, which contains the putative activation domain, is not needed to confer this peculiar ‘memory effect’ that allows continued transcription after loss of Abf1 binding.
INTRODUCTION
Abf1 and Rap1 are multipurpose, abundant, site-specific DNA-binding proteins that function in transcriptional activation, silencing and replication in the budding yeast Saccharomyces cerevisiae (1–3). Rap1 and Abf1 are essential for cell viability, and their multifunctional character has caused them to be termed General Regulatory Factors (GRFs). Binding sites for Abf1 and Rap1 are found in a large number of promoters, and have been shown to be important for activation of many of the corresponding genes. Genome-wide localization, or ‘Chromatin immunoprecipitation (ChIP)-on-chip’ experiments indicate that Rap1 and Abf1 bind ∼200–300 promoters each, and Rap1 has been found to bind with 122 out of 137 ribosomal protein (RP) genes (4–6). Genes containing Rap1 or Abf1 sites principally comprise RP genes and genes encoding proteins involved in amino acid biosynthesis, regulation of carbon source and sporulation. Many of these genes are among those most highly expressed in the yeast genome.
The broad scope of promoters bounded by Abf1 and Rap1, and the importance of such promoters in basic cellular metabolism, underscore the key role that these GRFs play in gene control in yeast. Consistent with this notion, genome-wide interaction studies have found both Abf1 and Rap1 to be network ‘hubs’, with the implication that they play central and coordinating roles in cell function (7,8). Thus, identifying functional targets of Abf1 and Rap1 is important in constructing a ‘circuit diagram’ of gene control in yeast. To this end, we report here the effect of loss of Abf1 and loss of Rap1 binding on genome-wide transcription. By combining our microarray results with genome-wide location results and sequence analyses, we identify probable direct targets of Rap1 and Abf1. Our results also indicate that many promoters that are controlled by Abf1 do not require continued binding by Abf1 for ongoing transcription, consistent with investigations of individual promoters (9,10). In contrast, loss of Rap1 at Rap1 binding promoters more commonly leads to significantly decreased transcription. Furthermore, we find that Rap1 activates genes from more distant sites than does Abf1, suggesting a previously unrecognized mechanistic difference between these two GRFs. Taken together, the results presented here provide new insight into the roles of Abf1 and Rap1 in genome-wide transcriptional regulation.
MATERIALS AND METHODS
Plasmids
The expression vector for abf1(1–592) ts was constructed by replacing the small NheI–XhoI fragment from pRS315abf1-1 with the corresponding fragment from pRS315Abf1(1–592), and the resulting plasmid shuttled into TMY86 as described previously (2).
Yeast strains and growth
The S.cerevisiae strains used in this study are listed in Table 1; all are derived from W303a. Transformations were done using a standard lithium acetate protocol (11), and yeast cells were grown at 30°C in complete synthetic medium (CSM) [6.7 g/l yeast nitrogen base without amino acids, 2% glucose and CSM dropout mixture (Bio101)] when selection was required, or in rich medium (YPD) (1% Bacto–yeast extract, 2% Bacto–peptone and 2% glucose). For temperature shift experiments, yeast were grown to mid-log phase in YPD medium at 23–25°C and an equal volume of medium at 50°C was added to bring the culture rapidly to 37°C; growth was then continued for 1–2 h (as indicated in the text) at 37°C.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| 1–731 | Mat a ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 pRS416-Abf1(1–731) | (9) |
| abf1 ts | Mat a ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 pRS416-abf1-1 | (9) |
| abf1(1–592)ts | Mat a ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 pRS416-abf1-1(1-592) | This study |
| Abf1-FLAG | Mat a ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 pRS416-Abf1-FLAG | (36) |
| abf1-1-FLAG | Mat a ade2-1 his3-11,15, leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 pRS416-abf1-1-FLAG | (36) |
| RMY28Δa | MATa, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1, rps28aΔabf1 | This study |
| YDS2 | MATa, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1 | (18) |
| YDS408 | MATa, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1, rap1-2ts | (18) |
Strain RMY28Δa, in which the Abf1 binding site in the promoter of RPS28A is mutated (replacing CGTCTAGAGTGAC with CATGG), was constructed by two-step replacement in GA208 using the URA3 gene (12). The modified RPS28A promoter from the resulting strain was verified by PCR amplification and sequencing of the promoter region.
ChIP and RNA analysis
ChIP and northern analysis were performed as described previously (9). For FLAG IP, 1.2 μl (6 μg) of antibody (Sigma) were used per IP reaction. Probes were prepared by PCR. For analysis of RPS28A mRNA, purified RNA was reverse transcribed with primers for RPS28A (GACGAGCTTCACGTTCAGATTCCATTAG) and PYK1 (TTTCGTGGTTTGGTGGGATT), using Bioscript reverse transcriptase (Bioline USA Inc., Randolph, MA). The resulting cDNA was amplified by PCR for 15–20 cycles, after pilot experiments to determine the range over which exponential amplification was observed. For quantification, a low number of cycles (usually ∼15) was used, and the products separated by gel electrophoresis and analyzed by Southern blotting and Phosphorimager analysis, as described previously (9). A control experiment using an rps28aΔ yeast strain yielded no product using this protocol.
Microarray analysis
RNA was prepared from exponentially growing yeast cells (A600 = 0.8 − 1.2) grown in YPD using the Masterpure yeast RNA purification kit (Epicenter Technology, Madison, WI). RNA was further purified using the RNeasy purification kit (Qiagen). Processing and hybridization using Affymetrix S98 microarrays (Affymetrix, Santa Clara, CA) were performed as per manufacturer's instructions. Changes in gene expression were calculated by averaging log2 expression changes, and false discovery rates (FDRs) (13) were calculated in Genespring, after normalizing within each experiment (i.e. wild-type and ts mutant analyzed in parallel). Comparative analyses were done using Excel (Microsoft) and sequence analysis was performed using MEME (http://meme.sdsc.edu/meme/) and regulatory site analysis tools) (RSAT) (http://rsat.ccb.sickkids.ca/) (14,15). Sequence logos (16) were generated from position specific weight matrices (derived using RSAT) using the webserver at http://weblogo.berkeley.edu/ (17).
Microarray accession number
Microarray gene expression data are available at the Gene Expression Omnibus under accession no. GSE6073, and are also available at www.wadsworth.org/resnres/bios/morse.htm and as Supplementary Data.
RESULTS
Genome-wide effects of loss of Abf1 or Rap1 binding
To assess the genome-wide effect of loss of Abf1 binding on transcription, we performed microarray analysis of mRNA from yeast harboring the abf1-1 ts mutation as well as from a congenic wild-type yeast strain after growth in rich medium (YPD) for 1 h at 37°C. RNA samples from three replicates were analyzed using Affymetrix microarrays. Similarly, genome-wide expression of wild-type and congenic rap1-2ts yeast incubated for 1 h at 37°C was also compared by microarray analysis. Neither abf1-1 nor rap1-2 yeast exhibit phenotypes that are characteristic of specific cell cycle arrest at the restrictive temperature (18,19). Consistent with this, no enrichment of 807 genes that exhibit cell cycle dependent regulation (20) was observed among genes showing altered transcription in abf1-1 yeast compared to wild-type at 37°C (data not shown). A modest enrichment was observed among genes showing decreased transcription in rap1 ts yeast compared to wild-type yeast at 37°C, with 76 out of 807 cell cycle regulated genes being among the 100 genes showing most strongly decreased transcription. However, this enrichment evidently did not reflect an altered cell cycle, as these genes showed dispersed peak expression patterns in the cell cycle, with peaks in S, G1/S, G1 and G2/M (20) (data not shown). Furthermore, the large majority of genes regulated by the cell cycle were not enriched among those showing altered expression in rap1 ts yeast compared to wild-type. Thus, cell cycle effects do not represent a major contribution to the observed changes in gene expression in abf1-1 or rap1 ts yeast.
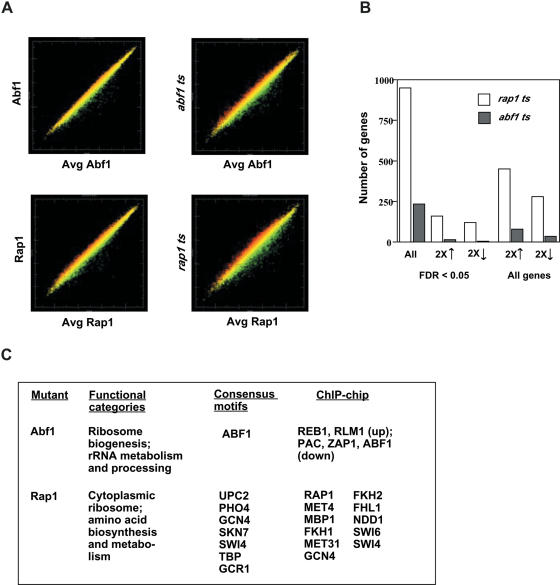
Analysis of the microarray data indicates that global gene expression is affected much more by the loss of Rap1 binding than by loss of Abf1 binding (Figure 1). We found 235 genes (215 after removing cell cycle regulated genes) having changed expression upon loss of Abf1 binding with FDR <0.05, compared to 947 genes (833 after removing cell cycle regulated genes) with FDR <0.05 upon loss of Rap1 binding. Similarly, >10 times as many genes within these sets showed changed expression by 2-fold upon loss of Rap1 binding than upon loss of Abf1 binding. These results are not due to more genes having Rap1 than Abf1 binding sites, as genome-wide location analysis (ChIP-chip analysis) indicates a similar number of promoters binding Rap1 and Abf1 under our growth conditions (4–6) as well as in vitro (21). Another possible explanation is that the quality of the data for the Rap1 experiments is better than for the Abf1 experiments, resulting in a higher fraction of ‘true’ positives having low FDRs for the former. However, even ignoring FDR values, we still observe considerably more genes showing changed expression upon loss of Rap1 than upon loss of Abf1 (Figure 1B). Analysis of this data to identify likely direct targets of Abf1 and Rap1 indicates that the larger effect genome-wide of loss of Rap1 compared to loss of Abf1 binding arises in part from Rap1 exerting a larger indirect effect on gene expression, and in part from fewer Abf1 binding than Rap1 binding targets being affected by loss of factor binding (see below).
Figure 1.
Microarray analysis of gene expression in rap1 ts or abf1 ts yeast compared to wild-type strains at 37°C. (A) Scatter plots of gene expression at 37°C in single mRNA preparations from wild-type or ts yeast strains, as indicated, plotted against average gene expression from three independent preparations from wild-type strains. Each point represents the expression of an individual gene. Only genes indicated by Affymetrix software as being present (i.e. expressed above background levels of detection) are shown. (B) Number of genes showing changed expression with FDR <0.05 and subsets with increased or decreased expression by 2-fold (left), and the total number of genes showing 2-fold increased or decreased expression without regard for FDR (right). (C) Major functional categories, consensus motifs, and transcription factors associated with differentially expressed genes from ChIP-chip data (6), derived from T-profiler (22). Only motifs or factors having E-value <0.01 are shown, and all were associated with decreased expression for Rap1.
We used the Funspec and T-profiler web tools to examine functional categories, promoter motifs and transcription factor binding sites derived from ChIP-chip data that were enriched in the gene sets responsive to loss of Abf1 or Rap1 binding (Figure 1C) (22,23). Funspec assigns genes in a specified list (in this case, genes most down-regulated upon loss of Abf1 or Rap1 binding) to functional categories and determines P-values for enriched categories, using Fisher's exact test (i.e. based on a hypergeometric distribution). T-profiler models gene activity as arising from a linear combination of transcription factor binding sites (consensus motifs) or transcription factors (ChIP-chip data) at each promoter, and models the entire set of microarray data on this basis, yielding an output of motifs or factors showing greatest contribution to gene expression changes. In both cases, the Abf1 data were enriched in mitochondrial ribosome biogenesis and the Rap1 data in ribosome biogenesis and function, and amino acid biosynthesis and metabolism. These represent known functions of Abf1 and Rap1 [(9) and references therein] and, therefore, this result indicates that gene expression changes measured by microarray analysis upon loss of binding accurately reflects the contribution of these GRFs to gene regulation.
In addition, the only consensus motif predicted to contribute to the change in expression in the Abf1 data set was that for Abf1, and Abf1 was one of only a few factors identified as contributing on the basis of ChIP-chip data (Figure 1C). In contrast, Rap1 was one of several factors identified as contributing to expression in the Rap1 dataset, with other identified factors (Met4, Fkh1, Met31, Gcn4, Fkh2 and Fhl1) or motifs (Pho4 and Gcr1) reflecting the known role of Rap1 in ribosomal biogenesis and amino acid biosynthesis (3,24). Several of these factors are indeed known to operate in conjunction with Rap1 in gene expression (25–29), and it is possible that expression of some genes that bind Rap1 but do not suffer significant loss in expression upon loss of Rap1 binding is maintained by these or other auxiliary transcription factors. However, the finding of few such factors or motifs contributing to expression of the Abf1 data set argues against the idea that a small set of auxiliary factors that bind together with Abf1 at gene promoters is responsible for the continued expression of Abf1 binding genes upon loss of Abf1 binding. Interestingly, no Rap1 consensus motif emerged from T-profiler analysis, possibly reflecting the wide variety of sequences capable of binding to Rap1 (30). Alternatively, the Rap1 consensus motif may have been obscured by noise if a large proportion of affected genes were not direct targets of Rap1 (see below).
Finally, we note that we did not observe any significant telomeric clustering of genes showing increased expression upon loss of Rap1 or Abf1 binding using Pyxis, which employs nearest neighbor probability calculations to determine significant gene clustering (31) (data not shown). Although such an effect might have been predicted, based on the well known role of Rap1 in telomeric silencing (32), it has also been shown that native yeast telomeres do not exhibit gene silencing properties identical with those of the truncated telomeres used in many telomeric silencing studies (33). It is also possible that subtelomeric heterochromatin remains stable following the loss of Rap1 binding and therefore, exerts a continued repressive effect.
Identification of direct targets of Rap1 and Abf1
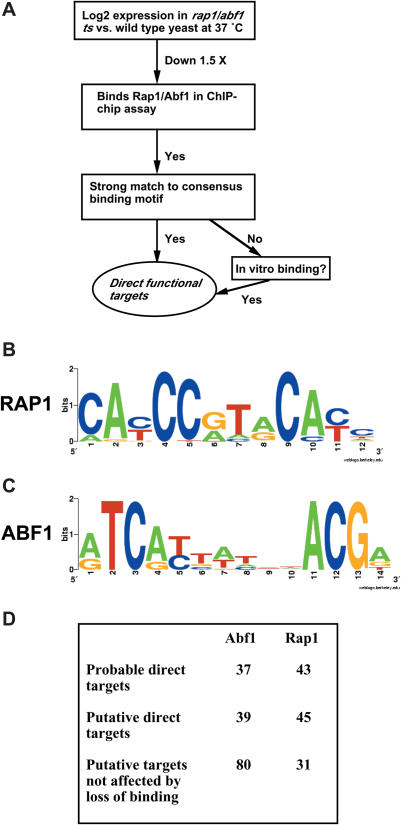
To identify genes likely to be directly controlled by Rap1, we first selected those genes identified as showing decreased expression by >1.5× (log2 change less than −0.59) in rap1 ts yeast compared to wild-type at 37°C (averages of three replicate microarray experiments) and binding Rap1 with P < 0.001 in asynchronous yeast grown in rich medium (YPD), the same conditions used in our microarray analyses [(6) and Figure 2]. Thirty-four such genes were found, 3 of which shared common promoters, leaving 31 unique promoters. These 31 promoters were analyzed for common sequence motifs using MEME (15) and two motifs identified. One of these was a T-rich motif, and the other was a clear match to previously reported Rap1 binding sites [(6,21,30) and Figure 2B]. We next used the position specific weight matrix (PSWM) representing the identified Rap1 binding site to search for Rap1 binding sites in these 31 promoters, and found 26 with significant matches to the Rap1 binding sequence. Using a protein binding microarray (PBM) protocol, 23 of these promoters were found to bind Rap1 in vitro (21), further supporting their assignment as highly probable direct targets of Rap1. Impressively, 18 of these 23 promoters contain Rap1 binding sites that are conserved among five closely related S.cerevisiae species, including 11 of 14 RP gene promoters (21). It seems likely in this case that the criterion of orthologous conservation employed is too stringent and excludes valid targets of Rap1. In addition, 2 of our original 31 promoters, CPA1 and ERG5, lacking a Rap1 binding site or having a marginal sequence match, bind Rap1 in the PBM assay but again are not conserved among related Saccharomyces species.
Figure 2.
Identification of gene targets of Abf1 and Rap1. (A) Flow chart for identification of direct functional targets. (B) Logo for Rap1 site. (C) Logo for Abf1 site. Total height of letters in each column reflects the total information content for that position, and the height of each individual letter reflects the relative frequency of a specific base at that position. (D) Summary of identified targets, as discussed in text and listed in Supplementary Tables 1–6.
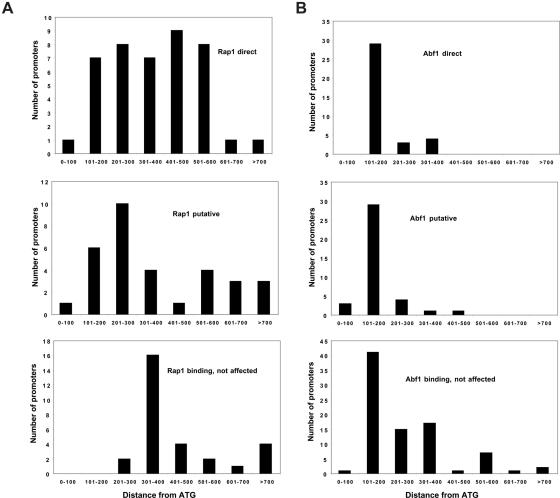
We similarly analyzed an additional 21 promoters showing decreased transcription by 2× or greater in rap1 ts compared to wild-type yeast at 37°C, and identified as binding Rap1 by Lieb et al. but not by Harbison et al. (5,6), using the PSWM defined above. (We used a slightly more stringent criterion for decreased transcription—2× instead of 1.5×—because P-values were not available as a quantitative criterion in the data of Lieb et al.) Twelve were found to have significant matches to the Rap1 binding sequence, and eight of these bound Rap1 in the in vitro binding PBM assay. In addition, 3 of the original 21 promoters bound Rap1 in vitro but did not have strong matches to the consensus site. Taking all these data together, we assigned 43 genes as probable direct targets of Rap1. These genes, together with their expression levels (34) and the location and sequence of the identified binding site(s), are listed in Supplementary Table 1. As expected, many of these genes are RP genes, and many are highly expressed. Interestingly, 27 of these 43 genes contain Rap1 sites >300 bp from the starting ATG, indicating that Rap1 exerts its activating effect on transcription over a relatively long distance compared to many yeast activators (Figure 3A). It is also notable that the large majority of highly expressed Rap1 targets identified here have their Rap1 binding site in the same orientation (as shown in Figure 2B).
Figure 3.
Frequency of Abf1 and Rap1 sites in target genes as a function of distance from ATG. Positions are given for the first nucleotide in the recognition sequence as defined in Figure 2; for promoters with more than one binding site, only the closest site to the ATG was used, so that each promoter was counted only once. Sites are grouped according to the categories discussed in the text and listed in Supplementary Tables 1–6. (A) Rap1 targets; (B) Abf1 targets.
We next identified additional putative targets of Rap1. These were defined as genes that (i) show decreased transcription by >2× in rap1 ts compared to wild-type yeast, and (ii) have a signficant match to the Rap1 binding sequence, as defined by its PSWM from above, or bind to Rap1 in vitro (PBM assay), but (iii) do not bind Rap1 in ChIP-chip studies. Of 391 unique promoters that showed at least 2-fold decreased transcription in rap1 ts compared to wild-type yeast but did not bind Rap1 in ChIP-chip studies (5,6), 30 contained sequences matching the Rap1 binding sequence and 16 of these bind Rap1 in vitro (PBM assay). An additional 15 promoters do not contain significant matches to the Rap1 binding site, but do bind Rap1 in vitro. Together, we categorize these 45 promoters as putative direct functional targets of Rap1 (Supplementary Table 2). Since these promoters derived from a set of 391 promoters showing 2-fold decreased transcription upon loss of Rap1 binding, this result suggests that a substantial fraction of genes affected upon loss of Rap1 binding are affected indirectly.
We also identified genes whose promoters are likely to bind Rap1 but that are unaffected upon loss of Rap1 binding. We selected 37 unique promoters for which the log2 change fell between −0.2 and 0.2, and which were identified as binding Rap1 with P < 0.001 (6). We eliminated from consideration four promoters for which a divergent transcript showed a decrease of at least 1.5-fold in rap1 ts yeast at 37°C. Significant matches to the Rap1 binding motif (Figure 2B) were found in 29 of the remaining 33 promoters, and 27 of these 29 also bind Rap1 in vitro (21). In addition, two sequences apparently lacking significant Rap1 binding sites, RPS26A and RPL33B, were found to bind Rap1 in vitro (21), yielding 31 genes identified as binding Rap1 but not affected by loss of Rap1 binding (Supplementary Table 3).
A similar analysis for Abf1 revealed that of 38 unique promoters showing decreased transcription (>1.5×) and binding Abf1 in ChIP-chip analysis (P < 0.001) (6), 37 had strong matches to the Abf1 motif identified from this set (Figure 2C and Supplementary Table 4). As with Rap1, the identified Abf1 motif is consistent with earlier work (6,21,35). Surprisingly, only 10 of these promoters were found to bind Abf1 in vitro using the PBM assay; eight of these promoters contained sites conserved in related Saccharomyces species (21). Furthermore, in contrast to promoters targeted by Rap1, the large majority (32 out of 37) of these promoters contained Abf1 binding sites within 300 bp, and most typically (29 out of 37) between 100 and 200 bp from the starting ATG, indicating that Abf1 may not activate transcription from distant locations as efficiently as Rap1 (Figure 3B). In addition, we found strong Abf1 binding sites in 37 of 66 unique promoters showing decreased transcription by 2× or greater but not identified as binding Abf1 in ChIP-chip studies (6); an additional two of these promoters lacking strong Abf1 binding sites were found to bind Abf1 in vitro (PBM) (21). These 39 genes are assigned as putative targets of Abf1 (Supplementary Table 5). Lastly, we identified genes whose promoters are likely to bind Abf1 but which are unaffected upon loss of Abf1 binding. We examined 92 genes for which the log2 change fell between −0.2 and 0.2, and which were identified as binding Abf1 with P < 0.001 (6). After removing from consideration five promoters associated with divergent genes that showed decreased (by >1.5-fold) transcription in abf1-1 ts yeast at 37°C, significant matches to the Abf1 PSWM (Figure 2C) were found in 79 of the remaining promoters, of which 47 bind Abf1 in vitro (21). In comparison, searching a set of 49 promoters binding Rap1 for matches to the Abf1 binding sequence yielded only three signficant matches. One additional gene that did not yield a significant match to the consensus binding site was found to bind Abf1 in vitro (21), giving a total of 80 genes that are likely to bind Abf1 but not be affected upon loss of Abf1 binding (Supplementary Table 6).
Expression of the RPS28A gene depends on an Abf1 binding site but is not affected by loss of Abf1 binding
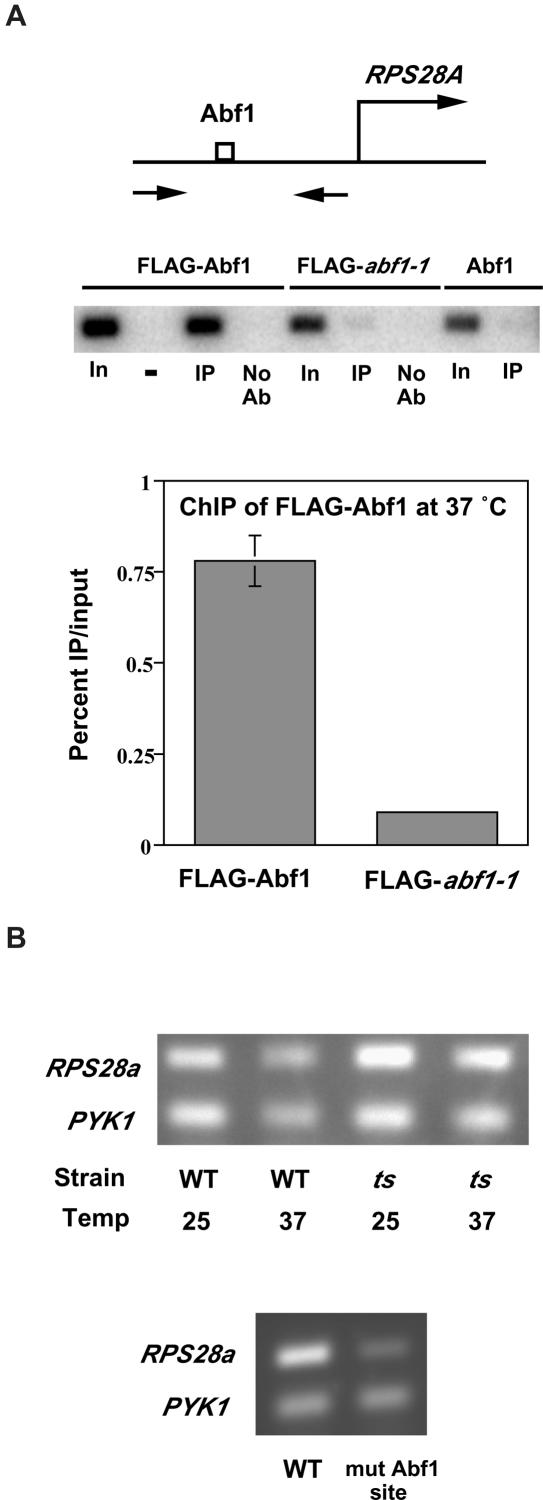
Our microarray analysis indicated that only four genes showed >2-fold increased expression in abf1-1 ts yeast at 37°C compared to wild-type, and one of these is the ABF1 gene itself, consistent with results of Miyake et al. (36). This suggests that a possible explanation for the modest effect on global gene expression in abf1-1 ts yeast at 37°C is that the decreased affinity of the mutant Abf1 protein is offset by its increased abundance. To examine this possibility, we used ChIP to test Abf1-FLAG binding in the ts mutant compared to wild-type at 37°C. We observed substantially reduced Abf1 binding in the abf1-1 ts mutant compared to wild-type at 37°C at the RPS28A and SPT15 loci (Figure 4A and data not shown), consistent with our earlier results examining the HIS4-Abf1-MEL1 reporter gene, as well as with results of Miyake et al. (9,36). Expression of both RPS28A and SPT15 were found to be unaffected in abf1-1 compared to wild-type yeast at 37°C in our microarray experiments, and we confirmed this for the individual transcripts (Figure 4B and data not shown). Expression of SPT15, which encodes TATA-binding protein (TBP), was also shown by Schroeder and Weil to be unaffected by loss of Abf1 binding, although SPT15 promoter activity depends strongly on the Abf1 binding site (10,37). Deletion of the Abf1 binding site from the RPS28A promoter driving a GUS reporter gene resulted in about a 10-fold decrease in expression (38). We found that replacement of the Abf1 binding site with a mutated site in the chromosomal RPS28A gene resulted in 2- to 4-fold decreased expression (Figure 4B and data not shown). Thus, the RPS28A gene, like SPT15 and the modified HIS4 promoter (9), depends on the Abf1 binding site for normal levels of transcription, and yet loss of Abf1 binding from this site does not reduce its expression.
Figure 4.
Loss of Abf1 binding in abf1-1 yeast does not result in decreased RPS28A expression. (A) ChIP of FLAG-tagged wild-type or ts Abf1 after 1 h at 37°C. Immunoprecipitated and input DNA were amplified using primers spanning the Abf1 site in the RPS28A promoter (upper panel) and the PCR fragments visualized by gel electrophoresis followed by Southern blotting (middle panel). Lanes are labeled as containing PCR products amplified from input DNA (In), immunoprecipitated DNA (IP), or mock IP without antibody (No Ab). The lane labeled ‘−’ is empty. Quantified results from this and an independent ChIP experiment are depicted at the bottom; SDs (too small to be visible for the FLAG-abf1-1 sample) are indicated. (B) RNA was prepared from wild-type or abf1-1 ts yeast grown at 25°C or for 1 h at 37°C (upper panel), and also from yeast having the Abf1 binding site in the RPS28A promoter mutated (lower panel). RPS28A and PYK1 mRNA were reverse transcribed and amplified by PCR, electrophoresed and visualized by ethidium bromide staining. These experiments were performed at least three times with similar results.
The C-terminal region of Abf1 is not required for continued transcription of Abf1-dependent genes after loss of Abf1 binding
One way in which an Abf1 binding site might contribute to gene activation without requiring continued occupancy by Abf1 would be if Abf1 recruited another protein that modified chromatin, or that remained bound to DNA even after loss of Abf1 binding. Since the C-terminal region of Abf1 can activate transcription and remodel chromatin in the context of protein fusions, this region is a logical candidate for effecting such hypothesized recruitment. We therefore tested whether the C-terminal region of Abf1 was necessary for ongoing transcription after loss of Abf1 binding.
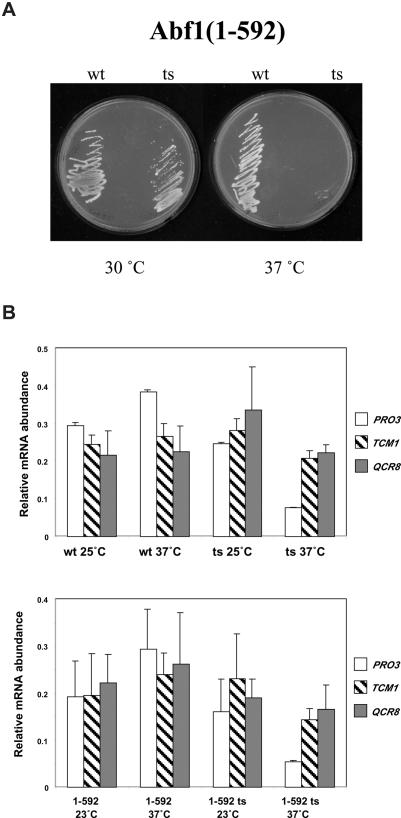
We first introduced the abf1-1 point mutation into the abf1(1–592) expression plasmid. When this plasmid was shuttled into yeast to replace the wild-type ABF1 gene, the resulting strain grew slowly and was temperature sensitive (Figure 5A and data not shown). We next examined transcripts of several genes that are known to depend on Abf1 binding sites for full expression in yeast expressing full-length Abf1, the ts mutant abf1-1, Abf1(1–592), which lacks the C-terminal region, or the corresponding ts mutant from yeast grown at 23–25°C or 1 h following a rapid shift to 37°C (Figure 5B). PRO3 showed a marked drop in transcript level in abf1-1 yeast at the restrictive temperature in the work of Miyake et al. (36) as well as in our microarray experiments, and so was examined as a positive control. Northern analysis showed that PRO3 mRNA levels decreased considerably after 1 h at 37°C in the ts mutants both in the background of full-length and the C-terminal deletion of Abf1, but not in the corresponding wild-type strains (Figure 5B). In contrast, neither QCR8 nor TCM1/RPL3, both of which depend on Abf1 binding sites for transcription (39,40), showed significantly decreased transcript levels at 37°C in either abf1-1 mutant, consistent with previous work (10), nor in the abf1(1–592) ts mutant (Figure 5B). Similar results were also seen with SPT15 and RPS28A (data not shown), both of which rely on Abf1 binding sites for their expression [(10) and Figure 3B]. Furthermore, only PRO3 showed significantly reduced expression in yeast expressing Abf1(1–592) compared to full-length Abf1. We conclude that whatever property Abf1 possesses that allows continued gene expression following loss of Abf1 binding does not reside exclusively in its C-terminal region.
Figure 5.
Transcript levels of Abf1-dependent genes at 25°C and after 1 h at 37°C in yeast expressing Abf1(1–592) or the corresponding ts mutant. (A) Growth of yeast expressing Abf1(1–592) or the corresponding ts mutant after 2 days at the indicated temperature on YPD plates. (B) Transcript abundance, measured by northern analysis and normalized to PYK1 mRNA, of TCM1/RPL2, QCR8 and PRO3, from yeast expressing full-length Abf1 (wt) or Abf1(1–592) or the corresponding ts mutants, as indicated, grown at 25°C or for 1 h at 37°C. SDs(n = 2–4) are indicated. Note that relative abundances of transcripts from yeast expressing full-length Abf1 (top) or Abf1(1–592) (lower panel) can be directly compared, as transcript levels were measured on the same blots (see Materials and Methods).
DISCUSSION
We report here the effects of loss of Abf1 binding and loss of Rap1 binding on global gene expression. As both of these factors are essential, gene expression was compared in wild-type and rap1 ts or abf1-1 ts mutants after 1 h growth at 37°C following a rapid temperature shift. Previous work has shown by DMS footprinting and chromatin IP (for Abf1) that both mutant proteins rapidly dissociate from their DNA targets at this temperature in vitro and in vivo (9,10,18,19,36,41). Genes showing decreased expression upon loss of Abf1 or Rap1 binding were enriched in the expected functional categories, and were also enriched in Abf1 or Rap1 binding genes (although only slightly for the latter; see below) (Figure 1), attesting to the quality of the microarray data. In addition, our microarray results on gene expression changes in the abf1-1 mutant compared to wild-type yeast agreed well with previous work; 39 of the 51 (P < 10−40) genes identified previously as showing decreased expression in abf1-1 yeast at the restrictive temperature also showed decreased expression by at least 1.5× in our study. However, our study differed from the previous work in that it included three biological replicates of both wild-type and mutant yeast, whereas the earlier work used three technical replicates from single mRNA isolations. The data in the current study therefore provides a better basis for statistical analysis of the microarray results. Regarding Rap1, although a previous study used microarrays to examine gene expression of yeast lacking the C-terminal silencing domain (42), to our knowledge this is the first work to examine the effect of loss of Rap1 binding on global gene expression.
To identify direct functional targets of Abf1 and Rap1, we used our microarray results together with motif analysis and published data on genome-wide in vivo and in vitro binding (5,6,21). This analysis led us to identify highly probable and putative direct targets for Abf1 and Rap1. Similar numbers of direct targets were found for both of these GRFs (Figure 2D), in agreement with the finding that roughly similar numbers of promoters bind both Abf1 and Rap1 in vitro and in vivo (6,21). However, a considerably smaller fraction of genes showing decreased transcription upon loss of Rap1 binding were identified as direct targets than of those showing decreased transcription upon loss of Abf1 binding (∼9% compared to 29%). The reason for Rap1 apparently exerting a larger indirect effect than Abf1 on genome-wide transcription is not clear, but it does provide a possible explanation for the Rap1 consensus not emerging in the T-profiler analysis (Figure 1), as it may have been obscured by the greater amount of sequence ‘noise’ caused by indirect effects on transcription. Interestingly, this analysis also yielded evidence that Rap1 binding sites typically function from a considerably greater distance from the transcription start site than do Abf1 sites; the mechanistic basis for this is completely unknown.
We also identified promoters likely to bind Abf1 or Rap1 that are not affected upon loss of binding (Supplementary Tables 3 and 6). Some of the Abf1 binding promoters in this class are probably not under control of Abf1 because the sites are too distant from the proximal promoter (Figure 3B), but this is probably not the case for Rap1 binding promoters, as Rap1 appears capable of activating promoters from sites far upstream (Figure 3A). It is also possible that some promoters bind Abf1 or Rap1 but are nevertheless not controlled by these GRFs, perhaps because other factors binding these promoters dominate their activation. However, such hypothetical promoters have not to our knowledge yet been verified (by mutagenesis of a validated binding site resulting in no change in transcription levels). Furthermore, there is no evident correlation between gene expression level and the effects of loss of Abf1 or Rap1 binding (Supplementary Tables 1–6). In contrast, several promoters that depend on Abf1 binding sites for fully activated levels of transcription have been shown not to yield decreased mRNA levels upon loss of Abf1 binding, including RPS28A (Figure 4) (10). What the mechanism is behind this ‘memory effect’ [see references (9,10) for discussion] remains a topic for future study.
Several Abf1 binding sites that have been shown to function in transcriptional control and to bind Abf1 in vivo by DMS footprinting (SPT15, QCR8 and TCM1/RPL) (10) do not show statistically signficant Abf1 binding in ChIP-on-chip experiments, nor were their promoters found to bind Abf1 in vitro (6,21). Thus, these genes were not identified as having Abf1 binding promoters in our analysis, indicating that the criteria we used were perhaps overly stringent and likely to lead to a conservative estimate of Abf1 and Rap1 binding promoters. Indeed, transcription factor binding and contribution to gene expression may better be represented as a continuum than as a discrete On/Off function (43). Much additional work will be needed to achieve a quantitative model of genome-wide transcription factor binding and contribution to gene expression for individual transcription factors.
The persistent transcription induced by Abf1 does not require the protein's C-terminal region, which contains the putative activation domain, as we found the effect undiminished in yeast expressing Abf1 that lacks this region [Figure 5 and (2)]. Interestingly, expression of some but not all Abf1 binding genes is affected by loss of this region, suggesting that Abf1 may activate transcription by more than one mechanism [Figure 5 and (9,36)]. Our analysis (using MEME) of Abf1 binding promoters did not yield any motifs that distinguished those affected by loss of Abf1 binding from those that were not.
At present we do not know whether any Rap1-dependent promoters may also show persistent transcription following loss of Rap1 binding. Fewer promoters that are likely to bind Rap1 but that are not affected by loss of Rap1 binding were identified than for Abf1, and none have thus far been verified by mutagenesis of Rap1 binding sites, as has been done for Abf1 (37–40). Promoters known to depend on Rap1 binding sites, such as HIS4 and many RP gene promoters (3,28) show decreased transcription in our microarray experiments, suggesting that at least most promoters that depend on Rap1 binding sites for full expression also require continued Rap1 binding. A more detailed investigation will be required to address this issue.
In summary, we have examined the effect on global gene expression of loss of binding of two GRFs, Abf1 and Rap1. Both of these GRFs bind to on the order of 200–300 promoters; of these, we have identified 76 and 88 promoters likely to be direct functional targets of Abf1 and Rap1, respectively. Since both Abf1 and Rap1 have been identified as ‘hubs’ in genome-wide interaction networks (7,8), these results should be useful in construction of circuit diagrams of the yeast transcriptome. We have also identified 80 promoters that are likely to bind Abf1 but are unaffected by loss of binding, and 31 such promoters for Rap1. At least some, and perhaps many, of the promoters that bind Abf1 but whose transcription is unaffected by loss of binding nonetheless depend on an Abf1 binding site (37–40). The mechanism by which Abf1 ‘marks’ such promoters remains enigmatic, but the phenomenon is reminiscent of regulation of many of the genes responsible for pattern formation in Drosophila, for which repressed or active states are maintained by PcG or Trx complexes even after the initial transcriptional repressors or activators have dissipated (44). It seems likely that further investigation into the mechanism by which Rap1 and Abf1 contribute to gene activation will provide new insights into fundamental aspects of transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors gratefully acknowledge assistance from the Wadsworth Center Microarray and Molecular Genetics Cores. The authors also thank Tsuyoshi Miyake, Rong Li (University of Virginia Health Sciences Center) for generously providing yeast strains. This work was supported by grant RO1 GMS51993 from the National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by state funds appropriated by the New York State Department of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Morse R.H. RAP, RAP, open up! new wrinkles for RAP1 in yeast. Trends Genet. 2000;16:51–53. doi: 10.1016/s0168-9525(99)01936-8. [DOI] [PubMed] [Google Scholar]
- 2.Miyake T., Loch C.M., Li R. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 2002;22:505–516. doi: 10.1128/MCB.22.2.505-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 4.Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 5.Lieb J.D., Liu X., Botstein D., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nature Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 6.Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Liu C., Skogerbo G., Zhu X., Lu H., Chen L., Shi B., Zhang Y., Wang J., Wu T., et al. Dynamic changes in subgraph preference profiles of crucial transcription factors. PLoS Comput. Biol. 2006;2:e47. doi: 10.1371/journal.pcbi.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luscombe N.M., Babu M.M., Yu H., Snyder M., Teichmann S.A., Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 9.Yarragudi A., Miyake T., Li R., Morse R.H. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder S.C., Weil P.A. Genetic tests of the role of Abf1p in driving transcription of the yeast TATA box binding protein-encoding gene, SPT15. J. Biol. Chem. 1998;273:19884–19891. doi: 10.1074/jbc.273.31.19884. [DOI] [PubMed] [Google Scholar]
- 11.Hill J., Donald K.A., Griffiths D.E., Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [published erratum appears in Nucleic Acids Res 1991 Dec 11;19(23):6688] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston M., Riles L., Hegemann J.H. Gene disruption. Meth. Enzymol. 2002;350:290–315. doi: 10.1016/s0076-6879(02)50970-8. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 14.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 16.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz S., Shore D. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 1991;5:616–628. doi: 10.1101/gad.5.4.616. [DOI] [PubMed] [Google Scholar]
- 19.Rhode P.R., Elsasser S., Campbell J.L. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nature Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boorsma A., Foat B.C., Vis D., Klis F., Bussemaker H.J. T-profiler: scoring the activity of predefined groups of genes using gene expression data. Nucleic Acids Res. 2005;33:W592–W595. doi: 10.1093/nar/gki484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson M.D., Grigull J., Mohammad N., Hughes T.R. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 25.Rudra D., Zhao Y., Warner J.R. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 27.Wade J.T., Hall D.B., Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- 28.Devlin C., Tice-Baldwin K., Shore D., Arndt K.T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol. Cell. Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tornow J., Zeng X., Gao W., Santangelo G.M. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 1993;12:2431–2437. doi: 10.1002/j.1460-2075.1993.tb05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pina B., Fernandez-Larrea J., Garcia-Reyero N., Idrissi F.Z. The different (sur)faces of Rap1p. Mol. Genet. Genomics. 2003;268:791–798. doi: 10.1007/s00438-002-0801-3. [DOI] [PubMed] [Google Scholar]
- 31.Chang C.F., Wai K.M., Patterton H.G. Calculating the statistical significance of physical clusters of co-regulated genes in the genome: the role of chromatin in domain-wide gene regulation. Nucleic Acids Res. 2004;32:1798–1807. doi: 10.1093/nar/gkh507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell. Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 33.Pryde F.E., Louis E.J. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 35.Beinoraviciute-Kellner R., Lipps G., Krauss G. In vitro selection of DNA binding sites for ABF1 protein from Saccharomyces cerevisiae. FEBS Lett. 2005;579:4535–4540. doi: 10.1016/j.febslet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Miyake T., Reese J., Loch C.M., Auble D.T., Li R. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:34865–34872. doi: 10.1074/jbc.M405156200. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder S.C., Weil P.A. Biochemical and genetic characterization of the dominant positive element driving transcription of the yeast TBP-encoding gene, SPT15. Nucleic Acids Res. 1998;26:4186–4195. doi: 10.1093/nar/26.18.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lascaris R.F., Groot E., Hoen P.B., Mager W.H., Planta R.J. Different roles for Abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Res. 2000;28:1390–1396. doi: 10.1093/nar/28.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Winde J.H., Grivell L.A. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae: ABF1 and CPF1 play opposite roles in regulating expression of the QCR8 gene, which encodes subunit VIII of the mitochondrial ubiquinol-cytochrome c oxidoreductase. Mol. Cell. Biol. 1992;12:2872–2883. doi: 10.1128/mcb.12.6.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Della Seta F., Ciafre S.A., Marck C., Santoro B., Presutti C., Sentenac A., Bozzoni I. The ABF1 factor is the transcriptional activator of the L2 ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2437–2441. doi: 10.1128/mcb.10.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drazinic C.M., Smerage J.B., Lopez M.C., Baker H.V. Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p) Mol. Cell. Biol. 1996;16:3187–3196. doi: 10.1128/mcb.16.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyrick J.J., Holstege F.C., Jennings E.G., Causton H.C., Shore D., Grunstein M., Lander E.S., Young R.A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 43.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cernilogar F.M., Orlando V. Epigenome programming by Polycomb and Trithorax proteins. Biochem. Cell. Biol. 2005;83:322–331. doi: 10.1139/o05-040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.