Abstract
RmInt1 is a self-splicing and mobile group II intron initially identified in the bacterium Sinorhizobium meliloti, which encodes a reverse transcriptase–maturase (Intron Encoded Protein, IEP) lacking the C-terminal DNA binding (D) and DNA endonuclease domains (En). RmInt1 invades cognate intronless homing sites (ISRm2011-2) by a mechanism known as retrohoming. This work describes how the RmInt1 intron spreads in the S.meliloti genome upon acquisition by conjugation. This process was revealed by using the wild-type intron RmInt1 and engineered intron-donor constructs based on ribozyme coding sequence (ΔORF)-derivatives with higher homing efficiency than the wild-type intron. The data demonstrate that RmInt1 propagates into the S.meliloti genome primarily by retrohoming with a strand bias related to replication of the chromosome and symbiotic megaplasmids. Moreover, we show that when expressed in trans from a separate plasmid, the IEP is able to mobilize genomic ΔORF ribozymes that afterward displayed wild-type levels of retrohoming. Our results contribute to get further understanding of how group II introns spread into bacterial genomes in nature.
INTRODUCTION
Group II introns are both catalytic RNAs and mobile retroelements initially found in organelles of plants and lower eukaryotes, but later identified in bacteria and in some archaeal genomes (1–3). A typical group II intron consists of a highly structured RNA in six distinct double-helical domains (DI to DVI) and an internally encoded (ORF within DIV) reverse transcriptase–maturase (RT) that is required in vivo to fold the intron RNA into a catalytically active structure (4–8). Intron mobility may occur either to intronless alleles (retrohoming) or to ectopic sites (retrotransposition) and is mediated by RNP (ribonucleoprotein) complexes consisting of the Intron Encoded Protein (IEP) and the excised lariat RNA.
Three main phylogenetic classes (IIA, IIB and IIC) of group II introns have been described based on the IEP and conserved intron RNA structures (2,5,9–12). The well-characterized Lactococcus lactis Ll.LtrB and yeast aI1 and aI2 introns belongs to the IIA class, the IEPs of which contain an N-terminal RT domain homologous to retroviral RTs, followed by domain X, associated with RNA splicing or maturase activity, and C-terminal DNA binding (D) and DNA endonuclease domains (En) (13–16). Retrohoming of these group IIA introns occurs by a target-DNA-primed reverse transcription mechanism in which the intron RNA reverse splices directly into one strand of a double-stranded DNA target site, whereas the IEP cleaves the opposite strand through its endonuclease domain and uses the 3′ end as a primer for reverse transcription of the inserted intron RNA (3,17,18).
Group II intron retrotransposition occurs at low frequency and it is considered one of the mechanisms for intron dispersal in nature. Similar to retrohoming, retrotransposition also occurs via RNA intermediate that reverse splices into DNA targets (19–21). Retrotransposition of the Ll.LtrB intron in L.lactis is biased toward the template for lagging-strand DNA synthesis and occurs in an endonuclease-independent manner, which suggests that the intron reverse splices into transiently single-stranded DNA at the replication fork (21,22). In Escherichia coli retrotransposition of Ll.LtrB frequently occurs into double-stranded DNA in an endonuclease-dependent manner similar to the retrohoming pathway. Thus, Ll.LtrB retrotransposes through two distinct mechanisms and the host environment seems to influence the choice of the integration pathway (23). Lateral inter- and intra-species transfer mediated by conjugation has been recently assessed for the Ll.LtrB intron (24,25) showing that upon acquisition by bacteria this intron is able to spread into the chromosome by retrotransposition, invading non-homologous sites. Nevertheless, the low level of ltrB transcript and relatively inefficient splicing of the intron may limit Ll.LtrB mobility and dissemination in nature (26).
Strikingly, over half of bacterial group II IEPs annotated to date lack the En domain and many other like those belonging to the IIB3 subclass (2) also lack the D domain. It has been proposed that such group II introns are ancestral and they target DNA with minimal sequence constraints to be recognized by its IEP (3,18,19,21). The best studied intron of this subclass is the Sinorhizobium meliloti RmInt1, a very efficient mobile element (27). Recently, it was determined that RmInt1 has two retrohoming pathways for mobility with a preferred pathway consistent with reverse splicing of the intron RNA into single-stranded DNA at a replication fork, using a nascent lagging DNA strand as the primer for reverse transcription (28). Thus, the mechanism of retrohoming of RmInt1 resembles that of ectopic transposition of Ll.LtrB in L.lactis.
Most bacterial group II introns are located within other mobile elements and outside the genes [(2) and references therein]. The natural homing site of RmInt1 is the insertion sequence (IS) ISRm2011-2 (29), which is usually present in high copy number in the genome of virtually all S.meliloti isolates. RmInt1 is present in 90% of the S.meliloti isolates (30) and also in other Sinorhizobium and Rhizobium species, where the intron homing sites correspond to IS elements of the ISRm2011-2 group (31). Thus, possibly this intron has been acquired by Sinorhizobium and Rhizobium species by vertical and horizontal inheritance (31). The absence of RmInt1 in ∼10% of the S.meliloti isolates is intriguing, since in these isolates there is no restriction to intron mobility. RmInt1 is therefore a model system to study group II intron survival, evolution and dissemination in nature.
In this work we investigate the retrohoming ability and dispersion pathway of RmInt1 in the S.meliloti genome upon intron acquisition by conjugative transfer. We used wild-type and engineered intron-donor constructs based on ribozyme coding sequence (ΔORF)-derivatives with increased retrohoming efficiency. Our data show the extraordinary ability of RmInt1 for its dispersal in nature by retrohoming and reveal the pathway followed by the intron to colonize the natural homing sites in this bacterial genome.
MATERIALS AND METHODS
Bacterial strains and growth conditions
S.meliloti strains used in this work were 1021 (genome-sequenced strain) (32) and RMO17 (RmInt1 intronless strain) (33). E.coli DH5α was used for cloning and maintenance of plasmid constructs. Rhizobial strains were routinely grown at 28°C on complete TY or defined minimal medium (MM) (34) and E.coli DH5α at 37°C on Luria–Bertani (LB) medium. Media were supplemented when required with antibiotics at the following concentrations: kanamycin 200 μg/ml for rhizobia and 50 μg/ml for E.coli; tetracycline 10 μg/ml and ampicillin 200 μg/ml.
RmInt1-donor constructs for in vivo assays
Schematics of all the constructs described below for delivery of RmInt1 and derivatives are depicted in Figures 1A and 2B. Plasmids expressing the wild-type RmInt1 intron were all pBBR1MCS-2 derivatives (Kmr). pKG2 was generated from pKG2.5 (27) by trimming the exon sequences to −50/+146 as described (35) whereas pBB2.5, pBB20/5H and pBB15/10H were generated in vivo by invasion of the corresponding target-recipient plasmids (27,36). To construct RmInt1-ΔORF-derivatives a DNA fragment deleted of intron 611–1759 nt was generated as described previously from pKG2 (35) and subsequently inserted in the BamHI site of pKGIEP (37) just upstream the IEP coding sequence to yield pKGΔORFIEP. ΔORF intron variants flanked by exon sequences −20/+5 were then generated by PCR amplification of the BamHI ΔORF fragment using pKGΔORFIEP as template and the appropriate primers to yield DNA products that were inserted as BamHI or SacI fragments in pKGIEP upstream (pKGΔORFIEP20/5) or downstream (pKGIEPΔORF20/5) the IEP coding sequence, respectively. pKGEMA4 was used as a donor plasmid for delivery of RmInt1 into the S.meliloti RMO17 and 1021 genomes were generated by engineering PmlI and BlnI sites internal to SacI in pKGIEPΔORF20/5.
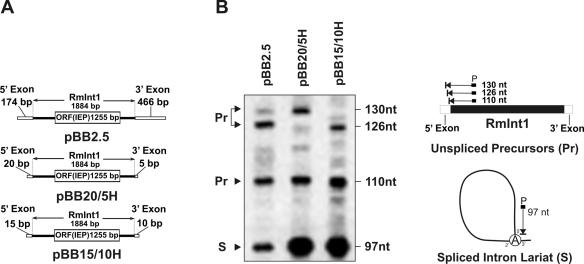
Figure 1.
Effect of exon lengths on RmInt1 splicing in vivo. (A) Schematics of constructs used in the primer extension analysis. (B) Primer extension. Analysis was performed on total RNA (20 μg) from RMO17 cells harboring intron-donor plasmids pBB2.5, pBB20/5H or pBB15/10H. The 97 nt cDNA product corresponds to the excised intron RNA (S) whereas the larger products of 110, 126 and 130 nt are derived from unspliced precursor RNA molecules (Pr). Schematics of primer extension products are shown to the right of the panel.
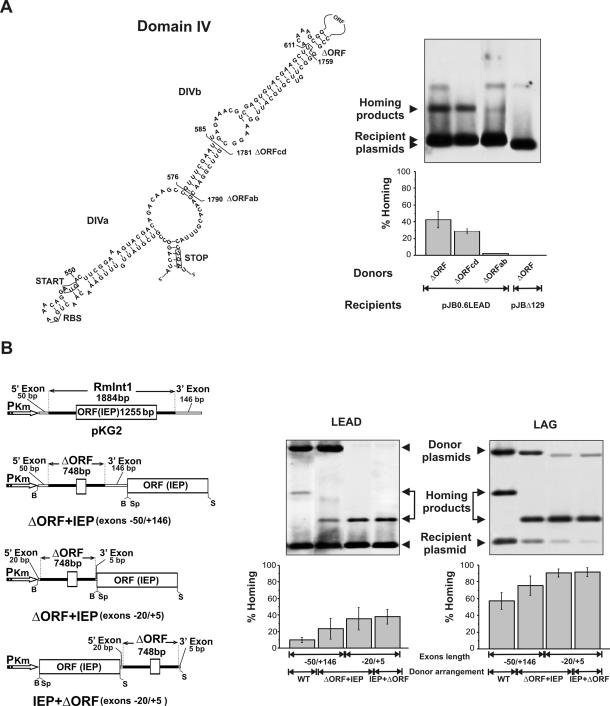
Figure 2.
Homing efficiency of wild-type RmInt1 and ΔORF-derivatives. (A) Deletion analysis of RmInt1 DIV. The predicted secondary structure of intron DIV is shown with indication of subdomains DIVa and DIVb. Start and stop codons of the IEP ORF are boxed and brackets demarcate the internal deletions (ΔORF, ΔORFcd and ΔORFab) cloned downstream of the IEP coding sequence in the donor constructs. For homing assays plasmid pools from RMO17 cells harbouring donor and recipient (pJB0.6LEAD) plasmids were analyzed by Southern hybridization with a DNA probe specific to the insertion sequence ISRm2011-2. Recipient plasmid pJBΔ129 was used as negative control in the assays. Target invasion rates in each homing assay were calculated as described in Materials and Methods and plotted in the graph-bar shown below the blot. (B) Effect of different donor genetic arrangements on intron homing. The configuration of the donor constructs is diagrammed to the left. PKm, promoter of Km resistance gene; B, BamHI; Sp, SpeI; S, SacI. Homing efficiency on recipient plasmids pJB0.6LEAD and pJB0.6LAG was quantified as described in A. The Southern blots are shown to the right along with the corresponding histograms. Features of the donor constructs are indicated bellow the plots.The hybridization signal corresponding to the donor plasmid with short flanking exons is variable and depends on the specific activity of the probe.
Further deletions of intron DIV extending positions 585–1781 (ΔORFcd) and 576–1790 (ΔORFab) were also generated as described by inverse PCR using primer pairs C (5′-GCCGCTCGAGATTCGAAACGGCTTGTCTGT-3′) and D (5′-GCCGCTCGAGTTC-GGAACGAACACGTTTA-3′); and A (5′-GCCGCTCGAGCT-TGTCTGTCGTACTTTCC-3′) and B (5′-GCCGCTCGAGAACACGTTTACCTGA-TGGGA-3′), respectively and assayed in a IEP+ΔORF (−20/+5) configuration for intron homing.
In vivo splicing and homing assays
RmInt1 splicing in vivo was assessed by primer extension analysis on total RNA from RMO17 cells expressing the wild-type intron from plasmids pBB2.5, pBB20/5H and pBB15/10H as described previously (37). cDNA-bands corresponding to the resolved extension products were quantified with the Quantity One software package (Bio-Rad Laboratories) and intron splicing was measured as 100[S/(S+Pr)], were S is the spliced RNA and Pr the precursor RNA species. In vivo homing efficiency of the wild-type and ΔORF-derivatives was determined by the double-plasmid assay in S.meliloti strain RMO17 using pJB0.6LEAD or pJB0.6LAG as recipient plasmids. The homing efficiency was calculated as the percentage of the ratio of homing product to the addition of homing product and non-invaded recipient plasmid. Data are the average of at least four independent determinations with their corresponding standard deviation (28,36,37). Recipient plasmid pJBΔ129 that lacks the RmInt1 target was used as negative control in the assays (27).
Analysis of RmInt1 dispersal in the S.meliloti genome and mobility of the inserted introns
pKG2.5 and pKGEMA4 were independently mobilized to RMO17 and 1021 strains by triparental mattings using pRK2013 as helper plasmid. Retrohoming events were then assessed in individual transconjugants from each cross (6–7 colonies) by Southern hybridization of BamHI- or SalI-digested genomic DNA (AquaPure Genomic DNA Isolation Kit, Bio-Rad Laboratories) with DIG-labeled probes specific to either the ISRm2011-2 or the RmInt1 ΔORF as described previously (38). pKG2.5x (27) and pKGEMA4x were used as controls in the assays carry a frameshift within the IEP coding sequence generated by eliminating the internal EcoRI in the intron sequence.
To assess homing efficiency in vivo of the newly acquired introns, two individual RMO17 derivatives containing genomic insertions of the wild-type (four insertions) or ΔORF (five insertions) intron, were cured of the donor plasmids pKG2.5 or pKGEMA4 by subjecting bacteria to successive cultures in MM broth without the antibiotic Km thus generating strains RMO17-2.5c2 and RMO17ΔIc50. Plasmid pJB0.6LAG containing the RmInt1 target (28) was then mobilized to the cured strains and homing efficiency was assessed as described previously on plasmid DNA of individual transconjugants (36).
pKGIEP (37) expressing a functional IEP and pKG4dV that expresses a non-functional ribozyme were also independently mobilized by conjugation to RMO17ΔIc50 harboring plasmid pJB0.6LAG. pKG4dV is a pKGEMA4 derivative in which nucleotides GTT (RmInt1 nucleotide positions 1840–1842) within the ΔORF wild-type fragment were replaced by CGA using the Altered Sites II in vitro Mutagenesis System (Promega). Homing efficiency of complemented ΔORF variants was determined on plasmid DNA of the transconjugants as described. Plasmid pKGIEPYAHH, a pKGIEP derivative that expresses a non-functional IEP containing a mutation in the RT domain (37) was used as negative complementation control in these experiments.
RESULTS
Engineering RmInt1-donor constructs for intron delivery
Recently, we reported that RmIn1 self-splices in vitro, a reaction that was favored by removing most of the protein coding sequence from the large terminal loop of DIV and by trimming the 5′ exon to its last 20 and 15 nt (35,39). Furthermore, in a previous work, we reported that a target site extending from positions −20 to −1 of the 5′ exon and +1 to +5 of the 3′ exon was sufficient to ensure efficient homing of intron RmInt1 (36).
In order to engineer a highly efficient RmInt1-donor construct for intron delivery, we first analyzed the influence of the length of the exons on intron splicing in vivo (Figure 1). The splicing reaction was characterized by primer extension using a primer P complementary to a sequence located 80–97 nt from the 5′ end of the intron. In these assays, the excised intron RNA (S) is detected as an extension product of 97 nt, along with larger bands derived from unspliced precursor RNA molecules (Pr) (40). As shown in Figure 1B, the extension product corresponding to the excised intron was detected with all the RNA samples from cells harboring constructs (Figure 1A) that transcribe the intron with either long (−174/+466) or short stretches (−20/+5, −15/+10) of the wild-type flanking 5′ and 3′ exons sequences along with additional sequences from the plasmid. The estimated relative amount of excised intron was ∼32% when transcribed with long exon sequences (pBB2.5), but it did increase 2-fold when only short stretches of the original exons were retained (−20/+5 and −15/+10 in pBB20/5H and pBB15/10H, respectively). Our results therefore indicate that exons trimming favors RmInt1 splicing in vivo as also occurs in vitro.
It has been previously reported that mobility frequencies of L.lactis group II intron Ll.LtrB can be significantly increased by using donor constructs expressing the LtrA protein from a position either downstream or upstream of the ΔORF, in which a large portion of the LtrA ORF within DIV had been deleted (41,42). To assess the effect of ORF deletions in RmInt1, homing efficiency (fraction of recipient targets invaded by the intron) was determined by double-plasmid mobility assays using a set of ΔORF- and wild-type donor plasmids all expressing the intron from the kanamycin resistance gene constitutive promoter. As shown in Figure 2A a RmInt1 ΔORF derivative deleted of intron 611–1759 nt (35) flanked by −20/+5 exons sequences and expressing the IEP upstream of the ribozyme (IEP+ΔORF) yielded invasion rates on pJB0.6LEAD of 40–42%. Larger deletions affecting subdomain DIVb reduced homing efficiency to ∼30% (ΔORFcd) and to <2% (ΔORFab). Donor constructs pKG2, in which the wild-type intron is flanked by exon sequences −50/+146, and three ΔORF-derivatives deleted of intron 611–1759 nt (35) and flanked by the same (−50/+146) or shorter (−20/+5) exon sequences, that express the IEP either downstream or upstream of the ribozyme (ΔORF+IEP and IEP+ΔORF, respectively) were further tested for homing efficiency (Figure 2B). The target-recipient plasmids used in the homing assays were pJB0.6LEAD and pJB0.6LAG, in which the RmInt1 target site is cloned in both orientations with respect to the direction of DNA replication (28). As shown in Figure 2B the wild-type intron (pKG2) exhibited ∼10% homing efficiency in pJB0.6LEAD whereas the use of a ΔORF+IEP variant flanked by the same exon sequences (−50/+146) resulted in a higher homing efficiency (∼23%). Interestingly, constructs ΔORF+IEP and IEP+ΔORF flanked by exon sequences −20/+5 displayed even higher homing efficiency (33 and 37%, respectively). Consistent with RmInt1 retrohoming bias toward the strand that serves as template for the nascent lagging-strand in DNA replication forks (28) the invasion rates in pJB0.6LAG recipient plasmid were markedly increased for all the donor constructs, but again the highest efficiency corresponded to ΔORF intron constructs flanked by short exons (∼90%, Figure 2B). In addition to an increased homing efficiency, the combination of ΔORF intron and short exons resulted even in rates of intron splicing close to 100% (data not shown). Taken together these data predict the ΔORF constructs flanked by short exon sequences (−20/+5) and expressing the IEP in cis within the same precursor transcript as the most efficient shuttle configuration for RmInt1 delivery in vivo.
Dispersion of RmInt1 in the S.meliloti genome
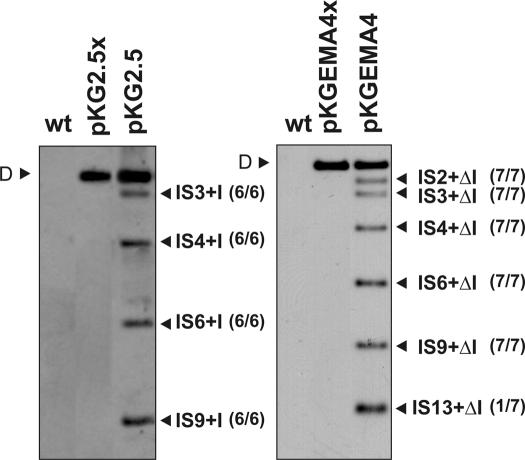
RMO17 is an RmInt1 intronless S.meliloti strain that nevertheless harbors 13 copies of the RmInt1 homing site (ISRm2011-2). To determine how RmInt1 colonizes the S.meliloti genome, the wild-type intron (pKG2.5) and an IEP+ΔORF derivative flanked by short (−20/+5) exons (pKGEMA4) were transferred to RMO17 by conjugation. Upon acquisition of either pKG2.5 or pKGEMA4, 100% of the transconjugants exhibited genomic retrohoming events revealing the extraordinary spreading ability of RmInt1. Retrohoming did not occur when the intron-donor plasmids encode an IEP truncated in the RT domain (pKG2.5x or pKGEMA4x; Figure 3), which is consistent with the requirement of reverse transcriptase activity and a functional IEP for intron mobility. We analyzed six independent RMO17 transconjugants harboring pKG2.5 and interestingly, in all these six clones the insertion of the wild-type intron occurred into four particular ISRm2011-2 genomic copies (IS3, IS4, IS6 and IS9) (the hybridization pattern of one of the clones is shown in Figure 3, left panel). Consistent with higher retrohoming efficiency, the ΔORF intron pKGEMA4 integrated into a larger number of homing sites. Seven independent transconjugants were analyzed, six displayed identical hybridization pattern to the RmInt1 probe sharing four retrohoming events with the RMO17 clones harboring pKG2.5 (IS3, IS4, IS6 and IS9) whereas insertion into IS2 was an additional retrohoming event. Retrohoming into IS13 was only detected in one clone of the seven analyzed (the hybridization pattern of this transconjugant is shown in Figure 3, right panel). These results suggest that certain homing sites favor RmInt1 retrohoming in the S.meliloti genome.
Figure 3.
Retrohoming of RmInt1 into the intronless RMO17 genome. Panels show Southern hybridizations of SalI-digested DNA from the RMO17 strain (wt) and derivatives harboring plasmids pKG2.5 (donor of the wild-type intron, left panel), pKGEMA4 (expressing a ΔORF variant, right panel), pKG2.5x or pKGEMA4x (both expressing non-mobile introns). The different lanes correspond to individual clones showing all the homing events, using as donor plasmids either pKG2.5 or pKGEMA4. Probe was specific to the 5′ end of the intron ribozyme sequence. D indicates the hybridization signal corresponding to the donor plasmid DNA. Genomic insertions of the wild-type intron (IS+I) or its ΔORF variant (IS+ΔI) are marked. IS were denoted with numbers (1–13) according to the hybridization pattern provided by SalI-digested total DNA using an IS probe numbering from the larger to the smaller hybridizing IS-containing fragment. Number of transconjugants with the observed events is indicated in brackets alongside the bands.
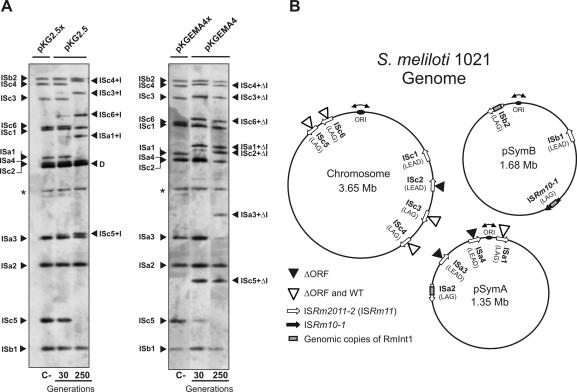
To get further insight into the dispersion pathway of RmInt1 in the S.meliloti genome, pKG2.5 and pKGEMA4 were transferred by conjugation into strain 1021, of which the genome has been sequenced (32). The tripartite genome of this S.meliloti strain is 6.7 Mb and contains a chromosome of 3.65 Mb, as well as two symbiotic megaplasmids known as pSymA (1.35 Mb) and pSymB (1.68 Mb). Strain 1021 harbors 13 homing sites for RmInt1 that include twelve copies of ISRm2011-2 and one copy of the closely related insertion sequence ISRm10-1. Two copies of ISRm2011-2 (one in pSymA and one in pSymB) are invaded by RmInt1, as occurs with the unique copy of ISRm10-1 located in pSymB (Figure 4B). The three homing sites invaded by RmInt1 reside on the template for the lagging-strand synthesis, which is consistent with the preferred retrohoming pathway for RmInt1 and other bacterial group II introns (21,24,28). It should be noted that the ISRm2011-2 intronless copies in strain 1021 all contain the characteristic nucleotide positions of RmInt1 wild-type target site. Analysis of independent 1021 transconjugants harboring pKG2.5 showed that the clones were not homogenous and the process of invasion of the homing sites by RmInt1 seems to occur in a gradual manner. This is revealed by the marked reduction in the intensity of hybridizing bands corresponding to intronless homing sites and the concomitant appearance of new ones containing the intron, with increased intensity with larger generations (compare hybridization patterns after 30 and 250 generations in Figure 4A, left panel). Our results revealed that upon acquisition of pKG2.5 five particular homing sites were gradually occupied by RmInt1. Likewise the ΔORF intron variant integrated into a higher number of homing sites as expected (Figure 4A, right panel). The wild-type intron expressed from pKG2.5 integrated into four chromosomal copies of ISRm2011-2 (ISc3, ISc4, ISc5 and ISc6) and one copy in pSymA (ISa1), all target-sites located on the template for the lagging-strand synthesis. The ΔORF variant inserted in the same homing sites and two more, one in pSymA (ISa3) and other in the chromosome (ISc2) (Figure 4B). Interestingly, these two additional retrohoming events occurred in target-sites located on the template for the leading-strand synthesis. Furthermore, some other clones carried one additional insertion into ISa4 also in the leading-strand orientation (Figure 4B). Taken together, these results suggest that the retrohoming pathway followed by RmInt1 for its dispersion in the S.meliloti genome is related to replication of the chromosome and symbiotic megaplasmids and prefers the template for lagging-strand synthesis as the target site.
Figure 4.
Retrohoming of RmInt1 into the genome of the S.meliloti reference strain 1021. (A) Southern hybridizations of BamHI-digested DNA from 1021 transconjugants harboring plasmids pKG2.5 or pKGEMA4 grown for 30 and 250 generations. Membranes were probed with a DNA fragment specific to the 3′ end of ISRm2011-2. First lane of each panel (C-) shows an ISRm2011-2 (IS) fingerprint of 1021 derivatives expressing non-functional introns from plasmids pKG2.5x or pKGEMA4x. Insertions of the wild-type intron (IS+I) or the ΔORF derivative (IS+ΔI) are indicated. D indicates hybridization signals from pKG2.5 and pKG2.5x DNA and the asterisk (*) a DNA fragment partially digested presumably containing ISb1. (B) Distribution of intron RmInt1 and the newly acquired intron copies (wt or ΔORF) within the 1021 genome. Schematic shows chromosome, pSymA and pSymB with indication of their replication origin, localization of RmInt1 targets, resident intron copies and new insertions upon acquisition of plasmids pKG2.5 or pKGEMA4 as deduced from the Southern analyses of all the transconjugants according to the S.meliloti 1021 genome annotation in (http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/). IS targets correspond to the following annotated 1021 genes: SMc02205 (ISc1), SMc00848 (ISc2), SMc02411 (ISc3), SMc01382 (ISc4), SMc03084 (ISc5), SMc03305 (ISc6), SMa3000 (ISa1), SMa3014 (ISa2), SMa3015 (ISa3), SMa3018 (ISa4), SMb21616 (ISb1), SMb21703-5 (ISb2) and ISRm10-1 (SMb21709-10).
Retrohoming ability of the genomic inserted intron copies and trans complementation of the ΔORF intron with the IEP coding sequence
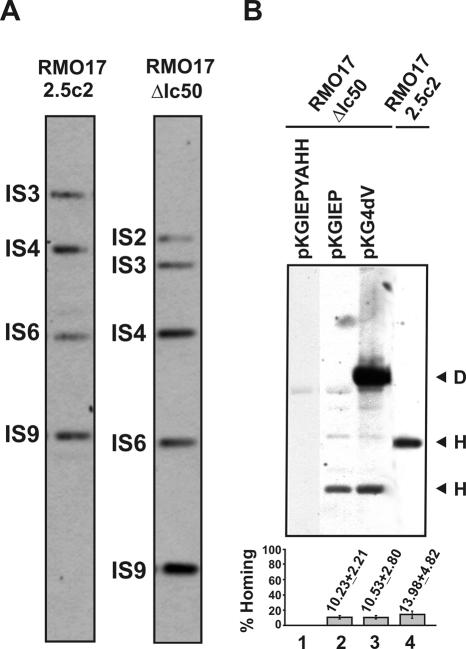
The results shown above suggest that the homing sites in the genome of S.meliloti will be gradually and sequentially colonized by RmInt1, as long as the intron is expressed in the bacterial cell. To test whether the newly acquired intron genomic copies retain retrohoming ability the RMO17 derivative strains were cured of donor plasmids (pKG2.5 or pKGEMA4) to generate RMO17-2.5c2 and RMO17-ΔIc50, respectively (Figure 5A) and a target-recipient plasmid pJB0.6LAG was then transferred by conjugation into these two strains. Retrohoming efficiency of the acquired wild-type introns in RMO17-2.5c2 was estimated to be 10–13% (Figure 5B, lane 4), whereas no insertion events into the target-recipient plasmid were detected in RMO17-ΔIc50, colonized by the ΔORF intron variant (data not shown). These results indicate that the acquired wild-type intron genomic copies retain retrohoming ability and therefore they could contribute to further intron dispersal. Furthermore, retrohoming of the intron genomic copies requires the IEP, which cannot be replaced by the host genetic background.
Figure 5.
Mobility of the genomic intron copies and trans complementation of the ΔORF derivative with the IEP. (A) Wild-type RmInt1 and ΔORF profiling in RMO17 derivatives cured from donor plasmids pKG2.5 (RMO17-2.5c2) or pKGEMA4 (RMO17-ΔIc50). Genomic DNA from both strains was SalI-digested and probed with a DNA fragment specific to the 5′ end sequence of the RmInt1 ribozyme. (B) In vivo homing of the acquired genomic introns. Target plasmid pJB0.6LAG was mobilized into RMO17-2.5c2 and RMO17-ΔIc50 strains. Compatible plasmids pKGIEP or pKG4dV expressing a functional IEP were then conjugated into RMO17-ΔIc50 containing pJB0.6LAG to complement the ΔORF insertions. Homing of wild-type RmInt1 and ΔORF derivative was assessed on plasmid pools from RMO17-2.5c2 and RMO17-ΔIc50, respectively, as described. pKGIEPYAHH was used as negative complementation control in the assays. Invasion rates were calculated as described in materials and methods and plotted in the histogram shown below. D, donor plasmid; H, homing product.
Intron Ll.LtrB maturase (LtrA) functions most efficiently when expressed in cis either from its normal location within the intron or from a position either upstream or downstream of the ΔORF. The maturase also functions when expressed in trans from a separate plasmid, but splicing is <10% of that for a cis configuration (43). To investigate the trans complementation of the RmInt1 IEP functions, plasmids pKGIEP and pKG4dV were individually conjugated into strain RMO17-ΔIc50 harboring the compatible target-recipient plasmid pJB0.6LAG. Both plasmids express the wild-type IEP from a kanamycin resistance gene constitutive promoter either alone (pKGIEP) or within the context of a splicing defective ribozyme that contains a mutation within the catalytic DV (GUU→CGA; pKG4dV). pKGIEPYAHH expressing a non-functional IEP with a mutation in the RT domain (YADD→YAHH) was used as negative complementation control. Interestingly, when complemented in trans with a functional IEP either alone or within the context of a defective ribozyme, the genomic copies of the ΔORF intron exhibited ∼10% homing efficiency in pJB0.6LAG, which is comparable to that displayed by the wild-type intron (Figure 5B, lanes 2 and 3). As expected, the RT mutant IEP was not able to mobilize the ΔORF intron (lane 1). These results presumably reflect the stability of the free protein and unspliced intron precursor RNA in the S.meliloti host and let us to hypothesize that inactive intron copies could still contribute to intron dispersal by supplying or being complemented in trans with an active IEP.
DISCUSSION
The dispersal of group II introns into bacterial genomes has been addressed mainly by analyzing retrotransposition of the Ll.LtrB intron (21–24,44). Dissemination by retrohoming has been also reported, but these studies were limited to the analysis of a reduced number of chromosomal events and the invasion of target-recipient plasmids (24,25) or to the use of engineered intron-donors (targetron) that could be influenced by many factors inherent to such experimental systems (45,46). In this study, we determine how a bacterial genome is colonized by a group II intron upon acquisition by conjugative transfer. This analysis was possible due to the existence of numerous unoccupied RmInt1 homing sites in the genome of some S.meliloti strains, thus providing further insights on intron dispersal in natural bacterial populations.
We first showed that RmInt1 splicing in vivo and its homing efficiency were greatly improved by removing most of the protein coding sequence from the large terminal loop of DIV and by using short flanking exon sequences extending 20 nt in the 5′ exon and 5 nt in the 3′ exon. This improvement on splicing was previously observed for the RmInt1 in vitro splicing reaction (35,39), although the influence of a shorter 3′ exon was not investigated. Furthermore, homing frequencies of a Ll.LtrB-ΔORF derivative expressing the IEP downstream or upstream of the ΔORF was greatly stimulated, presumably because a greater resistance of ΔORF intron to nucleolytic cleavage in DIV (41,42). For RmInt1, we determined that the IEP also functions efficiently either upstream or downstream the ΔORF intron. The ΔORF variant used in our assays still contains one DIVb stem of ∼30 nt pairings (Figure 2A). Deletion of Ll.LtrB DIVb inhibited splicing by ∼60% (43) and similar deletions in the yeast aI2 intron coding sequence strongly-inhibited maturase-promoted reverse splicing in an in vitro assay, but its effect was not observed in vivo. We found that a larger deletion of RmInt1 DIVb reduced homing efficiency and a full deletion almost inhibited homing (Figure 2A). Thus, DIVb is required for efficient homing of RmInt1 and presumably for IIB group II intron mobility.
Upon acquisition of the wild-type intron or an IEP+ΔORF variant with a marked increase of homing efficiency, the S.meliloti genome homing sites begin to be occupied by retrohoming events. This colonization occurs with an extraordinary frequency since these events were detected in 100% of the transconjugants analyzed. We previously reported that the preferred RmInt1 retrohoming pathway is consistent with reverse splicing of the intron RNA into single-stranded DNA at a replication fork, using a nascent DNA lagging-strand as the primer for reverse transcription (28) similar to the ectopic transposition of group II introns (21). The colonization of the homing sites in the S.meliloti genome corresponds to this preferred retrohoming pathway and our results predict that the homing sites located on the template for the lagging-strand synthesis are the first to be invaded by the intron, later the available homing sites on the leading strand template will be occupied as well. Interestingly, even though there is at least a proven ectopic site in S.meliloti named oxi1 (30) retrotransposition was not detected in our assays. Since this is a rather rare event, retrohoming seems to be the preferred mobility mechanism for intron dissemination in the S.meliloti genome. Once the homing sites are saturated, retrotransposition could contribute to further intron dispersion.
It has been reported that Ll.LtrB also retrotransposes into plasmid targets with a bias toward the template for the lagging-strand DNA synthesis even stronger than that for chromosomal events (22). It has been speculated that this bias may be related to the different location of chromosome and plasmids in the cell during replication. We have not observed such a bias for retrohoming of RmInt1 in the S.meliloti genome and in fact all the target sites located in the preferred strand for retrohoming into the chromosome were occupied. In S.meliloti, the two symbiotic plasmids pSymA and pSymB are considered chromosome-like replicons (32) and therefore they could share the same partioning properties as the main chromosome being located at similar sites during cell division and therefore equally accessible to intron RNPs. Furthermore, our results do not reveal a preference for integration of the intron into homing sites near the Ori domain as it has been observed for retrotransposition of Ll.LtrB (22,23) and retrohoming into E.coli genome of gene targeting vectors (targetron) based on Ll.LtrB intron (45).
We also showed that the retrohomed wild-type introns in the S.meliloti genome retain mobility, but not the ΔORF variant, which imply that the functions of the IEP on intron mobility cannot be substituted by any other factor in this host genetic background. Complementation in trans with LtrA has been achieved for a Ll.LtrB ΔORF derivative, but splicing is <10% of that in cis configuration (43). Interestingly, the RmInt1 ΔORF variant inserted in the genome of S.meliloti could be also mobilized by supplying a functional IEP in trans, either alone or within the context of a defective ribozyme, displaying a invasion rate on a plasmid-borne target site similar to that exhibited by the wild-type intron. These findings suggest that some mutant introns can still contribute efficiently to intron dispersal by supplying a functional IEP.
From an evolutionary perspective, it has been proposed that RmInt1 has been inherited vertically from a common ancestor in S.meliloti and related bacteria and also by independent horizontal-transfer events (31). Presently it is estimated that only 10% of the S.meliloti isolates lack the intron and that presumably, in this population there is no restriction to intron homing, i.e. strain RMO17. The extraordinary spreading ability of RmInt1 and the high number of homing sites available in these isolates predict that the intron might be disseminated into S.meliloti population by lateral transfer nowadays, with retrohoming being the main mechanism for the colonization of the recipient genomes.
Acknowledgments
This work was funded by the Spanish Ministerio de Educación y Ciencia (project no. BIO2005-02312). The authors are grateful to Vicenta Millán and Ascensión Martos for technical assistance. Funding to pay the Open Access publication charges for this article was provided by Spanish Ministerio de Educación y Ciencia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Michel F., Ferat J.L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 2.Toro N. Bacteria and Archaea group II introns; additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 3.Lambowitz A.M., Zimmerly S. Mobile Group II Introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 4.Carignani G., Groudinsky O., Frezza D., Schiavon E., Bergantino E., Slonimski P.P. An mRNA maturase is encoded by the first intron of the mitochondrial gene for the subunit I of cytochrome oxidase in S.cerevisiae. Cell. 1983;35:733–742. doi: 10.1016/0092-8674(83)90106-x. [DOI] [PubMed] [Google Scholar]
- 5.Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 6.Moran J.V., Mecklenburg K.L., Sass P., Belcher S.M., Mahnke D., Lewin A., Perlman P. Splicing defective mutants of the COX1 gene of yeast mitochondrial DNA: initial definition of the maturase domain of the group II intron aI2. Nucleic Acids Res. 1994;22:2057–2064. doi: 10.1093/nar/22.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saldanha R., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A.M. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 8.Matsuura M., Noah J.W., Lambowitz A.M. Mechanism of maturase-promoted group II intron splicing. EMBO J. 2001;20:7259–7270. doi: 10.1093/emboj/20.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toor N., Hausner G., Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerly S., Hausner G., Wu X.-C. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toro N., Molina-Sánchez M.D., Fernández-López M. Identification and characterization of bacterial class E group II introns. Gene. 2002;299:245–250. doi: 10.1016/s0378-1119(02)01079-x. [DOI] [PubMed] [Google Scholar]
- 12.Ferat J.L., Le Gouar M., Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 13.Mohr G., Perlman P.S., Lambowitz A.M. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–4997. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A.E. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shub D.A., Goodrich-Blair H., Eddy S.R. Amino acid sequence motif of group I endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 16.San Filippo J., Lambowitz A.M. Characterization of the C-Terminal DNA-Binding/DNA Endonuclease region of a group II Intron-Encoded Protein. J. Mol. Biol. 2002;324:933–951. doi: 10.1016/s0022-2836(02)01147-6. [DOI] [PubMed] [Google Scholar]
- 17.Lambowitz A.M., Caprara M.G., Zimmerly S., Perlman P.S. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 451–485. [Google Scholar]
- 18.Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M. Mobile introns: pathways and proteins. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA. 2nd edn. Washington DC: ASM Press; 2002. pp. 761–783. [Google Scholar]
- 19.Martínez-Abarca F., Toro N. Group II introns in the bacterial world. Mol. Microbiol. 2000;38:917–926. doi: 10.1046/j.1365-2958.2000.02197.x. [DOI] [PubMed] [Google Scholar]
- 20.Dickson L., Huang H-R., Liu L., Matsuura M., Lambowitz A.M., Perlman P.S. Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA. 2001;98:13207–13212. doi: 10.1073/pnas.231494498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichiyanagi K., Beauregard A., Lawrence S., Smith D., Cousineau B., Belfort M. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 2002;46:1259–1272. doi: 10.1046/j.1365-2958.2002.03226.x. [DOI] [PubMed] [Google Scholar]
- 22.Ichiyanagi K., Beauregard A., Belfort M. A bacterial group II intron favors retrotransposition into plasmid targets. Proc. Natl Acad. Sci. USA. 2003;100:15742–15747. doi: 10.1073/pnas.2536659100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coros C.J., Landthaler M., Piazza C.L., Beauregard A., Esposito D., Perutka J., Lambowitz A.M., Belfort M. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol. Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- 24.Belhocine K., Plante I., Cousineau B. Conjugation mediates transfer of the Ll.LtrB group II intron between different bacterial species. Mol. Microbiol. 2004;51:1459–1469. doi: 10.1111/j.1365-2958.2004.03923.x. [DOI] [PubMed] [Google Scholar]
- 25.Belhocine K., Yam K.K., Cousineau B. Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J. Bacteriol. 2005;187:930–939. doi: 10.1128/JB.187.3.930-939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Klein J.R., McKay L.L., Dunny G.M. Quantitative analysis of group II intron expression and splicing in Lactococcus lactis. Appl. Environ. Microbiol. 2005;71:2576–2586. doi: 10.1128/AEM.71.5.2576-2586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Abarca F., García-Rodríguez F.M., Toro N. Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol. Microbiol. 2000;35:1405–1412. doi: 10.1046/j.1365-2958.2000.01804.x. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Abarca F., Barrientos-Durán A., Fernández-López M., Toro N. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res. 2004;32:2880–2888. doi: 10.1093/nar/gkh616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Abarca F., Zekrí S., Toro N. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol. Microbiol. 1998;28:1295–1306. doi: 10.1046/j.1365-2958.1998.00894.x. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz E., Villadas P.J., Toro N. Ectopic transposition of a group II intron in natural bacterial populations. Mol. Microbiol. 2001;41:645–652. doi: 10.1046/j.1365-2958.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 31.Fernández-López M., Muñoz-Adelantado E., Gillis M., Willems A., Toro N. Dispersal and evolution of the Sinorhizobium meliloti group II RmInt1 intron in bacteria that interact with plants. Mol. Biol. Evol. 2005;22:1518–1528. doi: 10.1093/molbev/msi144. [DOI] [PubMed] [Google Scholar]
- 32.Galibert F., Finan T.M., Long S.R., Pühler A., Abola P., Ampe F., Barloy-Hubler F., Barnett M.J., Becker A., Boistard P., et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 33.Villadas P.J., Velázquez E., Martínez-Molina E., Toro N. Identification of nodule-dominant Rhizobium meliloti strains carrying pRmeGR4b-type plasmid within indigenous soil populations by PCR using primers derived from specific DNA sequences. FEMS Microbiol. Ecol. 1995;17:161–168. [Google Scholar]
- 34.Robertsen B.K., Aiman P., Darvill A.G., McNeil M., Albersheim P. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981;67:389–400. doi: 10.1104/pp.67.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa M., Michel F., Molina-Sánchez M.D., Martínez-Abarca F., Toro N. An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie. 2006a;88:711–717. doi: 10.1016/j.biochi.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Jiménez-Zurdo J.I., García-Rodríguez F.M., Barrientos-Durán A., Toro N. DNA-target requirement for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J. Mol. Biol. 2003;326:413–423. doi: 10.1016/s0022-2836(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 37.Muñoz-Adelantado E., San Filippo J., Martínez-Abarca F., García-Rodríguez F.M., Lambowitz A.M., Toro N. Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J. Mol. Biol. 2003;327:931–943. doi: 10.1016/s0022-2836(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-Abarca F., Toro N. RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res. 2000;28:4397–4402. doi: 10.1093/nar/28.21.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa M., Michel F., Toro N. Potential for alternative intron–exon pairings in group II intron RmInt1 from Sinorhizobium meliloti and its relatives. RNA. 2006;12:338–341. doi: 10.1261/rna.2240906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molina-Sánchez M.D., Martínez-Abarca F., Toro N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 2006;281:28737–28744. doi: 10.1074/jbc.M602695200. [DOI] [PubMed] [Google Scholar]
- 41.Guo H., Karberg M., Long M., Jones J.P., III, Sullenger B., Lambowitz A.M. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- 42.Plante I., Cousineau B. Restriction for gene insertion within the Lactococcus lactis Ll.LtrB group II intron. RNA. 2006;12:1–13. doi: 10.1261/rna.193306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui X., Matsuura M., Wang Q., Ma H., Lambowitz A.M. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Conlan L.H., Stanger M.J., Ichiyanagi K., Belfort M. Localization, mobility and fidelity of retrotransposed group II introns in rRNA genes. Nucleic Acids Res. 2005;33:5262–5270. doi: 10.1093/nar/gki819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong J., Karberg M., Lambowitz A.M. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 2003;31:1656–1664. doi: 10.1093/nar/gkg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao J., Zhong J., Lambowitz A.M. Gene targeting using randomly inserted group II introns (targetrons) recovered from an Escherichia coli gene disruption library. Nucleic Acids Res. 2005;33:3351–3362. doi: 10.1093/nar/gki649. [DOI] [PMC free article] [PubMed] [Google Scholar]