Abstract
Although ancient DNA (aDNA) miscoding lesions have been studied since the earliest days of the field, their nature remains a source of debate. A variety of conflicting hypotheses exist about which miscoding lesions constitute true aDNA damage as opposed to PCR polymerase amplification error. Furthermore, considerable disagreement and speculation exists on which specific damage events underlie observed miscoding lesions. The root of the problem is that it has previously been difficult to assemble sufficient data to test the hypotheses, and near-impossible to accurately determine the specific strand of origin of observed damage events. With the advent of emulsion-based clonal amplification (emPCR) and the sequencing-by-synthesis technology this has changed. In this paper we demonstrate how data produced on the Roche GS20 genome sequencer can determine miscoding lesion strands of origin, and subsequently be interpreted to enable characterization of the aDNA damage behind the observed phenotypes. Through comparative analyses on 390 965 bp of modern chloroplast and 131 474 bp of ancient woolly mammoth GS20 sequence data we conclusively demonstrate that in this sample at least, a permafrost preserved specimen, Type 2 (cytosine→thymine/guanine→adenine) miscoding lesions represent the overwhelming majority of damage-derived miscoding lesions. Additionally, we show that an as yet unidentified guanine→adenine analogue modification, not the conventionally argued cytosine→uracil deamination, underpins a significant proportion of Type 2 damage. How widespread these implications are for aDNA will become apparent as future studies analyse data recovered from a wider range of substrates.
INTRODUCTION
The study of post mortem DNA damage is critically important to help ensure the generation of accurate data from ancient or degraded sources of DNA. DNA damage not only rapidly reduces the length and number of PCR amplifiable starting template molecules within a biological sample, but also can lead to the generation of erroneous sequence. The better characterization of ancient DNA (aDNA) damage will help the development of new damage strategies to both extend the range of samples from which useful DNA can be recovered, and help monitor and account for potentially erroneous data, which can have disastrous consequences on any study that requires the recovery of accurate sequence, e.g. phylogenetic and population genetic studies (1), genomic data analyses (2) and environmental reconstructions (3).
Miscoding lesions are a defining characteristic of aDNA studies. Although usually observed as variations in individual sequences among datasets of cloned PCR products (4), they are sometimes noticeable within directly sequenced PCR products, conferring the impression of sequence heteroplasmy (5). Two mechanisms have been suggested as the underlying cause behind the observed miscoding lesions. The first is the result of regular PCR polymerase amplification errors (the misincorporation of erroneous nucleotide bases due to innate enzyme error). In situations where starting PCR template molecules are low (as with many aDNA studies), such errors can result in the modification of template molecules during early stages of the PCR, and thus will produce a significant (i.e. observable through cloning or direct sequencing) proportion of descendents with the modification. The alternative, probably complementary, mechanism is the generation of errors due to post mortem biochemical damage of the original starting template molecules. The chemical structure of these damage-derived miscoding lesions is such that they can be read by the PCR enzyme, although erroneously (4,6,7). Probably the most commonly accepted example of the latter is the hydrolytic deamination of cytosine to uracil or its analogues, which during subsequent enzymatic replication leads to the generation of cytosine to thymine miscoding lesions (4).
The existence of damage-derived miscoding lesions in DNA from fossil remains has previously been proven using several methods. Through statistical comparisons, it has been demonstrated that aDNA sequences are characterized by relatively high occurrences of transitions compared with sequences from contemporary specimens. Therefore, it has been argued that enzymatic error alone cannot explain the aDNA observations (7,8). The nature of these transitions themselves has however been a source of debate, both as to which types are truly associated with damage, and what the underlying cause of the miscoding lesions might be. Partly to blame for this lack of concordance is that the study of such damage is not trivial. Conclusions from most previous studies have been based on sequences generated from cloned PCR products. However, the nature of PCR causes any single original damage event to be viewed as two possible manifestations, dependent on whether a descendent of the damaged DNA strand, or of a complement sequence generated from the damaged strand, is sequenced. For example, consider an original cytosine to thymine transition (C→T) on a mitochondrial Light (L) strand molecule. L strand sequence generated post PCR will yield a C→T miscoding lesion. However, if the DNA is sequenced from the complementary Heavy (H) strand sequence, the transition will be read as a guanine to adenine (G→A) transition. Similarly, an original A→G damage event can be observed as either an A→G miscoding lesion, or the complementary T→C (7). Because the strand of origin of the miscoding event cannot be identified, Hansen et al. (7) suggest that miscoding lesions can be grouped into six complementary, effectively indistinguishable pairs: (A→C/T→G), (A→G/T→C), (A→T/T→A), (C→A/G→T), (C→G/G→C) and (C→T/G→A). Furthermore, the dominance of transitions in aDNA damage datasets has led the same authors to suggest that the two pairs of transitions be referred to as Type 1 (A→G/ T→C) and Type 2 (C→T/ G→A), respectively.
Although both types of transitions have been observed and commented on among aDNA datasets (4,7,8–12) controversy has raged as to whether both types truly represent damage [as argued by (7,8,10,12)] or whether Type 1 damage simply represents polymerase enzyme misincorporation errors at early stages of the PCR process (9,13). Furthermore, the debate does not stop there; the underlying causes of the damage are also under question. Though the few studies that attempt to examine miscoding lesions in detail have concurred that, as in vivo, Type 2 transitions arise from the deamination of cytosine to uracil (4,8,9). Those studies that argue for the existence of Type 1 damage also argue that the deamination of adenine to hypoxanthine, an analogue of guanine, is also important to aDNA (8).
The limitation of such arguments is that they are not to a large extent based on observations of the actual raw data, but rather on theoretical arguments drawn from what is known about in vivo damage systems, thus about what damage may exist, and how the polymerase enzymes therefore may react to them. A small number of studies have attempted to investigate damage directly using various biochemical experiments, for example, the treatment of aDNA extracts using uracil-N-glycosylase before PCR amplification, in order to investigate how the distribution of miscoding lesions varies as a result (4,8,9,14,15). However, such studies are subject to the limitations that they (i) can only provide information about damage types that are specifically targeted and (ii) assume a high degree of substrate specificity by the enzymes that may not necessarily be accurate. Thus, they both do not provide information about damage types that are not targeted by the assay and may in some cases lead to the incorrect assignment of an biochemical cause to an observed effect.
The recent development of the sequencing-by-synthesis technology (Genome sequencer GS20; Roche Applied Science) (16) offers a solution to these previously intractable problems. Specifically, the nature of the data-generation process is such that DNA sequence data can be unambiguously assigned to individual, original single-stranded molecules. In brief, during the initial stage of the data preparation DNA molecules are first fragmented, then denatured, and single-stranded molecules are emulsified with amplification reagents in a water-in-oil immersion, within which subsequent PCR occurs. During the subsequent emulsion-based clonal amplification (also known as emulsion PCR, emPCR), individual PCRs occur at a large scale, in parallel, and the descendant molecules of each individual reaction that are complementary to the originally isolated single-stranded molecule are bound by the capture bead. During the final stages of the data-generation process, the bound molecules on each individual capture bead are pyrosequenced as a single unit, in parallel with up to 0.8 million other beads from the same emPCR. Each contains PCR products from a different original, single-stranded DNA template molecule, and the data from each are recorded separately. The key benefit therefore is that each final sequence reaction is generated from a single single-stranded DNA molecule and, as such, provides a direct window into any damage-derived miscoding lesions that were present on the molecule, thus in an instant providing the critical information that has been lacking from previous aDNA damage studies. Further details of this process are outlined later in the manuscript and in Supplementary Data.
In light of these benefits, we have analysed a dataset of DNA sequences produced using the GS20 DNA sequencing platform to further explore the nature of aDNA damage-driven miscoding lesions. Through comparative analyses on 390 965 bp of chloroplast DNA (cpDNA) generated from fresh (thus not containing damage forms that arise through conventional aDNA degradation processes) yellow-poplar (Liriodendron tulipfera) chloroplasts, and 131 474 bp of ancient woolly mammoth (Mammuthus primigenius) mitochondrial DNA (mtDNA), we show that a clear difference exists between the miscoding lesion spectra of modern and ancient, permafrost preserved DNA. Through statistical analysis of the data we conclusively demonstrate that Type 2 (C→T/G→A) miscoding lesions represent the overwhelming majority (88% total miscoding lesions, 94% of transitions) of damage-derived miscoding lesions in aDNA from this specimen, in accordance with the hypothesis of Hofreiter et al. (9) and in contrast to others, including those postulated by some of the authors of this paper (7,8,12). Finally, using a simple logical argument based in principle on our observations on the GS20 data-generation process, we demonstrate how the strand of origin of the sequences can be identified, and further how the underlying cause of observed damage types on the aDNA data can be identified, thus removing the need to group miscoding lesions into complementary pairs. Through subsequent subdivision of the aDNA data into the different (L and H) strands of origin using this method, we demonstrate that the rate of occurrence and distribution of damage types is not significantly different between the two strands of the mitochondria. Finally, we explore the biochemical basis of the damage, and demonstrate that in this specimen at least (from the aDNA point of view a fairly standard, permafrost preserved bone), in addition to the conventionally argued (4,7–9,12) deamination of cytosine to uracil and its analogues, an as yet unidentified derivative of guanine that leads to the generation of G→A miscoding lesions underpins a significant proportion of aDNA damage-driven miscoding lesions. Our findings are derived from permafrost preserved bone material, and as such their applicability across other common aDNA substrates remains unknown. As future datasets that are based on other samples are released, analyses such as ours will help further in understanding of aDNA damage.
MATERIALS AND METHODS
aDNA sequence data
As a consequence of the relatively rare occurrence of aDNA damage-derived miscoding lesions, comparative studies require large quantities of DNA sequence data in order so that statistically supported conclusions can be drawn. Furthermore, genetic regions analysed require multiple sequence coverage, so that true damage can be discriminated from other sources of sequence variation, such as allelic variation or the co-amplification of nuclear-mitochondrial sequences (numts). In short, this explains why to date few studies have been able to investigate damage-derived miscoding lesion damage in detail (7–9,12). The GS20 sequencing platform has opened up the possibility of generating large amounts of aDNA sequence data (with the caveat that samples contain sufficient DNA, of a minimum quality, to enable successful analysis). Furthermore, because of their relatively high copy number, and thus overall cellular abundance in comparison with nuDNA, the initial GS20 analyses on aDNA extracts have characteristically produced large amounts of mtDNA (2). For our analysis, we have used a dataset of woolly mammoth ancient mtDNA sequence that comprises the sequences published in the first GS20 aDNA study (2), plus further mtDNA sequences from the same individual that have been generated since. These DNA sequences plus the full mitochondrial L strand consensus sequence for the specimen are provided in the Supplementary Data to this paper. Care was taken to avoid nuclear copies of mtDNA (numts), as follows. Analysis of numts in fully sequenced mammalian genomes showed that at most 3% of the reads aligning to the mitochondrial genome (at our criteria) could be expected to be numts. Requiring that a read be 98% identical to mammoth mtDNA sequence eliminated 15% of the aligning reads, most of which we believe to be low-quality data. Even if as much as 1% of the remaining reads are numts (so recent as to retain 98% identity), none of our broad conclusions would be materially affected. The large amount of sequence data, in contrast to the length of the mammoth mitochondria (16 770 bp for this individual; W. Miller, H.N. Poinar, J. Qi, C. Schwartz, L.P. Tomsho, R.D.E. MacPhee and S.C. Schuster, submitted for publication) results in up to 21 times coverage of some parts of the mtDNA genome, with a mean and modal coverage of 7.8 and 7 times, respectively.
The individual sequence reads were aligned with the pre-determined consensus sequence of the mtDNA genome using the blastz program (17), and miscoding lesions were extracted from the alignment and assigned to the six complementary pairs of miscoding lesions of Hansen et al. (7) using straightforward computer programs that we wrote for that purpose. See Table 1 for summary data.
Table 1.
Number of miscoding lesions observed within chloroplast and mammoth datasets
| Miscoding lesions originally derived from A and T nucleotides | Miscoding lesions originally derived from C and G nucleotides | |||||||
|---|---|---|---|---|---|---|---|---|
| A→G T→C | A→C T→G | A→T T→A | Total A+Ta | C→A G→T | C→G G→C | C→T G→A | Total C+Gb | |
| Chloroplast | 78 | 24 | 89 | 244 230 | 33 | 9 | 52 | 146 735 |
| Mammoth | 39 | 7 | 9 | 81 790 | 16 | 8 | 597 | 49 684 |
| Corrected Mammothc | 116 | 21 | 27 | 47 | 24 | 1763 | ||
| Nucleotide ratio | 2.99 | 2.95 | ||||||
aTotal number of adenine and thymine nucleotides in dataset.
bTotal number of cytosine and guanine nucleotides in dataset.
cCorrected Mammoth: the number of observed lesions among the mammoth sequence data, scaled to match the total chloroplast nucleotides sequenced. For example, corrected mammoth count for A→G/T→C pair was calculated as (Observed Mammoth A→G/T→C)*(Total Chloroplast A+T)/(Total Mammoth A+T) = 39*244 320/81 790 = 116.
Analysis 1: Statistical discrimination of damage from PCR enzyme misincorporation error
The GS20 emPCR process incorporates the use of the high-fidelity polymerase, Platinum Taq Hifidelity (Invitrogen), an enzyme mixture composed of recombinant Taq DNA polymerase, Pyrococcus spp. GB-D thermostable polymerase and Platinum Taq antibody. This enzyme is marketed partly on its very low misincorporation rate, 2 × 10−6 (Invitrogen). In this study we find the actual rate of misincorporation to be higher (≈7 × 10−4), similar to results from a previous aDNA study that has also specifically examined these properties of this enzyme (8). To discriminate between true aDNA damage and enzyme error or potential damage that may have arisen during the DNA extraction or that may have been present in the DNA before extraction, we analysed a further dataset of GS20 sequences, generated from a modern DNA extract, comprising 390 965 bp of L.tulipfera cpDNA. These data are part of the first chloroplast genome sequenced using the GS20 (J.E. Carlson, J.H. Leebens-Mack and D.G. Peterson, manuscript in preparation) and constitutes all the sequence reads between np 45 000 and 90 000 of the genome (J.E. Carlson, J.H. Leebens-Mack and S. Schuster, unpublished data). Although we are aware that in theory some complications may be envisioned when comparing cpDNA with mtDNA, at the current time there is a paucity of available datasets that contain sufficiently large amounts of sequence data to enable meaningful statistical comparisons. Thus this dataset provides the most suitable information at this time. The data analysed here have maximal coverage of 36 times, with a mean and modal coverage of 8.7 and 8 times, respectively. The L.tulipfera cpDNA sequences are available at the NCBI Trace Archives (Trace Identifiers 1367656065–1367659980). Analysis of the genomic data produced indicates that levels of heteroplasmy in the sample are negligible, thus unlikely to effect the analyses (J.E. Carlson, J.H. Leebens-Mack and S. Schuster, unpublished data). Furthermore, as DNA from this sample was freshly extracted from modern tissue, miscoding lesions observed in the data are unlikely to be due to anything other than PCR or other sequencing error that arises during the GS20 data production process. The miscoding lesion spectrum was extracted from the data in the same manner as applied to the mtDNA data. For data summary see Table 1.
A χ2-test of independence was used to investigate whether the distribution of miscoding lesions was the same in the mammoth and chloroplast sequence data. The data were first summarized into six complementary damage pairs (Table 1). Subsequently, because nucleotide usage is different between the mammoth and chloroplast data, tests were performed separately on those miscoding lesions that originated from an A or T (A+T), and those that originated from a G or C (G+C).
Analysis 2: Determination of which complementary miscoding lesion pairs represent true damage
To identify which of the six complementary pairs of miscoding lesions represent true damage in the mammoth mtDNA data as opposed to enzyme misincorporation errors, the data were modelled using the Poisson distribution with rates derived from the chloroplast data, i.e. making the assumption that the observed miscoding lesion rates from the chloroplast data represent the true enzyme rates of lesions (Table 2). For example, the number of A→G/T→C miscoding lesions in the mammoth data were assumed to follow a Poisson distribution, with rate [Observed chloroplast A→G/T→C miscoding lesions]*[(Total Mammoth A+T nucleotides)/(Total Chloroplast A+T nucleotides)] = 78*81 790/244 230 = 26.12. Subsequently, the test-probability P(X ≥ Observed, or X ≤ Observed − Expected) = P(X ≥ 39, or X ≤ 39 − 26.12) was calculated, where X represents the number of lesions. If P is low, then it is likely that another mechanism than enzyme failure is accountable for the observed number of lesions in mammoth. The test-probability was made two-sided, because a priori we do not know the direction of deviation from the chloroplast data.
Table 2.
Number of observed and expected miscoding lesions in mammoth dataset
| A→G T→C | A→C T→G | A→T T→A | C→A G→T | C→G G→C | C→T G→A | |
|---|---|---|---|---|---|---|
| Observeda | 39 | 7 | 9 | 16 | 8 | 597 |
| Expectedb | 26.12 | 8.04 | 29.81 | 11.17 | 3.05 | 17.61 |
| P-valuec | 0.011 | 0.86 | 8 × 10−4 | 0.17 | 0.013 | <1 × 10−5 |
| Occurrence per bp sequenced | 1.5 × 10−4 | 0 | 0 | 9.7 × 10−5 | 9.9 × 10−5 | 0.01 |
aAbsolute number of miscoding lesions observed.
bExpected number of miscoding lesions, modelled using the Poisson distribution with rates derived from the chloroplast data.
cWhen using a 5% Bonferroni corrected significance level P-values below 5%/6=0.0084 are significant, leaving only (A→T/T→A) and (C→T/G→A) significant.
Under the assumption that the chloroplast data represent the true enzyme error, a basic rate of damage occurrence can be calculated for the six miscoding lesion types with the formula Max(Observed − Expected,0)/(Total source nucleotides).
Identification of the original template source and orientation for each DNA sequence
As mentioned above, the GS20 data production differs from conventional PCR and sequencing methods, in so far as each individual DNA sequence is ultimately derived from a single single-stranded DNA molecule. Although the DNA preparation and manipulation involves many steps, with regards to our analyses three key steps occur.
The original DNA molecules are rendered single-stranded and physically isolated from each other. All subsequent steps retain this isolation.
emPCR is performed on the original molecules, in isolated, parallel reactions. At the end of this process the final double-stranded emPCR products are themselves rendered single-stranded. Only the DNA molecules that are complementary to the original single-stranded molecule are retained.
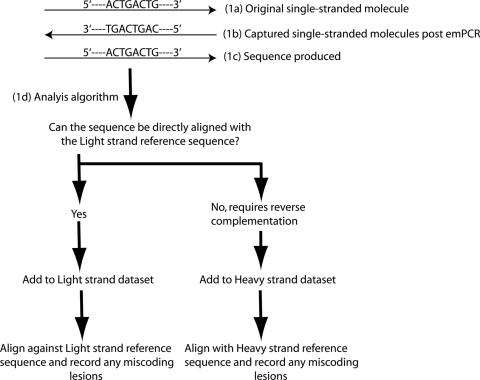
These molecules are pyrosequenced (still in isolated, parallel reactions). The sequence produced from each individual reaction is the complement of the emPCR products, thus identical to the original DNA molecule (Figure 1).
Figure 1.
Orientation of the DNA molecule at different steps of the data production process, and the algorithm used to subsequently segregate and analyse the sequence data. The GS20 emPCR and pyrosequencing process occurs in isolated, parallel reactions (up to 0.8 million per GS20 run). This figure illustrates key stages of the data-generation and analytical process for each individual reaction within a single GS20 run. (1a) Before emPCR original single-stranded DNA molecules are isolated. (2a) Post-emPCR, only descendents of the original molecule that are in the complementary orientation are retained. (1c) These molecules are pyrosequenced, generating sequence in the identical orientation to the originally isolated single-stranded DNA molecule. (1d) A simple analytical algorithm is then applied to the sequence to identify the orientation of, and any miscoding lesions in, the original single-stranded DNA molecule.
The details of this process are expanded upon in Supplementary Data. With the knowledge that the sequence directly describes the orientation of the original source single-stranded DNA molecule, it is possible to identify the strand of origin of each generated DNA sequence, by following a simple algorithm as demonstrated in Figure 1. Furthermore, once the strand of origin of the sequence is identified, the sequence can be directly compared to a strand-specific ‘reference sequence’ (in our analyses, L strand molecules are compared against the mtDNA consensus L strand sequence for this mammoth, and H strand molecules are compared against the complementary H strand sequence). As the sequence directly represents the original template molecule, any miscoding lesions observed directly represent original damage events. Therefore, in contrast to previous studies that have been limited to analysing the 12 potential miscoding lesions as 6 complementary pairs, all 12 original miscoding lesion frequencies can be obtained.
Using the above algorithm, we separated the ancient mtDNA sequences into two datasets, those derived from original mitochondrial H strand molecules, and those derived from original L strand molecules. Subsequently, we obtained the miscoding lesion frequencies for the two datasets for use in further analyses detailed below.
Analysis 3: Investigation for differences in strand damage accumulation rates
It has previously been speculated that the miscoding lesion damage rates on the different mtDNA strands (i.e. the L and H) may vary due to base composition, secondary structure or other reasons (8,18). To identify this phenomena, the miscoding lesion distributions of the separate L and H strand datasets were statistically compared using a χ2-test. The data are shown in Table 3, classified according to the type of the original base. A separate χ2-test was performed for each type of nucleotide to account for the differences in nucleotide compositions between the two strands, because they are complementary and because the two strands are sampled randomly.
Table 3.
Absolute number of damage events underlying observed miscoding lesions, subdivided by Light and Heavy template molecules
| A→Na | C→N | G→N | T→N | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | G | T | A | C | G | T | A | C | G | T | A | C | G | T | |
| Light | 22 613 | 1 | 9 | 0 | 2 | 15 808 | 1 | 290 | 54 | 2 | 8727 | 6 | 0 | 4 | 2 | 19 116 |
| Heavy | 18 816 | 0 | 15 | 3 | 0 | 8417 | 0 | 141 | 112 | 5 | 16 111 | 8 | 6 | 11 | 4 | 21 190 |
| Total | 41 429 | 1 | 24 | 3 | 2 | 24 225 | 1 | 431 | 166 | 7 | 24 838 | 14 | 6 | 15 | 6 | 40 306 |
aWhere N refers to four possible derived nucleotide states, as listed in subsequent subcolumns.
Analysis 4: The underlying causes of the complementary damage pairs
Owing to limitations in conventional PCR and sequencing technologies, it has previously not been possible to discriminate between the relative contributions of the two potential causes of each of the six complementary miscoding lesion pairs. As we are able to segregate the data into the 12 original miscoding lesions, we are able to approach this issue for the first time. A χ2-test of independence was used to test whether there are any differences in the miscoding lesion frequencies between the constituent miscoding lesions for each of the pairs (e.g. whether the rate of A→G occurrences differs from the rate of T→C occurrences within the A→G/T→C complementary pair). The test relies on the assumption that there are no differences between the rates of occurrence on the two strands (see results of Analysis 3). The data from the two strands were therefore pooled and classified according to the source type (A+T or G+C) of the original base. If there are no differences in the rates of damage, miscoding lesions would happen at the same rates from A as from T, and likewise at the same rates from C as from G. Therefore two χ2 tests of independence were performed on the two datasets.
RESULTS
Analysis 1: Statistical discrimination of damage from PCR enzyme misincorporation error
Two χ2 tests of independence were used to compare whether the distribution of miscoding lesions from the ancient mtDNA (Table 1) differs significantly from that of the modern cpDNA (Table 1). This enables the identification of whether any of the miscoding lesions in the mtDNA dataset are over-represented, thus can be attributed to post mortem damage. The χ2 analyses provide strong statistical support for the notion that the miscoding lesion spectra in the modern and ancient DNA are different (miscoding lesions from A and T: P-value <1.3 × 10−4; miscoding lesions from C and G: P-value = <2.2 × 10−16). As such, it can be concluded that a significant part of the lesions within the aDNA dataset are derived from damage.
Analysis 2: Determination of which complementary miscoding lesion pairs represent true damage
To discriminate between which of the six complementary miscoding lesion pairs within the ancient mtDNA data represent damage and which represent enzymatic error, χ2 analyses were performed comparing the observed rates of the miscoding lesion pairs with the expected rates as calculated using the modern cpDNA data (Table 2). Once corrected for multiple comparisons, the χ2 analysis provides statistical support that only two of the pairs cannot be attributed to enzymatic error. Specifically, Type 2 transitions (C→T/G→A) are exceedingly over-represented, constituting 88% of the observed miscoding lesions. This provides strong support of previous arguments that they form the dominant form of aDNA damage-derived miscoding lesions (4,7–9,12). However, in contrast to some of our previous observations, and in agreement with the arguments of Hofreiter et al. (9), Type 1 transitions (A→G/T→C), which here constitute <6% of the total miscoding lesions, and just over 6% of the total transitions observed, appear to play little or no role in aDNA damage-derived miscoding lesions in this study. The overall Type 1/Type 2 ratio of approximately 1:15 is considerably lower than that observed in all the previous studies [≈1:2 (8), ≈1:3 (12) and ≈1:6 in the data used in the study of Hofreiter et al. (9); M. Hofreiter, personal communication]. Further, we observe that A→T/T→A transversions are unusually underrepresented in the mammoth aDNA data. As this clearly cannot be a result of damage, it seems likely that this observation is the result of the small number of A→T/T→A observations overall. In contrast, the much larger number of observations, and much stronger statistical support (much smaller P-value) suggest that this is not the case for the Type 2 transitions.
Analysis 3: Investigation for differences in strand damage accumulation rates
mtDNA sequences were separated into the distinct L and H strand datasets, and miscoding lesions were tabulated for both datasets using the simple algorithm outlined in Figure 1. A χ2 analysis of the miscoding lesions within the two datasets representing the L and H strand sequences (Table 3) shows that when corrected for multiple tests (actual P-value required for 5% significance level of 0.05/4 = 0.0125), there is no significant difference between the distributions (miscoding lesion from A: P-value = 0.03, from C: P-value = 0.53, from G: P-value = 0.80, from T: P-value = 0.03). Therefore, there is no evidence to support previous hypotheses (8) that the damage distribution may vary significantly by strand.
Analysis 4: The underlying causes of the complementary damage pairs
Previous studies have argued that although 12 potential damage-derived transitions and transversions exist, only a few of these actually contribute to aDNA damage-derived miscoding lesions (4,6–14). To investigate the relative contribution of the different transitions and transversions, the miscoding lesion counts from the separate L and H strand datasets were pooled together and analysed. The statistical analysis of the constituent damage types within the six complementary miscoding lesion pairs (Table 4) shows that although there is no evidence for a bias in contribution by the various damage events for A+T miscoding lesions (P > 0.08), there is significant support for a bias within C+G miscoding lesions (P < 2.2 × 10−16), arising due to the over-abundance of C→T over G→A transitions. With an occurrence per C nucleotide sequenced of 0.01779 in comparison to 0.00668 per G nucleotide (2.7 times more common), C→T modifications clearly represent the bulk of aDNA miscoding lesion damage. However, G→A transitions also constitute a significant number of miscoding lesions (nearly 7 times more common than A→G transitions, the next most frequently observed miscoding lesion). This finding is in stark contrast to all previously published hypotheses that have concurred that it is cytosine to uracil deamination, resulting in C→T miscoding lesions that is the predominant, if not sole, cause of Type 2 transitions (4,7–9,12).
Table 4.
Contribution of individual damage events to observed miscoding lesion pairs
| Original damagea | i j | A→G T→C | A→C T→G | A→T T→A | C→A G→T | C→G G→C | C→T G→A |
|---|---|---|---|---|---|---|---|
| Mammoth dataset | i | 24 | 1 | 3 | 2 | 1 | 431 |
| j | 15 | 6 | 6 | 14 | 7 | 166 | |
| Complementary pair total | i+j | 39 | 7 | 9 | 16 | 8 | 597 |
| Per cent of total mammoth observations | 5.8 | 1.0 | 1.3 | 2.4 | 1.2 | 88.3 |
aConstituent damage events within each of the six complementary miscoding lesion pairs, identified as i and j, respectively. Subsequent rows of the table describe observed number of i and j for each dataset, plus total (i + j).
As the C14 age of the mammoth sample is known [27 740 ± 220 years, (2)], the damage rate (r) can be calculated for the Type 2 transitions, both as the complementary pair and individually, using the following equation:
where t is time (e.g. in years or seconds) and x is the damage occurrence per base sequenced (either adjusted to take into account the assumed error misincorporation rate (rate of occurrence of Type 2 complementary pair) or unadjusted, thus reflecting the total miscoding lesion rate (individual C→T and G→A observations), which as it does not account for PCR enzyme error, represents a slight overestimate of the true rate. The damage rates are shown in Table 5. Although this damage rate is clearly relevant only under the preservation history of the specimen examined, this damage rate is likely to be more accurate than rates that could be calculated using previous aDNA data, as they face the limitation of being unable to discriminate whether multiple miscoding lesions observed at a single position within cloned sequences from a single PCR product, are actually independent damage events, or simply descendents of a single damaged molecule (8,11). Although no other data exists with which to compare the rates calculated here, as other datasets from dated samples become available these rates can be compared to investigate whether there is any universal aspect to the rates of occurrences.
Table 5.
Type 2 damage rate, nucleotides per unit time
| Per year | Per second | |
|---|---|---|
| Type 2 damagea | 4.2 × 10−7 | 1.3 × 10−14 |
| C→Tb | 6.2 × 10−7 | 2.0 × 10−14 |
| G→Ab | 2.3 × 10−7 | 7.4 × 10−15 |
aAdjusted to account for enzyme contribution to miscoding lesions.
bUnadjusted for enzyme contribution, therefore overestimate of true rate.
DISCUSSION
We have demonstrated, with DNA sequence data generated using the GS20 emPCR and sequencing platform, that in this particular dataset and in agreement with the arguments of Hofreiter et al. (9), Type 2 transitions are the overwhelmingly dominant cause of post mortem damage-derived miscoding lesions. This is in contrast to other studies (published by several authors of this study) that report significantly higher levels (than the 6% reported here) of Type 1 miscoding lesions within aDNA datasets (7,8,12). Although it is tempting to explain this discrepancy as laboratory-specific phenomena, it is worth noting that the conclusions of the aforementioned studies were based on both new data generated in the respective studies, plus data from a number of previous aDNA studies. Therefore, Type 1 transitions appear to be a true phenomena of at least some aDNA sequence data, and we are therefore left in the difficult position of how to explain the discrepancy in the findings.
Caveat about conclusions drawn from the modern cpDNA data
The modern cpDNA dataset can be expected to contain some innate levels DNA damage, for example, DNA that had not been repaired before extraction or DNA that was damaged during the extraction. Furthermore, cpDNA damage spectra may differ from that found in modern mtDNA, due to the differences between the structure of the genomes and the organelle biology. Thus, as the miscoding lesions observations on the cpDNA may represent the sum of the enzymatic error plus prior damage, enzymatic error may be overestimated in this dataset. However, the observed increase in the ratio of Type 1 to Type 2 transitions between the modern and aDNA datasets is so great (1:1 in the modern DNA versus 1:15 in the aDNA) that any overestimation of cpDNA damage is unlikely to significantly affect the conclusions of this study.
Explanations for the lack of Type 1 transitions
One potential explanation is that differences in DNA extraction methodologies may play a significant role in the observed results. For example, the dataset of Hofreiter et al. (9) was generated from DNA extracted and purified using a silica-based methodology [modified from Boom et al. (19)], and thus the nucleic acids were exposed to both acidic conditions and high concentrations of guanidinium chaotropes. In contrast, however, the majority of data from the conflicting studies was generated using a buffered digestion mixture at neutral pH, followed by organic purification of the nucleic acids [in general modified from Sambrook et al. (20)]. Thus it might be argued that the conditions of the silica method might somehow result in the fragmentation of DNA at positions where the underlying cause of Type 1 transitions have occurred. Unfortunately, however, a major problem with this explanation is that the DNA analysed in this study was initially purified using the non-silica method, thus rendering this explanation unlikely.
An alternative explanation that has been proposed previously is that Type 1 transitions derive from innate enzyme error at early stages of the PCR process (9,13), giving rise to what have been described as ‘singleton’ miscoding lesions, in contrast to ‘consistent’ miscoding lesions among cloned data (9). In light of our data, this explanation is equally problematic, as unlike previously generated data, our sequences all stem from a single single-stranded template molecules. This places us in an optimal position to observe the true enzymatic misincorporation behaviour, and as the enzyme used in the emPCR (Invitrogen's Platinum Taq Hifidelity) is that used in many of the previously studied aDNA datasets, it is difficult to support the argument.
A third explanation is that Type 1 transitions, if they had existed in the original data, may have been removed through strand fragmentation at the site of Type 1 transitions during the multiple preparation steps that DNA is required to go through during the GS20 process. Specifically the DNA is first fragmented through physical shearing. Subsequently, the DNA must be polished through blunt ending and phosphorylation using T4 DNA polymerase (exhibits 3′–5′ exonuclease activity), Escherichia coli DNA polymerase (Klenow fragment) (fills in recessed 3′ ends), and T4 polynucleotide kinase (phosphorylating 3′ ends). These treatments do not suggest a good reason for why miscoding lesion damage in the middle of a template molecule may be removed. The next stage however may provide a key as the enzyme-treated DNA is subsequently re-purified using silica spin columns (e.g. Qiagen's QIAquick columns). As this involves the use of further guanidinium containing buffers it could be that Type 1 damage is removed at this stage.
The fourth explanation is that the damage observed in this sample is simply the true damage spectra, but owing to as yet undetermined factors the spectra vary by individual ancient sample. This specimen is unique to some extent as it was recovered in a frozen state directly from frozen Siberian permafrost, and subsequently retained at sub-zero (predominantly −15°C) conditions before DNA extraction (2). The damage spectra therefore do not directly reflect the DNA damage within all other specimens, perhaps through an unusually limited access of the DNA to free water molecules.
The dual causes of Type 2 transitions
The most intriguing finding of this study is that our data demonstrate that in contrast to all previous statements on the subject (4,8,9,12,13) the cause of Type 2 transitions is not only cytosine to uracil (or analogues) deamination, but also the degradation of guanine to a derivative that is misread by the PCR enzyme as an adenine. Cytosine to uracil deaminations clearly are important—the resultant manifestation of C→T transitions are observed here at a highly significant rate (in comparison to the enzyme misincorporation rates), and cytosine to uracil deamination has been experimentally identified through previous UNG treatment assays of purified aDNA (4,8,9). It is worth noting here that in our experience UNG treatment of aDNA extracts sometimes leaves some remaining C→T transitions in the resultant PCR amplified and cloned sequences (M.T.P.G., unpublished data). Although we have previously thought that these simply result from incomplete enzymatic cleavage of all the damaged sequences a more likely explanation is that the remaining C→T lesions were in fact derived from guanine derivatives. Naturally we caution again that due to the recent advent of the GS20 technology, limited aDNA data are publicly available for study; thus, our conclusions are based on the data from a single sample. Therefore until further studies are undertaken on additional samples, conclusions as to how widespread this phenomenon is cannot be drawn. That guanine degradation appears to also be a significant cause of Type 2 transitions is interesting, in so far as a previous study has also remarked on the dominance of other guanine modifications among oxidative forms of aDNA damage. Specifically, using gas chromatography/mass spectrometry (GC/MS), to identify PCR-blocking hydantoins, Höss et al. (14) report that guanine modifications dominate in the majority (3/5) of samples from which they can successfully PCR amplify DNA.
Clearly, the major outstanding question arising from this study is what exactly the damage to guanine is that can give rise to G→A miscoding lesions. Although various modifications of guanine are known to cause miscoding lesions, during enzymatic replication these result in the generation of transversions; for example, a common product of oxidative degradation, 8-oxoguanine, generates G→T transversions (21), whereas other guanine products such as 8-methyl-2′-deoxyguanosine generate both G→C and G→T transversions (22). Furthermore, the mutagen ethylene dibromide, a once common insecticide, fumigant and anti-knock agent for gasoline has been observed to indirectly cause G→A miscoding lesions in living cells (23). However, it is unlikely that this is the cause in this situation, as the process requires enzymatic conjugation to form the half-mustard S-(2-bromoethyl)glutathione (GSCH2CH2Br), which clearly could not happen in post mortem systems. Therefore, we are unaware of an explanation as to what causes G→A transitions in this situation, although the damaged form would of course have to result in the misincorporation of a thymine opposite. However, at this point we note that a problem with previous studies on damage is that they have attempted to draw explanations from what is known about damage in in vivo systems in order to explain post mortem observations. There is no good reason why the two systems need to be identical, indeed it might be expected that key differences exist between metabolically active and the deceased environments.
Re-evaluation of previous conclusions in light of current findings
Under the assumption that Type 1 transitions do not represent true aDNA damage, and that Type 2 transitions may originate from both C→T and G→A events, several past statements with respect to aDNA need to be evaluated as follows.
Damage hotspots
The existence of particular post mortem DNA damage hotspots has been argued based on observations of the distribution patterns of miscoding lesions (predominantly Type 1 and Type 2 transitions) (8,11). ‘Hotspots’ are defined as ‘specific nucleotide positions that appear to undergo damage at a rate significantly above that expected under the hypothesis that damage is equally likely to affect all positions’. Although the possibility has been raised that the hotspot observations originally made on human DNA sequences may be flawed due to contamination of the samples (13), this seems unlikely as a second study on bison that specifically investigated, and ruled out as not significant, the potential role of sample contamination, produced similar findings (11). However, if Type 1 damage is not a true phenomena, but simply represents PCR enzyme misincorporation, then while the underlying observation of miscoding lesions observed at specific non-random nucleotide positions does not change, the argument that damage may preferentially occur at these positions does. Data used in such studies warrant reanalysis, to remove Type 1 transitions, then statistical tests require recalculation in order to investigate whether support still exists for damage hotspots. A recent study has reported the existence of DNA sequencing error hotspots (24); thus, these may also play some effect in the original damage hotspot observations. However, we stress that one of the original conclusions of the damage hotspot papers is that aDNA sequence authenticity may be challenged by predominance of miscoding lesions at specific, phylogenetically informative nucleotide positions, remains unchanged whether the cause of hotspots is damage or position-specific sequencing errors.
Jumping PCR
Based on the hypothesis that single damage events can explain the origin of Type 1 and Type 2 transitions, respectively (cytosine to uracil and adenine to hypoxanthine deamination, respectively), Gilbert et al. (8) have argued that this provides a tool for identifying recombinant aDNA sequences that may have arisen through jumping PCR (25). For example, if, as previously hypothesized, Type 2 transitions could only arise through cytosine to uracil deaminations, then in an absence of jumping PCR the resultant two damage phenotypes (C→T and G→A) transitions should never be observed within the same individual cloned DNA sequence (as the C→T observation must have arisen from an original cytosine deamination on a template molecule in the same orientation as the final read sequence, whereas the G→A observation must have arisen on a different template molecule of the complementary orientation). In light of the findings that Type 1 transitions may not represent true damage, and that Type 2 transitions may originate from both cytosine and guanine degradation, the theory behind this argument does not hold. Therefore, we advise that jumping PCR analyses cannot be performed in the described manner.
CONCLUSIONS
Through statistical comparisons of large amounts of DNA sequence data generated using the GS20 platform we demonstrate conclusively that it is both C→T and G→A transitions that generate the majority of aDNA miscoding lesions. The advent of the GS20 and other high-throughput DNA sequencing techniques will rapidly increase the data available for aDNA damage analyses. As these data become available, we should be able to investigate how general the conclusions of this study are. In combination with improved methods for the efficient recovery of aDNA, and newly developed biochemical assays that have started to overturn conventional damage dogma [e.g. the dominance of DNA cross-linking in some aDNA sources (15)], our understanding about the extent and biochemical basis behind aDNA damage should rapidly increase, enabling future expansion on what samples are available for aDNA analyses, and what can be done with the recovered DNA.
STATEMENT OF AUTHOR CONTRIBUTIONS
The study was conceived by M.T.P.G. and S.C.S. S.C.S. and W.M. generated and assembled the data. M.T.P.G., J.B., W.M., S.C.S., C.W. and E.W. developed and performed the data analysis and wrote the manuscript. J.E.C., J.H.L.M. and H.P. provided the DNA extractions.
NOTE ADDED IN PROOF
This manuscript is a modified version of the original that previously appeared online as a preprint at Nucleic Acids Research. Shortly after online publication of the original manuscript M.T.P.G. noted a problem in the analytical process used to subdivide the sequence data into individual H and L strands. Specifically the data as originally presented represented the complement of the true situation. This change affected one original conclusion of the study, that G→A transitions occur at a higher frequency than C→T transition. However, the true observation is the opposite. However, our novel claim for the existence of G→A transitions per se as a form of aDNA damage is not changed. Other changes to the manuscript that were warranted in light of this error were the re-assignment of the data within Tables 3–5 to different headings. Further, discussion of the methods in the main text and Supplementary Data have been significantly modified to enhance clarity. M.T.P.G. assumes full responsibility for the error, and would like to thank M. Hofreiter for his helpful discussion, and the Executive Editor, R. Roberts, for his assistance in modifying the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
M.T.P.G. was supported by the Marie Curie FP6 Actions ‘FORMAPLEX’ grant. J.B. and E.W. were supported by the Wellcome Trust, UK, the Carlsberg Foundation, DK, and the National Science Foundation, DK. W.M. was supported by HIN grant HG002238. This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds appropriated by the legislature. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. Funding to pay the Open Access charges for this article was provided by Marie Curies FP6 Actions ‘FORMAPLEX’ grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shapiro B., Drummond A.J., Rambaut A., Wilson M., Sher A., Pybus O.G., Gilbert M.T.P., Barnes I., Binladen J., Willerslev E., et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 2.Poinar H.N., Schwarz C., Qi J., Shapiro B., MacPhee R.D.E., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 3.Willerslev E., Hansen A.J., Binladen J., Brandt T.B., Gilbert M.T.P., Shapiro B., Bunce M., Wiuf C., Gilichinsky D.A., Cooper A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science. 2003;300:791–795. doi: 10.1126/science.1084114. [DOI] [PubMed] [Google Scholar]
- 4.Pääbo S. Ancient DNA: extraction, characterization, molecular cloning and enzymatic amplification. Proc. Natl Acad. Sci. USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderung C., Bouwman A., Persson P., Carretero J.M., Ortega A.I., Elburg R., Smith C., Arsuaga J.L., Ellegren H., Götherström A. Prehistoric contacts over the Straits of Gibraltar indicated by genetic analysis of Iberian Bronze Age cattle. Proc. Natl Acad. Sci. USA. 2005;102:8431–8435. doi: 10.1073/pnas.0503396102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Hansen A., Willerslev E., Wiuf C., Mourier T., Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert M.T.P., Hansen A.J., Willerslev E., Rudbeck L., Barnes I., Lynnerup N., Cooper A. Characterisation of genetic miscoding lesions caused by post mortem damage. Am. J. Hum. Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofreiter M., Jaenicke V., Serre D., von Haeseler A., Pääbo S. DNA sequences from multiple amplifications reveal artefacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4693–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert M.T.P., Willerslev E., Hansen A.J., Rudbeck L., Barnes I., Lynnerup N., Cooper A. Distribution patterns of post mortem damage in human mitochondrial DNA. Am. J. Hum. Genet. 2003;72:32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert M.T.P., Shapiro B., Drummond A., Cooper A. Post mortem DNA damage hotspots in Bison (Bison bison and B.bonasus) provide supporting evidence for mutational hotspots in human mitochondria. J. Archaeol. Sci. 2005;32:1053–1060. [Google Scholar]
- 12.Binladen J., Wiuf C., Gilbert M.T.P., Bunce M., Larson G., Barnett R., Hansen A.J., Willerslev E. Comparing miscoding lesion damage in mitochondrial and nuclear ancient DNA. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pääbo S., Poinar H., Serre D., Jaenicke-Despres V., Hebler J., Rohland N., Kuch M., Krause J., Vigilant L., Hofreiter M. Genetic analyses from ancient DNA. Ann. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 14.Höss M., Jaruga P., Zastawny T., Dizdaroglu M., Pääbo S. DNA damage and DNA sequence retrieval from ancient tissue. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen A.J., Mitchell D.L., Wiuf C., Paniker L., Brand T.B., Binladen J., Gilichinsky D.A., Ronn R., Willerslev E. Crosslinks rather than strand breaks determine access to ancient DNA sequences from frozen sediments. Genetics. 2006;173:1175–1179. doi: 10.1534/genetics.106.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z.T., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with Blastz. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyer E., Zietkiewicz E., Rochowski A., Yotova V., Puymirat J., Labuda D. Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees. Am. J. Hum. Genet. 2001;69:1113–1126. doi: 10.1086/324024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 21.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 22.Kohda K., Tsunomoto H., Kasamatsu T., Sawamura F., Tershima I., Shibutani S. Synthesis and miscoding specificity of oligodeoxynucleotide containing 8-phenyl-2′-deoxyguanosine. Chem. Res. Toxicol. 1997;10:1351–1358. doi: 10.1021/tx970111k. [DOI] [PubMed] [Google Scholar]
- 23.Foster P.L., Wilkinson W.G., Miller J.K., Sullivan A.D., Barnes W.M. An analysis of the mutagenicity of 1,2-dibromoethane to Escherichia coli: influence of DNA repair activities and metabolic pathways. Mutat. Res. 1988;194:171–181. doi: 10.1016/0167-8817(88)90019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandstätter A., Sanger T., Lutz-Bonengel S., Parson W., Beraud-Colomb E., Wen B., Kong Q.P., Bravi C.M., Bandelt H.-J. Phantom mutation hotspots in human mitochondrial DNA. Electrophoresis. 2005;18:3414–3429. doi: 10.1002/elps.200500307. [DOI] [PubMed] [Google Scholar]
- 25.Pääbo S., Irwin D., Wilson A. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.