Abstract
Controlled proteolytic cleavage of membrane-associated transcription factors (MTFs) is an intriguing activation strategy that ensures rapid transcriptional responses to incoming stimuli. Several MTFs are known to regulate diverse cellular functions in prokaryotes, yeast, and animals. In Arabidopsis, a few NAC MTFs mediate either cytokinin signaling during cell division or endoplasmic reticulum (ER) stress responses. Through genome-wide analysis, it was found that at least 13 members of the NAC family in Arabidopsis contain strong α-helical transmembrane motifs (TMs) in their C-terminal regions and are predicted to be membrane-associated. Interestingly, most of the putative NAC MTF genes are up-regulated by stress conditions, suggesting that they may be involved in stress responses. Notably, transgenic studies revealed that membrane release is essential for the function of NAC MTFs. Transgenic plants overexpressing partial-size NAC constructs devoid of the TMs, but not those overexpressing full-size constructs, showed distinct phenotypic changes, including dwarfed growth and delayed flowering. The rice genome also contains more than six NAC MTFs. Furthermore, the presence of numerous MTFs is predicted in the whole transcription factors in plants. We thus propose that proteolytic activation of MTFs is a genome-wide mechanism regulating plant genomes.
INTRODUCTION
Transcriptional responses are regulated at multiple steps, including expression of genes encoding transcription factors and translocation of transcription factors from the cytoplasm to the nucleus. In many cases, transcription factors are stored in their dormant forms in the cytoplasm, and upon stimulation, they are activated and localized into the nucleus. Regulated activation of preexisting dormant transcription factors certainly provides an efficient way of gene regulation and is considered to be an adaptive strategy that undertakes prompt responses to environmental changes (1–4).
There are several mechanisms for activating dormant transcription factors. In one example, they are activated by post-translational modifications, such as protein phosphorylation. This has been proven in the case of the STAT transcription factors that are activated through JAK-mediated phosphorylation (2,3). In another example, such as the NF-κB/Rel transcription factors that are anchored to IκBα, the dormant transcription factors are activated after the degradation of the cytoplasmic anchors (5). The most striking example is the activation of membrane-associated transcription factors (MTFs) that are expressed as dormant precursors and integrated into the intracellular membranes. They are activated by controlled proteolytic cleavage through either one of two distinct, but biochemically related pathways. In regulated ubiquitin (Ub)/proteasome-dependent processing (RUP), they are ubiquitinated and partially degraded by the 26S proteasome in a tightly controlled manner, resulting in the release of transcriptionally active forms (6). In regulated intramembrane proteolysis (RIP), active forms are released by specific membrane-associated proteases (2,4).
It is remarkable that Ub-mediated protein degradation is directly linked to the activation of dormant proteins as well as to the inactivation of unnecessary proteins. One example of RUP is the activation of the SPT23/MGA2 transcription factor in yeast. SPT23/MGA2 is necessary for OLE1 expression in the OLE pathway that regulates membrane fluidity (6). It is expressed as a dormant ER/nuclear membrane-associated precursor (p120), from which a transcriptionally active form (p90) is released by RUP. The RUP-mediated p120 processing is abolished by unsaturated fatty acids. However, RUP-mediated MTF activation has not yet been reported in higher eukaryotes.
Several MTFs have been shown to be activated by RIP in prokaryotes and animals, among which the SRE-binding protein (SREBP) transcription factor has been most extensively studied (1). The RIP activity has also been implicated in the activation of an Arabidopsis NAC MTF, NTM1 (www.plantcell.org/cgi/doi/10.1105/tpc.106.043018) (7). After release from the ER/nuclear membranes by calpain or its functional homolog, the activated NTM1 form enters the nucleus and activates a subset of CDK inhibitor genes, e.g. KRPs, thereby resulting in reduced cell division (8). One additional NAC MTF has been recently characterized in Arabidopsis. A transcription factor AtbZIP60 has been predicted to be membrane-associated and regulate ER stress responses (9). Although the nature of the activation process is unknown, it is apparent that membrane release is essential for the AtbZIP60 function. To our knowledge, NTM1 is the only plant MTF whose activation mechanism and physiological role have been studied in detail (7).
NTM1 belongs to the NAC transcription factor family that are unique to plants (10,11). The NAC proteins contain a highly conserved NAC DNA-binding domain (DBD) that consists of approximately 160 residues in their N-terminal regions. The transcriptional activities reside in their C-terminal regions, although the C-terminal sequences are quite diverse. The NAC family is one of the largest transcription factor families in plant genomes. There are approximately a hundred of NAC transcription factors in each of the Arabidopsis and rice genomes (12,13). Several NAC members have been functionally studied in floral development (14), apical meristem formation (15), growth hormone signaling (16,17), ER stress responses (9) and cell-cycle control (7). However, most NAC transcription factors have not yet been functionally characterized, and the protein structures of the NACs have not been thoroughly examined.
In this study, we analyzed the protein structures of the Arabidopsis and rice NACs through the ARAMEMNON membrane protein database (18) and found that more than 13 NAC members in Arabidopsis and 6 NAC members in rice possess strong α-helical TMs like the NTM1 structure. The putative membrane-associated NACs were designated NTLs for NTM1-like in this report. Molecular and transgenic studies revealed that membrane release is a prerequisite for the NTL function and that the NTLs play regulatory roles in diverse plant growth and developmental processes, such as stress signaling and flowering initiation. Furthermore, a significant number of plant transcription factors were predicted to be anchored to the intracellular membranes, thus indicating that proteolytic activation of MTFs is a regulatory scheme that occurs widely in plant genomes.
MATERIALS AND METHODS
Bioinformatics softwares
The amino acid sequences of the NTLs were compared using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/), and their phylogenetic relationships were inferred using PHYLIP (http://evolution.genetics.washington.edu/phylip.html). Protein domain predictions were performed using the softwares available in the ExPASY database (http://us.expasy.org/tools/). TMs were predicted from the NAC proteins and other transcription factors using the ARAMEMNON membrane protein database (http://aramemnon.botanik.uni-koeln.de/) (18). The NTL gene structures were obtained from the TAIR and MIPS databases (http://www.arabidopsis.org/, http://mips.gsf.de/).
Plant materials, growth conditions and Arabidopsis transformation
The Arabidopsis thaliana lines used were in the Columbia ecotype (Col-0). Plants were grown in a controlled culture room at 23°C and a relative humidity of 60% under long day (LD) conditions (16 h light and 8 h dark) with white light illumination (110 μmol photons m−2s−1) provided by fluorescent FLR40D/A tubes (Osram, Seoul, Korea). Agrobacterium-mediated transformation of Arabidopsis plants was carried out by a modified floral dip method (19).
Transcript level analysis
Semiquantitative RT–PCR was employed for analysis of transcript levels. Total RNA samples were extracted from plant materials using the RNeasy Plant Total RNA Isolation Kit (Qiagen). Prior to use, the total RNA samples were extensively pretreated with RNase-free DNase I to eliminate any contaminating genomic DNA. The first-strand cDNA was synthesized from 1–2 μg of total RNA in a 20 μl reaction volume using the Superscript II reverse transcriptase (Invitrogen), and 2 μl of the reaction mixture was subsequently used for RT–PCR runs in a 50 μl reaction volume. The RT–PCR runs consisted of 15–35 cycles, depending on the linear range of PCR amplification for each gene. Each PCR cycle included incubations at 94°C for 1 min, at 55°C for 30 s, and at 72°C for 3 min. One additional cycle at 72°C for 7 min was run after the last cycle to allow trimming of incomplete polymerizations. Whenever possible, positive and negative control genes were included in the reactions to assure the feasibility of the assay conditions.
RT–PCR-based Southern blot hybridizations were carried out using gene-specific probes that had been labeled with digoxigenin-UTP by the DIG RNA Labeling Kit according to the procedure provided by the manufacturer (Roche Applied Science).
Abiotic stress treatments
Wild-type Arabidopsis plants that were grown for 2 weeks on Murashige and Skoog (MS)-agar plates were used for all abiotic stress treatments. For cold and heat treatments, plants were incubated at either 4°C for 24 h or at 37°C for 1 h before harvesting plant materials, respectively. For drought stress treatments, plants were dehydrated on dry 3 MM filters for the indicated time periods. To examine the effects of NaCl on the NTL expression, plants were transferred to a hydroponic growth medium containing 100 or 200 mM NaCl and gently shaken for 5 h before harvesting plant materials.
For methylmethanesulfonate (MMS) treatments, plants were transferred to a hydroponic growth medium containing 100 p.p.m. MMS and incubated for 1–19 h. For hydrogen peroxide treatments, plants were transferred to a hydroponic growth medium containing 50 mM H2O2 and subjected to vacuum for 5 min followed by sudden release. They were subsequently incubated under normal growth conditions for the indicated time periods.
Flowering time measurements
The NTL8 transgenic plants were grown under LD conditions for flowering time measurements. The number of rosette leaves at floral initiation and the days to the first floral bud formation were used to measure flowering times.
Tissue-specific expression of NTL6
A GUS (β-glucuronidase)-coding sequence was transcriptionally fused to the NTL6 promoter (pNTL6, base positions +1 to −1980). The pNTL6-GUS construct was transformed into Arabidopsis plants, and homozygotic plants were subjected to GUS staining. The stained plants were examined by bright-field microscopy.
Analysis of MTF processing
Six copies of the myc-coding sequences were in-frame fused to the 5′ ends of the full-size Arabidopsis and rice NTL genes. The myc-NTL gene fusion constructs were directly infiltrated into Nicotiana benthamiana leaves as described previously (20), and the leaves were incubated at room temperature for 24 h. The plant leaves were then ground in liquid nitrogen, and total proteins were extracted in 1× SDS–PAGE buffer. The protein samples were analyzed on 12% SDS–PAGE gels and blotted onto Hybond-P+ membranes (Amersham-Pharmacia Biotech). The blots were hybridized with a polyclonal anti-myc antibody (Santa Cruz Biotechnology).
Transcriptional activity assays and cell fractionation analysis
The transcriptional activity assays were carried out as described previously (7) using the pGBKT7 vector and the yeast strain AH109 (Clontech, Palo Alto, CA). The cell fractionation assays were carried out as described previously using transgenic Arabidopsis plants overexpressing a myc-NTL8 gene fusion under the control of the CaMV 35S promoter.
RESULTS
Arabidopsis genome contains more than thirteen NAC MTFs
RIP-mediated MTF activation has been characterized in several prokaryotic and animal transcription factors (1). RUP mediates the activation of the SPT23/MGA2 transcription factor in yeast (6). In plants, NTM1 is the only MTF that has been characterized at the molecular level (7). NTM1 belongs to the plant-specific NAC transcription factor family. However, it is unique among the characterized NAC proteins in that it contains a strong TM and is associated with the intracellular membranes. Meanwhile, a variety of genome-wide gene expression analysis have shown that a number of NACs are influenced by various biotic and abiotic stresses (21–23), suggesting that they may be involved in plant stress responses and signaling. Considering the physiological nature of the MTF activation to be an adaptive strategy for quick responses to adverse growth conditions, we anticipated that other NAC members might also be regulated by the membranes like NTM1.
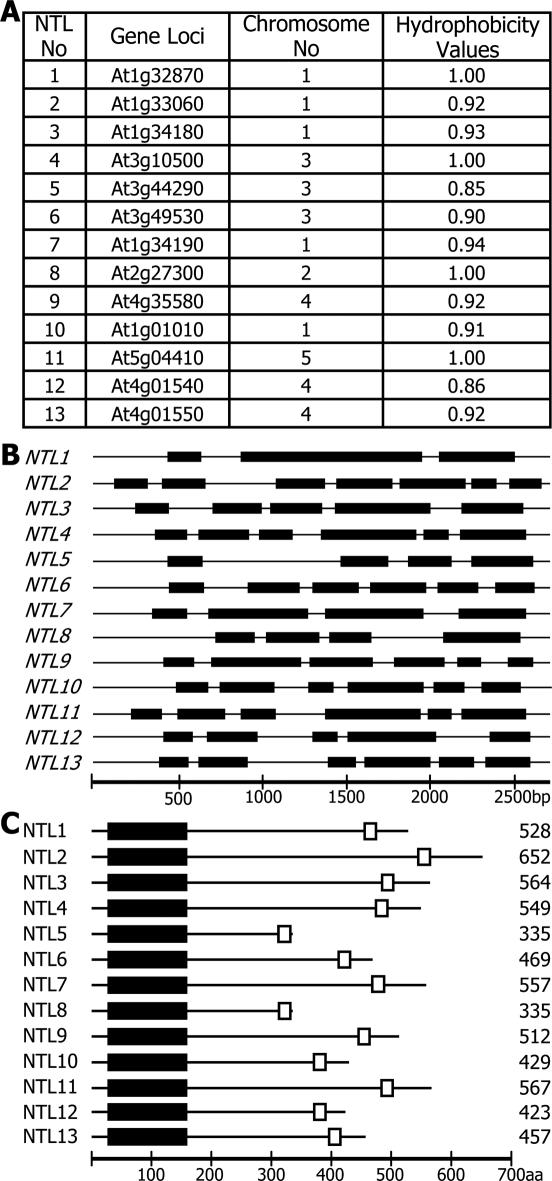
To examine this hypothesis, the protein structures of all the Arabidopsis NAC proteins were analyzed using the ARAMEMNON membrane protein database (18). Interestingly, a significant number of NAC proteins had a structural organization similar to that of NTM1: a NAC domain was present in their N-terminal regions and a strong α-helical TM was found in their C-terminal regions. For further analysis, we selected 13 members whose TMs had the hydrophobicity values of >0.85 (Figure 1A). The NTM1-like (NTL) genes are distributed throughout the five chromosomes with the exception of NTL12 and NTL13 that are located close together on chromosome 4. Furthermore, the NTL gene structures are quite diverse and have varying numbers of exons, ranging from three to seven (Figure 1B).
Figure 1.
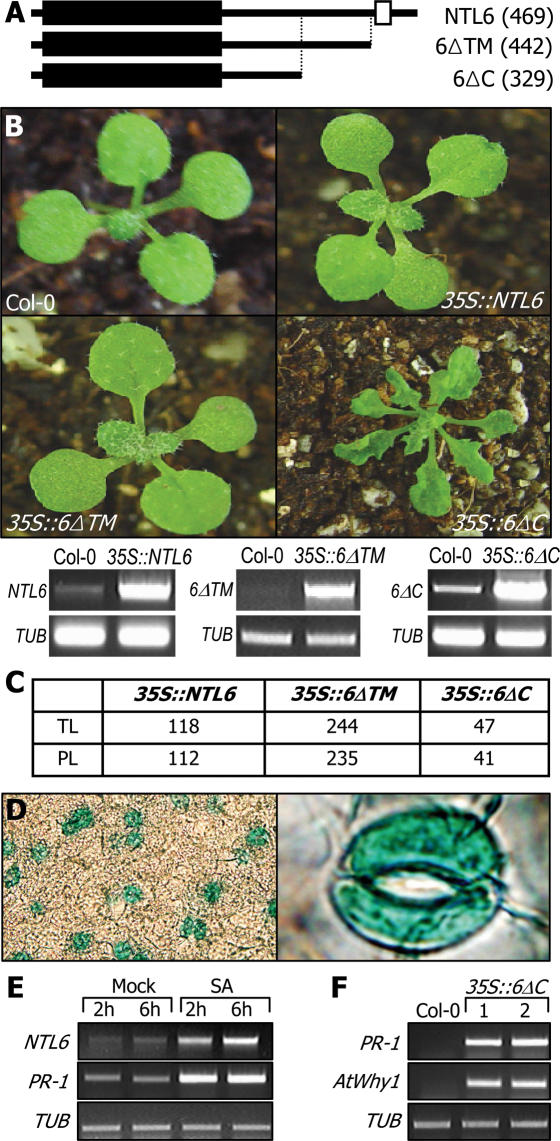
NTL genes and protein structures of NTLs. (A) Arabidopsis NTL genes. The presence of the TMs in the Arabidopsis NAC transcription factors was examined in the membrane protein database ARAMEMNON. At least thirteen NAC members were predicted to be MTFs. NTL12 and NTL13 are equivalent to NTM1 and NTM2, respectively (7). (B) NTL gene structures. They consist of 3–7 exons. (C) Protein structures of NTLs. The highly conserved NAC domains are present in their N-terminal regions (black boxes). The α-helical TMs (open boxes) are located in their far C-terminal regions.
Although the sizes of the NTL proteins are quite variable, consisting of 335–652 residues, they all have a common structural organization in which the NAC domains are located in their N-terminal regions and the TMs are located in their far C-terminal regions (Figure 1C). However, the sequences that lie between the NAC domains and the TMs differ in amino acid sequences and apparently represent the size variations among the NTL proteins. Membrane-free NACs that have been functionally characterized so far consist of ∼320–330 residues (24–26). A transcriptionally active form of NTM1 also contains 322 residues (7). These indicate that the C-terminal sequences removed by the cleavage events vary among the NTLs.
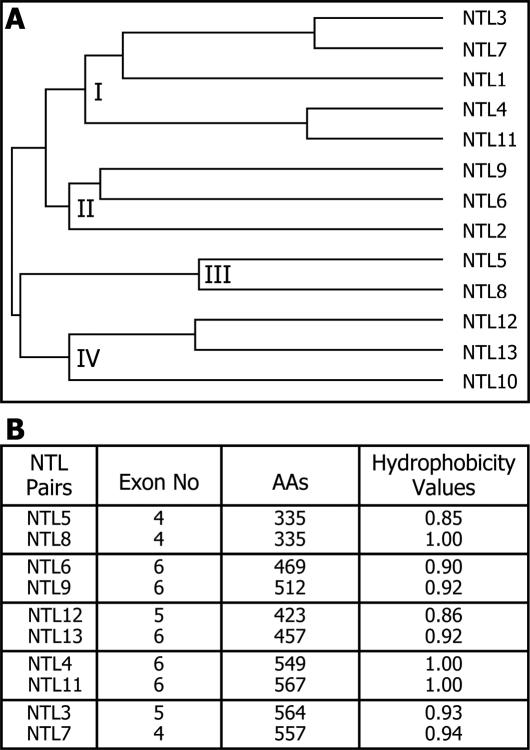
NTLs are classified into four phylogenetic subgroups
To determine the phylogenetic relationships among the thirteen NTLs, their amino acid sequences were analyzed using PHYLIP. Since the sequence similarities are restricted to the NAC domains in most NTLs, the phylogeny would mainly reflect the sequence similarities within their NAC domains. The NTLs were evidently classified into four phylogenetic subgroups (Figure 2A). However, the phylogenetic relationships were not strictly related to the protein sizes, gene structures or chromosomal locations of the NTLs (Figure 2B). Furthermore, the NTL pairs that showed close phylogenetic relationships did not always exhibit similar expression profiles (see below). Collectively, these observations suggest that each NTL may have a distinct role, although some degree of functional redundancy among them cannot be excluded.
Figure 2.
Phylogenetic analysis of NTLs. (A) Phylogenetic relationships. The phylogenetic relationships were inferred using the PHYLIP program. The NTL proteins are apparently classified into 4 phylogenetic subgroups (I-IV). (B) NTL pairs with close relationships.
The whole NAC proteins in Arabidopsis and rice have been classified into two major groups, I and II, that include 14 and 4 subgroups, respectively (10). Interestingly, the majority of the NTLs are clustered within a few subgroups of group I, such as NAC2, TIP, and OsNAC8 (Supplementary Figure S1). The NTLs may be related with the functional distinction of these NAC subgroups.
Each NTL gene exhibits a distinct expression pattern
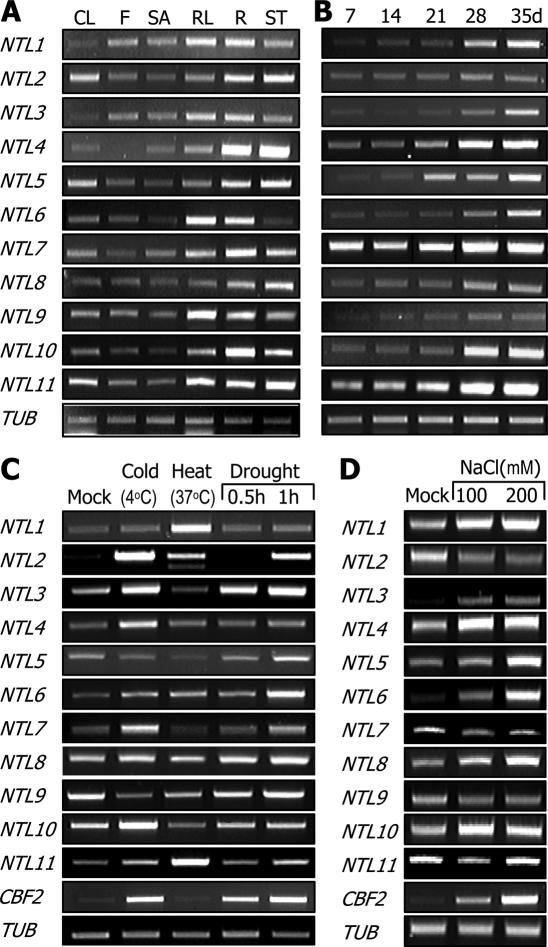
To obtain insights into the physiological roles of the NTLs, their expression patterns were examined in terms of tissue specificity and growth stage-dependence. The transcript levels of the NTLs were so low that they could not be discernibly detected by northern blot hybridization. We therefore employed semiquantitative RT–PCR to compare the NTL transcript levels. While most NTLs were expressed to relatively high levels in leaves, stems and in roots, the expression levels were low in flower and shoot apex tissues (Figure 3A). Together with their induction by stress conditions (see below), this observation suggests that NTLs may affect growth and developmental processes of vegetative organs in response to external signals.
Figure 3.
Expression patterns of NTLs and their responses to abiotic stresses. (A) Tissue-specific expression patterns. Total RNA samples were separately prepared from cauline leaves (CL), flowers (F), shoot apexes (SA), rosette leaves (RL), roots (R) and stems (ST). A tubulin gene (TUB) was included as a control for constitutive expression. (B) Growth stage-dependent expression patterns. Plants were harvested at the indicated time intervals throughout the life span for total RNA extraction. (C) Effects of abiotic stresses on NTL expression. Plants grown for 2 weeks on MS-agar plates were incubated at 4°C (cold) for 12 h or 37°C (heat) for 1 h before harvesting plant materials. To examine the effects of drought stress, plants were dehydrated on dry 3 MM paper for 30 or 60 min. CBF2 was included as a positive control. (D) NaCl effects on NTL expression.
NTL4, NTL7 and NTL11 were expressed to relatively high levels, and the expression levels were further elevated throughout the life span (Figure 3B). In contrast, the expression levels of NTL2 and NTL9 were low, and they were constantly expressed from germination to flowering. NTL3, NTL5, NTL6 and NTL10 were expressed to low levels in young seedlings, but the levels rapidly increased as plants grew. The diverse tissue-specific and growth stage-dependent expression profiles may represent distinct roles of individual NTLs.
NTL genes are affected by abiotic stresses
Numerous transcriptome studies have shown that many NAC genes are regulated by diverse biotic and abiotic stresses (21–23), suggesting that they may have a role in stress responses and signalings. Furthermore, the expression of many NTLs are altered in stressed plants (21,27,28). We observed that most NTLs are highly expressed in vegetative organs that are more susceptible to abiotic stress conditions (Figure 3A).
To further examine the effects of abiotic stresses on the NTL expression, wild-type plants were subjected to various stress conditions, including drought, cold, heat and high-salinity, and the NTL expression was analyzed by semiquantitative RT–PCR runs. Interestingly, NTLs responded differentially to different abiotic stresses (Figure 3C and D). NTL1 and NTL11 were induced primarily by heat (37°C), and NTL4 and NTL7 were induced specifically by cold (4°C). Meanwhile, NTL3 and NTL6 were dramatically up-regulated by NaCl in a similar kinetics to that of CBF2 (Figure 3D). Notably, some NTLs, such as NTL2 and NTL3, were broadly influenced by cold, drought, and NaCl. The various expression profiles may be associated with the distinct roles of individual NTLs in diverse abiotic stress responses.
Notably, most of the NTLs affected by various stress conditions (21–23) were also influenced in similar patterns in our assay conditions (Supplementary Figure S2), supporting the feasibility of our assay system and regulatory roles for NTLs in stress signaling in plants.
NTL genes are activated by MMS
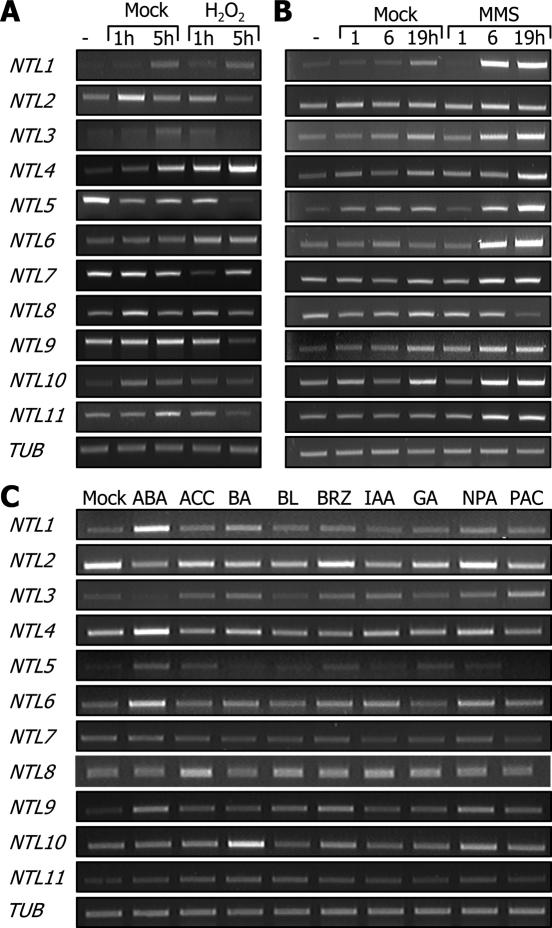
Responses of the NTL genes to hydrogen peroxide (H2O2) were not prominent except in the case of NTL4 that was slightly but reproducibly induced by this treatment. NTL2, NTL9 and NTL11 were only marginally repressed by the treatment (Figure 4A). It appears that NTLs are not directly involved in responses to reactive oxygen species (ROS).
Figure 4.
Regulation of NTL expressions by H2O2, MMS and growth hormones. (A) Effects of H2O2. Plants were subjected to a vacuum/release cycle in hydroponic MS media supplemented with 50 mM H2O2 and subsequently grown under normal growth conditions for the indicated time periods. (B) Effects of MMS. Plants were incubated in hydroponic MS media containing 100 p.p.m. MMS for the indicated time periods. (C) Growth hormone effects. Plants that were grown for 2 weeks on MS-agar plates were incubated in hydroponic MS media supplemented with appropriate growth regulators. They were used at the final concentrations of 100 μM for ABA and indole-3-acetic acid (IAA), 50 μM for 1-aminocyclopropane-1-carboxylic acid (ACC), BA and paclobutrazol (PAC), 20 μM for gibberellic acid (GA) and 1-N-naphthylphthalamic acid (NPA), and 1 μM for brassinolide (BL) and brassinazol (BRZ).
Plant responses to genotoxic agents, including MMS and cyclophosphamide (CP) that alkylate nucleophilic bioorganic molecules, are typically illustrated by cell cycle arrest, induction of DNA repair systems, and activation of stress signaling mediators (27,28). To examine the effects of MMS on the NTL expression, wild-type plants were treated with 100 p.p.m. MMS. Many NTLs, including NTL1, NTLs 3–6, NTL9 and NTL11, were significantly induced by MMS (Figure 4B). This result further supports the notion that NTLs regulate stress responses or signaling.
Consistent with the NTL induction by abiotic stress treatments, NTL1, NTL4, NTL6 and NTL9 were significantly induced by abscisic acid (ABA) (Figure 4C), a primary signaling molecule that mediates abiotic stress responses (29–32). Notably, NTL10 was induced by N-benzyladenine(BA), suggesting a role in cytokinin signaling. The NTL10 function is likely to be different from those of other NTLs.
Membrane release is essential for the NTL6 function
Our results strongly support the idea that the NTLs are intimately related to abiotic stress responses. To further examine this, we decided to produce a series of transgenic plants that overexpress NTLs and examine any phenotypic alterations. Based on the effects of abiotic stress conditions on the NTL expression patterns, NTL6 and NTL8 were chosen for transgenic studies. While NTL6 was prominently affected by diverse abiotic stresses and MMS, NTL8 was not discernibly affected by heat, cold or drought (Figures 3 and 4). It was induced only by high-salinity. In addition, NTL6 and NTL8 belong to different phylogenetic groups (Figure 2A). It was thus anticipated that NTL6 and NTL8 might play distinct roles.
To investigate whether membrane release is essential for the NTL6 function, two partial-size constructs were also included in the transgenic assays. One construct (6ΔC) was devoid of the TM and adjacent sequence region and thus assumed to be structurally equivalent to other nuclear NACs. The other partial-size construct (6ΔTM) lacked only the TM (Figure 5A). Transgenic plants overexpressing either a full-size NTL6 (35S::NTL6) or the 6ΔTM construct (35S::6ΔTM) did not show any detectable phenotypic changes (Figure 5B). Surprisingly, the transgenic plants overexpressing 6ΔC (35S::6ΔC) exhibited severe phenotypic alterations, such as dwarfed growth and curled leaves with slightly serrated margins. Most of the transgenic lines obtained from each transformation consistently displayed the phenotypes shown in Figure 5B and C, supporting that the phenotypes of the transgenic plants are caused by the overexpression of the NTL6 constructs. These observations indicate that membrane release is essential for the NTL6 function, similar to the cases of NTM1 and AtbZIP60 (7,9). Notably, the 35S::6ΔC transgenic plants resembled wild-type plants grown under abiotic stress conditions in that they exhibited reduced growth and morphological alterations (29,32). Furthermore, the phenotype of the 35S::6ΔC transgenic plants is also consistent with the responses of NTL6 to abiotic stresses (Figures 3 and 4). Meanwhile, a T-DNA insertional knockout mutant displayed a phenotype essentially identical to that of wild-type plants (data not shown). This may be either due to functional redundancy among the NTLs or related to the low transcription of NTL6 in normal growth conditions (see Discussion). Overexpression of the transgenes in the transgenic plants was confirmed by RT–PCR runs (Figure 5B).
Figure 5.
NTL6 transgenic plants. (A) NTL6 constructs used for Arabidopsis transformation. The numbers in parentheses are those of amino acid residues. (B) NTL6 transgenic plants. Ten-day-old plants are displayed. Overexpression of the transgenes was confirmed by RT–PCRs. (C) Statistics of transgenic plants. TL indicates the number of total transgenic lines obtained, and PL indicates the number of transgenic plants showing the phenotype shown in (B). (D) Localized expression of NTL6 in guard cells. (E) NTL6 induction by SA. Ten-day-old wild-type plants were treated with SA at a final concentration of 0.1 mM for 2 h or 6 h. (F) Defense gene induction in the 35S::6ΔC transgenic plants. Two representative transgenic plants (1 and 2) were analyzed.
To get more insights into the role of NTL6 at the cellular level, a GUS-coding sequence was transcriptionally fused to the promoter (pNTL6) of NTL6, and the pNTL6-GUS construct was transformed into Arabidopsis plants. Notably, the GUS activity was predominantly detected in the guard cells of the transgenic plants (Figure 5D) in addition to the vascular tissues (data not shown). Consistent with the high-level expression of NTL6 in root tissue (Figure 3A), the GUS activity was also detected to a significant level in the vascular cylinders of primary and lateral roots (data not shown). NTL6 may have a role in defending against dehydration occurring under abiotic stress conditions, such as drought, cold and high-salinity (32).
Some stress symptoms are commonly observed in plants grown under both biotic and abiotic stress conditions. These include growth retardation, morphological alterations and reduced metabolic activities (33). Furthermore, some NTLs, including NTL6, are also influenced by some bacterial infections (21). We therefore hypothesized that NTL6 might also be related to biotic stress responses. We first examined whether the NTL6 expression was influenced by SA, a critical signaling molecule that mediates biotic stress responses in plants (34). Interestingly, NTL6 was significantly induced by SA, like pathogenesis-related-1 (PR-1) (Figure 5E). Furthermore, the transcript levels of PR-1 and AtWhy1, which are activated by SA-mediated signals and play important roles in pathogen resistance (35), were greatly elevated in the 35S::6ΔC transgenic plants (Figure 5F). These observations indicate that NTL6 is linked to the SA-dependent signaling as well as to the ABA-mediated abiotic stress responses.
NTL8 regulates flowering time
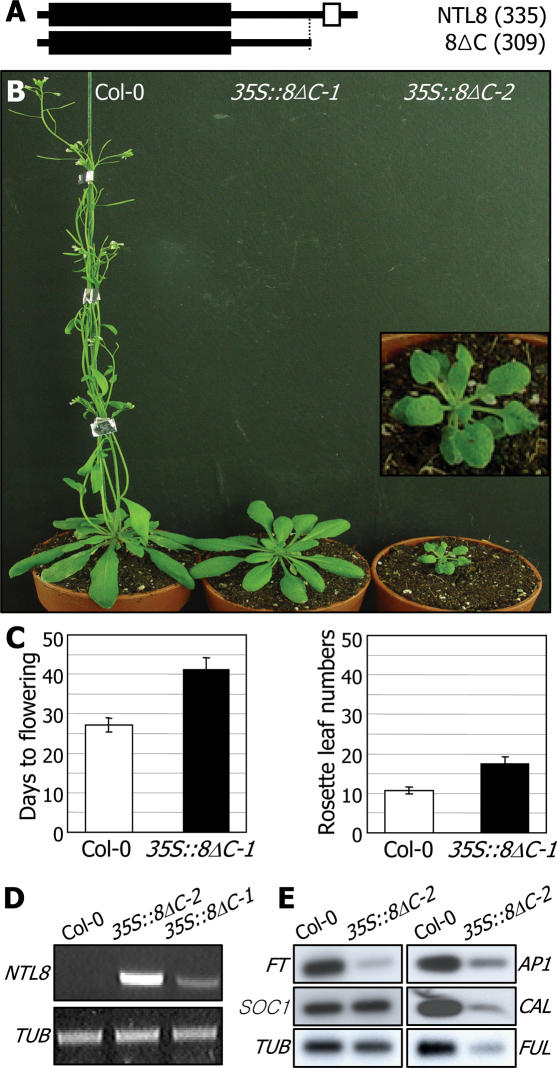
NTL8 was chosen for transgenic studies based on the following characteristics. It is distinct among the NTL genes in that it does not detectably respond to most of the stress treatments examined except for the slight induction by NaCl (Figure 3D). It may not be directly related to stress responses, although it is still possible that NTL8 may affect certain aspects of plant growth and development under specific stress conditions. In addition, NTL8 is the smallest of the thirteen NTLs (Figure 1C). It was thus impractical to discriminate between the 8ΔC and 8ΔTM constructs (Figure 6A). Therefore, we included only the 8ΔC construct in addition to a full-size construct in the transformation experiments.
Figure 6.
NTL8 transgenic plants. (A) NTL8 constructs used for Arabidopsis transformation. (B) NTL8 transgenic plants. Two types of the 35S::8ΔC transgenic plants were isolated: one type has a normal morphology but is late flowering (35S::8ΔC-1), and the other type shows reduced growth as well as severe late flowering (35S::8ΔC-2). Inlet shows an enlarged view of the 35S::8ΔC-2 transgenic plant. (C) Flowering time measurements of the 35S::8ΔC-1 transgenic plants. Thirty plants were measured and averaged for each plant group. Bars denote standard error of the mean. Statistical significance was determined using a student t-test (P < 0.01). (D) ΔC transgene expression. The severity of phenotypic alterations in the 35S::8ΔC transgenic plants is proportional to the level of the ΔC transgene expression. (E) Expression of flowering time genes. The transcript levels were analyzed by RT–PCR-based Southern blot hybridization.
We obtained two distinct homozygotic lines of the 35S::8ΔC transgenic plants with different phenotypes (Figure 6B). One line (35S::8ΔC-1) was late flowering with apparently normal leaf morphology (Figure 6C). In contrast, the other line (35S::8ΔC-2) exhibited severely dwarfed growth, similar to the 35S::6ΔC transgenic plants (Figure 5B), and delayed flowering. Although the molecular mechanism underlying the differential ΔC transgene expressions in these two lines is unclear, the transgene expression was significantly higher in 35S::8ΔC-2 (Figure 6D). These results suggest that NTL8 does not simply regulate flowering time but modulates it, probably in response to stress signals. The NTL8-mediated flowering time control may be related to the altered flowering phenotype frequently observed in stressed plants (31,32). Consistent with the delayed flowering phenotype, flowering time genes, including FLOWERING LOCUS T (FT), FRUITFUL (FUL) and CAULIFLOWER (CAL), were down-regulated in the 35S::8ΔC-2 transgenic plants (Figure 6E) as well as in the 35S::8ΔC-1 transgenic plants (data not shown).
Transgenic studies also suggest that the biochemical mechanism underlying the NTL8 function may be different from that of the NTL6 function. Transgenic plants overexpressing the full-size NTL8 (35S::NTL8) showed similar phenotypic changes to those of the 35S::8ΔC transgenic plants (Figure 6B). This is in contrast to that exhibited by the 35S::NTL6 transgenic plants (Figure 5). Growth was retarded, and flowering was significantly delayed in the 35S::NTL8 transgenic plants like the 35S::8ΔC transgenic plants (data not shown). This is probably related to the distinct nature of the NTL8 processing. While the NTL6 processing may be an inducible event, the NTL8 processing may be constitutive but blocked by certain endogenous or external signals. Consistent with this view, whereas the full-size NTL6 was predominantly detected in total cellular extracts, both the full-size and processed NTL8 forms were detected (Figure 7B).
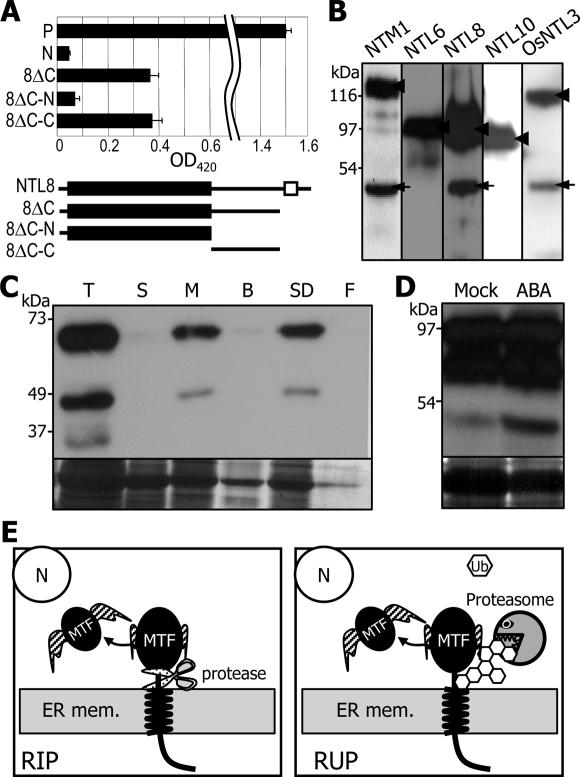
Figure 7.
Processing of Arabidopsis and rice NTLs. (A) Analysis of transcriptional activities. P, positive control (full-size GAL4); N, negative control (DNA-binding domain); 8ΔC-N and 8ΔC-C, N-terminal and C-terminal regions of 8ΔC, respectively. Three independent measurements were averaged (P < 0.01). (B) NTL processing. The myc-NTL gene fusion constructs were infiltrated into N.benthamiana leaves. The full-size (arrow heads) and the processed (arrows) NTLs were immunologically detected using an anti-myc antibody. (C) Membrane-association of NTL8. Aliquots of each fraction, which have been processed as described previously (7), were subject to western blot analysis (upper panel) using an anti-myc antibody. The Commassie-stained membrane is displayed at the bottom. T, total extract; S, soluble fraction; M, membrane fraction; B, buffer-extracted fraction; SD, SDS-extracted fraction; F, final membrane fraction. (D) ABA effects on NTL6 processing. Plants were treated with 100 μM ABA for 6 h. A part of Commassie-stained gel is shown at the bottom. (E) Two putative mechanisms for releasing MTFs. Controlled cleavage of MTFs either by intramembrane proteases (RIP) or ubiquitin/proteasome-dependent processing (RUP) is schematically shown. This diagram is modified from Hoppe et al. (2).
Processed NTL forms are transcriptional activators
Our observations suggest that NTLs are released from the membranes and localized into the nucleus, where they may function as transcriptional activator like other known NAC transcription factors.
To examine this hypothesis, the NTL8 gene constructs were in-frame fused to the GAL4 DBD in the yeast expression vector pGBKT7. The GAL4-NTL8 fusions were expressed in a yeast strain containing a reporter gene (LacZ), and α-galactosidase activity was measured. The yeast cells expressing a full-size NTL8 showed LacZ activation (data not shown). Notably, the 8ΔC and even the C-terminal region of 8ΔC also activated the LacZ reporter gene to a level even higher than that by the full-size NTL8 (Figure 7A). However, the NAC domain alone did not activate it, clearly demonstrating that NTL8, and possibly other NTLs too, is a transcriptional activator.
NTM1 is membrane-associated, and western blot analysis has shown that total cellular extracts contain two NTM1-specific polypeptides: the full-size, membrane-associated form and the processed, nuclear form (7).
To investigate whether the NTL proteins are processed in a similar manner, six copies of the myc-coding sequences were fused in-frame to the 5′ ends of some of the Arabidopsis and rice NTL genes. The myc-NTL gene fusions were transiently expressed in N.benthamiana leaves after direct infiltration. Western blot analysis using a polyclonal anti-myc antibody revealed that total cellular extracts prepared from the leaves infiltrated with the myc-NTL8 and myc-osNTL3 constructs contained two specific bands in each case (Figure 7B). Size estimations predicted that the upper bands were the full-size forms, and the lower bands were the processed forms. In contrast, only a single band, which is apparently the full-size NTL, was detected in each of the cellular extracts prepared from the myc-NTL6 and myc-NTL10 leaves. These results suggest that the NTL processing might be either repressible or inducible. The processing of NTL8 and osNTL3 may occur constitutively under normal growth conditions, but it would be blocked by certain incoming stimuli. In contrast, the processing of NTL6 and NTL10 would be an inducible event. This interpretation is also consistent with the phenotypes of the NTL6 and NTL8 transgenic plants (Figures 5 and 6). While transgenic plants with the full-size NTL6 construct were identical to wild-type plants (Figure 5B), those overexpressing the full-size NTL8 construct exhibited a similar phenotype to that of the 35S::8ΔC transgenic plants (Figure 6B).
Membrane release of NTL6 is induced by ABA
We further examined the membrane-association of NTLs through cell fractionation assays using transgenic Arabidopsis plants overexpressing a myc-NTL8 fusion. Total cellular extract contained two major NTL8-specific bands (Figure 7C). The upper band was predicted to be the full-size NTL8 protein and exclusively detected in the membrane fractions. The lower band was close to that of 8ΔC and other nuclear NACs, suggesting that it was a processed form. Unexpectedly, the processed form was not detected in the soluble fractions. This might be due to an unstable nature of the processed form, as has been observed with the soluble forms of other MTFs(2,7). Similar results were obtained with NTL6 (data not shown). These observations demonstrate that NTLs are associated with the membranes.
The next question was what signals trigger the membrane release of NTLs. To answer the question, transgenic plants overexpressing a myc-NTL6 fusion were treated with various stresses and growth hormones, and total cellular extracts were analyzed by western blot analysis. Whereas most of the treatments did not exhibit any discernible effects on the NTL6 processing, ABA significantly induced it (Figure 7D). It was unexpected that SA did not have any effects, since PR genes were up-regulated in the 35S::6ΔC transgenic plants (Figure 5). NTL6 may have a role in mediating ABA signals in biotic stress responses.
Some rice NACs are also membrane-associated
The rice genome contains 74 NAC transcription factors, which comprise one of the largest transcription factor families in this plant species (12). To examine whether the membrane-association of the NACs in rice, the protein structures of the rice NACs were analyzed in a similar way as with the Arabidopsis NACs using the ARAMEMNON database. At least six NAC members contained strong TMs, with hydrophobicity values of higher than 0.85, in positions similar to those of the Arabidopsis NTLs (Supplementary Figure S3). It is evident that membrane regulation of the NAC MTFs is conserved at least in two plant species, Arabidopsis and rice.
In this work, we analyzed the protein structures of the NAC transcription factors and studied the physiological roles of selected Arabidopsis NTLs. A similar regulatory mechanism is also envisioned for other MTFs in plants. Our data also suggest that membrane regulation of transcription factors is not an exceptional mechanism but a genome-wide strategy for gene regulation functioning widely in plant genomes. The MTFs are activated either by RIP or RUP (Figure 7E). It is also possible that both activities may be required for the regulation of a certain MTF. For example, a MTF may be released by intramembrane proteases, but the stability of the released form may further be regulated by the 26S proteasome, like the NTM1 processing (7).
DISCUSSION
MTF activation is an efficient way of achieving rapid transcriptional responses
Plants are constantly exposed to a variety of biotic and abiotic stresses in nature. Therefore, they have developed versatile systems for accurate stress signal perception and transduction. Taking this into account, the MTF activation strategy that ensures more rapid transcriptional responses to environmental fluctuations would be an efficient way to maximize plant survival under adverse growth conditions (1–3).
Here, we demonstrated through protein structural analysis and transgenic studies that a subset of plant-specific NAC transcription factors is membrane-associated. The NTL genes are possibly related to plant stress responses. They are influenced by diverse stress conditions, and transgenic plants overproducing constitutively active NTL forms exhibit typical phenotypic alterations that are frequently observed in plants grown under stress conditions.
Several NAC transcription factors have been functionally studied in diverse plant growth and developmental processes, including floral development (14), apical meristem formation (15) and growth hormone signaling (7,16,17). There are up to 13 NAC MTFs, including NTM1 and NTM2 (18), in Arabidopsis. The NTLs certainly belong to a distinct set of the NAC transcription factors whose functions are modulated by their release from the intracellular membranes. It is notable that NTLs are influenced by various stresses. The transgenic plants overexpressing constitutively active NTL6 or NTL8 forms (6ΔC or 8ΔC, respectively) exhibit reduced growth and altered leaf morphologies, indicative of hypersensitive responses to environmental stresses (29,32), suggesting that the NTLs may mediate diverse aspects of stress responses or signaling. It is envisioned that membrane regulation of the NTL activation is a unique biochemical scheme that plants use to efficiently defend against environmental stresses.
Arabidopsis genome contains numerous MTFs
The Arabidopsis and rice genomes contain 1533 and 1336 transcription factor genes, respectively, covering 5–6% of each genome (12,13). The Arabidopsis genome contains 109 members of the NAC transcription factors, and the rice genome contains 74 NAC members. It is therefore estimated that ∼10% of the NAC members are membrane-associated in each genome.
Surprisingly, ARAMEMNON-assisted protein structure analysis of the whole transcription factors in the Arabidopsis genome predicted that a considerable number of transcription factors from most of the major transcription factor families possess strong TMs in their C-terminal regions (Supplementary Figure S4) (18). For example, >4 members are predicted to be membrane-associated in each of the bHLH, C2H2, MADS and SBP families, and six members of the bZIP family contain the TMs. These observations are also consistent with the computational prediction study on transcription factors in eukaryotes (36). More than 76 transcription factors have been estimated to be membrane-associated. The wide occurrence of the TMs in most transcription factor families in Arabidopsis further supports that the MTF processing is a genome-wide mechanism. Furthermore, numerous proteins, which contain known DBDs but have not been classified as transcription factors because of the presence of the TMs, would also function as transcription factors. More extensive analysis of plant genomes may lead to the identification of a greater number of transcription factor genes than has originally been estimated.
MTF may be involved in various growth and developmental processes
The MTFs are released from the membranes by either one of the two activation mechanisms: RIP and RUP. In RIP, a set of intramembrane proteases, including S1P and S2P, work sequentially to release the MTFs in a stepwise manner (1,37,38) (Figure 7C). A few S1P and S2P homologues are also present in the Arabidopsis genome. Furthermore, NTM1 is cleaved by calpain, a membrane-associated cysteine protease, or its functional homologue (7). Notably, an Arabidopsis mutant in which a calpain protease gene had been inactivated showed a strikingly similar phenotype to that of the NTM1 mutant (7,39). The previous and our observations suggest that the membrane-bound calpain protease or its functional homologues may also have a role in the activation of the NTLs and other MTFs. Considering the large number of the MTFs in plant genomes, the battery of membrane-associated proteases that regulate the MTF activation would be much larger.
The SPT23/MGA2 transcription factor in yeast is the only example that has been unequivocally shown to be activated by RUP (6). Although the RUP pathway has not yet been reported in higher eukaryotes, the 26S proteasome activity is likely to be involved in the regulation of the MTF function. It has recently been demonstrated that NTM1 is released from the membranes by proteolytic cleavage but the protein stability of the active NTM1 form is further modulated by the 26S proteasome. It is also envisioned that the RUP pathway may activate at least some of the NTLs and other MTFs in plants. Recent studies have shown that Ub-mediated proteolytic cleavage exerts a role in various growth hormone signaling pathways (40–42). There are more than a thousand of E3 ligases that determine substrate specificities in the Arabidopsis genome (43). It is possible that individual NTLs or MTFs may be processed or the protein stability may be regulated by distinct E3 ligases in different growth hormone signaling pathways, particularly those mediated by SA and ABA.
NTM1 exerts a role in cell-cycle control (7). We showed that NTL6 regulates stress responses in plants. NTL8 regulates flowering time, apparently by repressing a floral integrator (FT) (44,45). Furthermore, the NTL8 function might also be linked to stress signaling pathways. The membrane-associated Arabidopsis transcription factor, AtbZIP60, regulates ER stress responses in a way unique to plants (9). These observations suggest that MTFs, including NTLs, may play various roles in a wide-variety of plant growth and developmental processes in plants.
Knockout mutants that had T-DNA insertions within the open reading frames of NTL6 and NTL8 were similar to wild-type plants. This would be caused by functional redundancy among the NTLs or between the NTL-mediated pathways and other stress signaling pathways. We observed that most NTLs are induced by stress conditions. Alternatively, this may be related to the expression patterns of the NTLs. They are expressed to very low levels in normal growth conditions but induced under stress conditions. Further works are required to discriminate these two possibilities. For example, multiple mutants would help answer the question. In addition, it will also be interesting to examine whether the knockout mutants exhibits reduced resistance to biotic and abiotic stresses compared to wild-type plants.
Taken together, our data indicate that NAC MTF activities are apparently regulated at the post-translational level as well as at the transcriptional level. We are under way to investigate how stress-related growth hormones, such as ABA and SA, and stress conditions affect the NTL processing and how they are released from the membranes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Mrs Younghee Choi for proofreading the manuscript. This work was supported by the Brain Korea 21, Biogreen 21 (20050301034456), National Research Laboratory (2005-01039) Programs, and by grants from the Plant Signaling Network Research Center and from KOSEF (R08-2004-000-10066-0 to Y.-S.K.) and KRF (2005-070-C00129). Funding to pay the Open Access publication charges for this article was provided by Korea Science & Engineering Foundation (KOSEF) and Korea Research Foundation (KRF).
Conflict of interest statement. None declared.
REFERENCES
- 1.Brown M.S., Ye J., Rawson R.B., Goldstein J.L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoppe T., Rape M., Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell. Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaffman A., O'Shea E.K. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell. Dev. Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 4.Vik Å., Rine J. Membrane biology: membrane-regulated transcription. Curr. Biol. 2000;10:R869–R871. doi: 10.1016/s0960-9822(00)00822-8. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh S., May M.J., Kopp E.B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 6.Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H.D., Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y.-S., Kim S.-G., Park J.-E., Park H.-Y., Lim M.-H., Chua N.-H., Park C.-M. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006 doi: 10.1105/tpc.106.043018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Veylder L., Beeckman T., Beemster G.T.S., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., MirandeNaudts M., Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwata Y., Koizumi N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl Acad. Sci. USA. 2005;102:5280–5285. doi: 10.1073/pnas.0408941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., Matsubara K., Osato N., Kawai J., Carninci P., et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 11.Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 12.National Institute of Agrobiological Science Rice Full-size cDNA Project Team. Collection, mapping and annotation of over 28 000 cDNA clones from Japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 13.Riechmann J.L., Heard J., Martin G., Reuber L., Jiang C.-Z., Keddie J., Adam L., Pineda O., Ratcliffe O.J., Samaha R.R., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 14.Sablowski R.W., Meyerowitz E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 15.Hibara K., Takada S., Tasaka M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003;36:687–696. doi: 10.1046/j.1365-313x.2003.01911.x. [DOI] [PubMed] [Google Scholar]
- 16.Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.-S.P., Yamaguchi-Shinozaki K., Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 17.Xie Q., Frugis G., Colgan D., Chua N.-H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.-I., Kunze R. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 20.Llave C., Kasschau K.D., Carrington J.C. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl Acad. Sci. USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 24.Ernst H.A., Olsen A.N., Skriver K., Larsen S., Lo Leggio L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duval M., Hsieh T.F., Kim S.Y., Thomas T.L. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002;50:237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- 26.Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Franke S.I.R., Prá D., Erdtmann B., Henriques J.A.P., da Silva J. Influence of orange juice over the genotoxicity induced by alkylating agents: an in vivo analysis. Mutagenesis. 2005;20:279–283. doi: 10.1093/mutage/gei034. [DOI] [PubMed] [Google Scholar]
- 28.Revenkova E., Masson J., Koncz C., Afsar K., Jakovleva L., Paszkowski J. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 1999;18:490–499. doi: 10.1093/emboj/18.2.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanton M.L., Roy B.A., Thiede D.A. Evolution in stressful environments. I. Phenotypic variability, phenotypic selection, and response to selection in five distinct environmental stresses. Evol. Int. J. Org. Evol. 2000;54:93–111. doi: 10.1111/j.0014-3820.2000.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J., Verslues P.E., Zheng X., Lee B.H., Zhan X., Manabe Y., Sokolchik I., Zhu Y., Dong C.H., Zhu J.K., et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl Acad. Sci. USA. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 1996;7:161–167. doi: 10.1016/s0958-1669(96)80007-3. [DOI] [PubMed] [Google Scholar]
- 33.Bohnert H.J., Nelson D.E., Jensen R.G. Adaptations to environmental stresses. Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah J. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 2003;6:365–371. doi: 10.1016/s1369-5266(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 35.Desveaux D., Subramaniam R., Després C., Mess J.N., Lévesque C., Fobert P.R., Dangl J.L., Brisson N. A ‘Whirly’ transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 36.Zupicich J., Brenner S.E., Skarnes W.C. Computational prediction of membrane-tethered transcription factors. Genome Biol. 2001;2:0050.1–0050.6. doi: 10.1186/gb-2001-2-12-research0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe M.S., Xia W., Ostaszewski B.L., Diehl T.S., Kimberly W.T., Selkoe D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 38.Zelenski N.G., Rawson R.B., Brown M.S., Goldstein J.L. Membrane topology of S2P, a protein required for intramembraneous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 39.Ahn J.-W., Kim M., Lim J.H., Kim G.-T., Pai H.S. Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 2004;38:969–981. doi: 10.1111/j.1365-313X.2004.02102.x. [DOI] [PubMed] [Google Scholar]
- 40.Itoh H., Matsuoka M., Steber C.M. A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 2003;8:492–497. doi: 10.1016/j.tplants.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Vierstra R.D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003;8:135–142. doi: 10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 42.Smalle J., Vierstra R.D. The ubiquitin-26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 43.Risseeuw E.P., Daskalchuk T.E., Banks T.W., Liu E., Cotelesage J., Hellmann H., Estelle M., Somers D.E., Crosby W.L. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003;34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- 44.Samach A., Onouchi H., Gold S.E., Gary S., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 45.Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.