Abstract
Many methods exist for genotyping—revealing which alleles an individual carries at different genetic loci. A harder problem is haplotyping—determining which alleles lie on each of the two homologous chromosomes in a diploid individual. Conventional approaches to haplotyping require the use of several generations to reconstruct haplotypes within a pedigree, or use statistical methods to estimate the prevalence of different haplotypes in a population. Several molecular haplotyping methods have been proposed, but have been limited to small numbers of loci, usually over short distances. Here we demonstrate a method which allows rapid molecular haplotyping of many loci over long distances. The method requires no more genotypings than pedigree methods, but requires no family material. It relies on a procedure to identify and genotype single DNA molecules, and reconstruction of long haplotypes by a ‘tiling’ approach. We demonstrate this by resolving haplotypes in two regions of the human genome, harbouring 20 and 105 single-nucleotide polymorphisms, respectively. The method can be extended to reconstruct haplotypes of arbitrary complexity and length, and can make use of a variety of genotyping platforms. We also argue that this method is applicable in situations which are intractable to conventional approaches.
INTRODUCTION
The discovery of abundant single-nucleotide polymorphisms (SNPs) in the human genome has driven the development of technologies for rapid and efficient genotyping. However, a more difficult challenge is to resolve the two haplotypes carried by a diploid individual, determining which alleles lie on each of the two homologous chromosomes. For instance, an individual may have the genotype AB/ab (heterozygous at each of loci A and B), but could carry haplotypes AB and ab or, conversely, Ab and aB.
It is increasingly appreciated that haplotypes, rather than genotypes alone, carry the richest data on human variation (1–5). Quantitative traits such as drug responsiveness or disease susceptibility may be more strongly correlated with certain haplotypes than with certain genotypes, particularly where several polymorphic loci fall within a single gene. Hence, both the discovery of an association between trait and polymorphism, and the implications of this association for an individual, depend on a knowledge of haplotypes. Haplotype structure is also important in understanding the evolution of a species and of populations within it, as haplotype blocks are shuffled in successive generations. The persistence of ancestral haplotypes can also be used to simplify genotyping experiments: the genotype at one locus may serve as a proxy for the genotypes of neighbouring loci if they lie within the same conserved haplotype block.
Classically, haplotypes have been inferred by genotyping several generations of a pedigree and tracing the segregation of markers. At a population level, conversely, the abundance of different haplotypes can be estimated from the combined genotype data for many unrelated individuals—one of the aims of the HapMap project [www.hapmap.org and Ref. (6), though see also Ref. (7)]. Population abundances of haplotypes can also be used to infer individual haplotypes from genotyping data, but only with certain assumptions and only over the very short distances within which recombination can be assumed not to have occurred. None of these approaches, however, can generally be applied diagnostically to resolve long-range haplotypes in a heterozygote in the absence of multi-generational family members.
To overcome these limitations, a number of methods have been proposed to physically resolve the diploid chromosomes of an individual into their haplotypes. Cloning in hybridomas or in bacterial or yeast cells, or the natural occurrence of hydatidiform moles arising from a single haploid gamete, can be used to isolate a single haplotype which can then be revealed by simple genotyping (8–11). Such approaches, however, are ill-suited to large-scale studies or to diagnostic screening of individuals. A variety of techniques based on allele-specific PCR, electrophoretic separation of haplotypes or allele-specific hybridisation have been used to detect different haplotypes or to separate haplotypes for later analysis (12–22). All of these approaches, however, are limited to the analysis of small numbers of loci over short distances (typically a few hundred base pairs), although it was suggested that some methods could be used to build up longer haplotypes step-wise (14).
Other methods have been based on the analysis by PCR of single DNA molecules which, of course, represent single haplotypes. The most direct implementation of this strategy is the genotyping of single sperm, (23,24) in which meiosis has done the job of isolating a single copy of each chromosome. Other approaches rely upon limiting dilution to isolate (statistically) single DNA molecules, followed by amplification and genotyping of two or more loci (25–28). However, haplotypes assembled by these techniques rarely exceed 20–30 kb in length (29–31), never involve more than a few loci and the methods are often inefficient, since only a few of the highly dilute samples which are genotyped will prove to contain informative molecules. An exception is the use of the ‘polony’ method to genotype intact chromosomal DNA molecules dispersed in an acrylamide film (32). This elegant technique is efficient and long-range, but requires metaphase cells, careful primer optimisation and again appears to be limited to the analysis of small numbers of loci.
We have devised a molecular haplotyping method which also relies upon limiting dilution but which, for the first time, demonstrates the reconstruction of haplotypes spanning large numbers of loci over long distances, from the DNA of a single individual (Figure 1). DNA is dispensed at extreme dilution into a panel of aliquots, each containing much less than one genome's worth of DNA. Hence, any given segment of the genome will be found in only a minority of the aliquots and, in those aliquots, only one of the two haplotypes is likely to be present. Importantly, an initial pre-screening reveals the precise molecular content of each aliquot. From these results, a small subset of aliquots can be selected which are likely to be informative, and only these aliquots need be genotyped for particular loci. By genotyping just a few aliquots at each locus, robust haplotypes involving multiple loci can be built up over distances which are not limited by the size of DNA fragments.
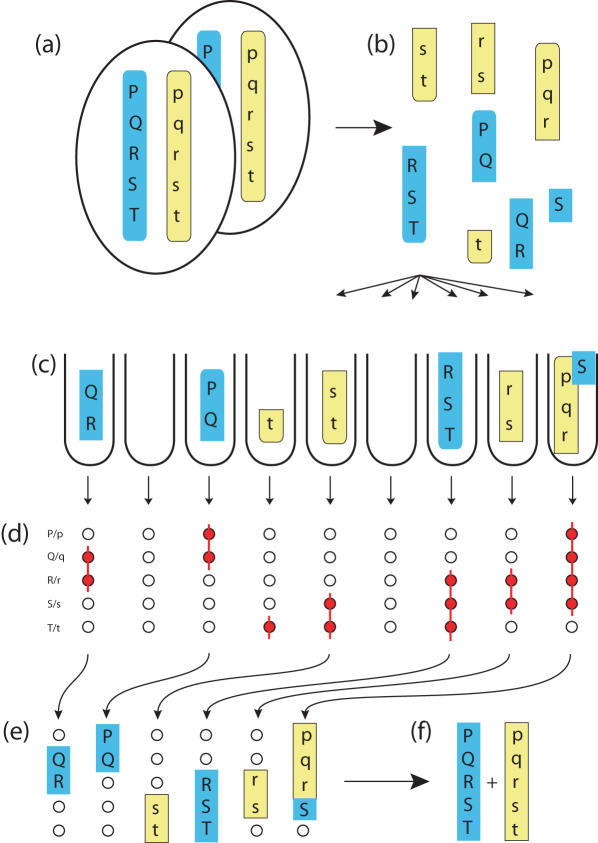
Figure 1.
Principle of the method. Diploid cells (a) are shown containing two haplotypes (upper-case, blue; and lower-case, yellow). DNA is prepared (b) with inevitable breakage through shearing, and dispensed at extreme dilution into aliquots (c), each containing much less than a complete genome. Initially, the samples are pre-screened by PCR to find out which loci they each contain, but without genotyping (d) red circles indicate a positive PCR result; red lines show the inferred extent of the fragments in each aliquot. Only a handful of aliquots then need to be genotyped for each locus (e). From these results, the complete haplotypes (f) can be reconstructed. Note that a few aliquots may contain mixed haplotypes. In this case, two of the aliquots give the partial haplotypes rs and RS, whilst the rightmost one gives the mixed haplotype rS—the correct linkage phase is inferred from the majority (in this case, rs/RS rather than rS/Rs).
The total number of genotypings required is comparable to—or less than—that needed in pedigree studies. Moreover, the panel of aliquots can be re-accessed many times, allowing large-scale haplotyping.
MATERIALS AND METHODS
Preparation of mapping panels
DNA embedded in agarose was prepared from two anonymized male blood donations (National Blood Service, Oxford, UK) as described previously (33), melted at 69°C in 0.5× PCR BufferII (Perkin-Elmer) in a volume calculated to give a final concentration of roughly 0.02–0.06 genomes (0.06–0.18 pg) of DNA per microlitre, and mixed by gentle inversion. Aliquots (5 μl, ∼0.1–0.3 genomes) of diluted DNA were dispensed into a 96-well microtitre plate and overlaid with 25 μl/well of light mineral oil (Sigma). The exact concentration of the DNA required was estimated based on the initial pre-screening results (below) for several loci and adjustments made as necessary to produce the desired concentration.
Aliquots were pre-amplified using primer-extension preamplification [PEP, Ref. (34)] by supplementing each well with 0.7 μl 10× PCR BufferII (Perkin-Elmer), 0.7 μl 25 mM MgCl2, 0.06 μl 25 mM dNTPs, 0.07 μl 1 mM N15 oligonucleotide (Operon Technologies), 0.28 μl Taq polymerase (Amplitaq, Perkin-Elmer, 5 U/μl) and 0.19 μl of water. Reactions were cycled (5 min at 93°C, followed by 50 cycles of 94°C for 30 s, 37°C for 2 min, 37–55°C ramp over 3 min, 55°C for 4 min). Products were diluted to 200 μl each, and 5 μl samples transferred to replicate plates and stored at −80°C.
A second panel was made from a mixture of two male-derived DNAs (see Results section). This panel was prepared by pooling equal amounts of agarose-embedded DNA from two different male donors, after the melting step.
Selection of loci and design of PCR primers
Single-nucleotide polymorphisms were chosen from the HapMap database (www.hapmap.org). Twenty SNP loci were selected from a region of 283 kb on Xp22.1, and 105 loci from a 993 kb region on 21q21. The distance between consecutive loci varied from 87 bp to 49.5 kb (average 9.8 kb). For each locus, hemi-nested PCR primers (forward-external, forward-internal and reverse) were designed using simple parameters [each primer ∼20 bases long, ∼50% (G+C); where possible, two G/C bases at 3′ end and one G/C base at 5′ end], such that the SNP was located within the internal amplimer (between forward-internal and reverse primers). Details of all loci and their PCR primers are given in Supplementary Data.
Initial panel pre-screening
Initial pre-screening of loci on the panels was performed using a hemi-nested PCR, in which the first phase was multiplexed. For first-phase PCRs, 5 μl samples of the mapping panel (above) were supplemented to give 10 μl total volume containing 1× PCR Gold buffer (Perkin-Elmer), 4 mM MgCl2, 200 μM each of dATP, dCTP, dGTP and dTTP, 0.2 μM of each forward-external and reverse primer for each of the loci to be typed (up to 105 primer pairs) and 0.5U Taq Gold DNA polymerase (Perkin-Elmer), then thermocycled (93°C × 9 min, then 28 cycles of 94°C × 20 s, 53°C × 30 s, 72°C × 60 s). Products of this reaction were diluted to 1000 μl with water, and 5 μl aliquots of this were used as template in a monoplex (single marker) second-phase PCR for each marker in question (10 μl volume, 1 μM each of the relevant forward-internal and reverse primers, 1× PCR Gold buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.2U Taq Gold DNA polymerase; 93°C × 9 min, then 35 cycles of 94°C × 20 s, 55°C × 30 s, 72°C × 60 s). Products were analysed (scoring presence or absence of the expected PCR product) either by high-density gel electrophoresis or by melting-curve analysis (ABI 7900HT, Applied Biosystems); in the latter case, the second phase PCRs were supplemented with 0.5× SyBr Green I (Molecular Probes) and 0.5 μM ROX (ABgene), and the concentration of MgCl2 was increased to 4 mM.
Genotyping
Where appropriate, PCR products generated as above were genotyped for the relevant SNP by sequencing. Unincorporated dNTPs and primer molecules were degraded by supplementing the 10 μl PCR product with 10 U Antarctic phosphatase (NEB) and 20 U Exonuclease I (NEB), incubating for 15 min at 37°C, and inactivating the enzymes for 15 min at 65°C. Products were then sequenced by standard protocols, using either the forward-internal or reverse PCR primer.
Statistical analysis
Statistical analysis is not necessary for reconstructing haplotypes using this approach (see Results section), but can provide evidence for the reliability of the haplotypes. We therefore give a brief statistical analysis here, and a more detailed analysis in Supplementary Data.
Previous analyses (35) have focused on situations in which multiple samples of dilute DNA are genotyped without prior selection, and in which many of the resulting genotypes are uninformative (for example, containing mixed alleles or no alleles at some loci). We have a simpler theory, in which the log-likelihood (LogL) of a given linkage phase between two loci (for example, A/a and B/b) is given by
| 1 |
where P (a ‘power factor’) is a function of the average DNA content of each aliquot in the panel and of the distance between loci A/a and B/b expressed as a fraction of the average size of DNA fragments in the aliquots; NH is the number of aliquots which were genotyped for both loci and found to agree with the supposed haplotypes; and NX is the number of aliquots genotyped for both loci and found to disagree with the supposed haplotypes. (Aliquots which contain mixed genotypes for either or both loci, or which do not to contain both loci, are uninformative and are simply ignored.) Full details of the analysis, and of the calculation of P, are given in Supplementary Data. However, for simplicity we give a simple look-up table by which the appropriate value of P can be estimated (Table 1).
Table 1.
Lookup table for P
| P | DNA content of aliquots (average genomic copies per aliquot) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | 0.3 | 0.35 | 0.4 | 0.45 | 0.5 | |
| Inter-locus distance divided by average fragment size | ||||||||||
| 0.01 | 5.61 | 5.31 | 5.14 | 5.02 | 4.93 | 4.86 | 4.80 | 4.74 | 4.70 | 4.66 |
| 0.02 | 5.01 | 4.71 | 4.54 | 4.42 | 4.33 | 4.26 | 4.19 | 4.14 | 4.10 | 4.06 |
| 0.05 | 4.21 | 3.91 | 3.74 | 3.62 | 3.53 | 3.46 | 3.40 | 3.35 | 3.30 | 3.26 |
| 0.1 | 3.61 | 3.31 | 3.14 | 3.02 | 2.93 | 2.86 | 2.80 | 2.75 | 2.70 | 2.66 |
| 0.2 | 3.00 | 2.71 | 2.54 | 2.42 | 2.33 | 2.26 | 2.20 | 2.15 | 2.11 | 2.07 |
| 0.3 | 2.65 | 2.36 | 2.19 | 2.07 | 1.98 | 1.91 | 1.85 | 1.80 | 1.76 | 1.72 |
| 0.4 | 2.40 | 2.11 | 1.94 | 1.83 | 1.74 | 1.67 | 1.61 | 1.56 | 1.52 | 1.48 |
| 0.5 | 2.20 | 1.91 | 1.71 | 1.63 | 1.55 | 1.48 | 1.42 | 1.37 | 1.33 | 1.29 |
| 1 | 1.59 | 1.30 | 1.15 | 1.04 | 0.96 | 0.90 | 0.85 | 0.81 | 0.77 | 0.74 |
The value of P (see text) is given for a range of DNA concentrations and for a range of inter-locus distances expressed as a fraction of the average size of DNA fragments in the aliquots. For details, see text and Supplementary Data.
For example, suppose a panel contains an average of 0.1 genome equivalents of DNA (∼0.3 pg for human) per aliquot, with an average fragment size of 50 kb. Seven aliquots are genotyped for two loci which are 10 kb apart (0.2 of the average fragment size). Of these aliquots, two give genotyping result AB, three give result ab, one gives result Ab, and one gives result AaB (i.e. mixed genotype for locus A/a). If we assume that the true haplotypes are AB and ab, then NH (the number of ‘concordant’ results) is 5 and NX (the number of ‘discordant’ results) is 1; and, from Table 1, P = 2.71. So,
So we can infer with a confidence of 1010.84:1 (∼100 billion to 1) that the true haplotypes are AB and ab.
RESULTS
Recovery of X-chromosome ‘haplotypes’ from a mixed DNA sample
We first sought to recover haplotypes which could be independently validated. Since all X-chromosome sequences (other than those lying in the pseudo-autosomal region) are present only in a single copy in the male genome, we were able to create a ‘synthetic’ genome, diploid with respect to the X-chromosome, by mixing equal amounts of DNA from two unrelated male individuals. We then sought to recover the two haplotypes for a portion of the X-chromosome, before validating our results by genotyping the original, unmixed samples.
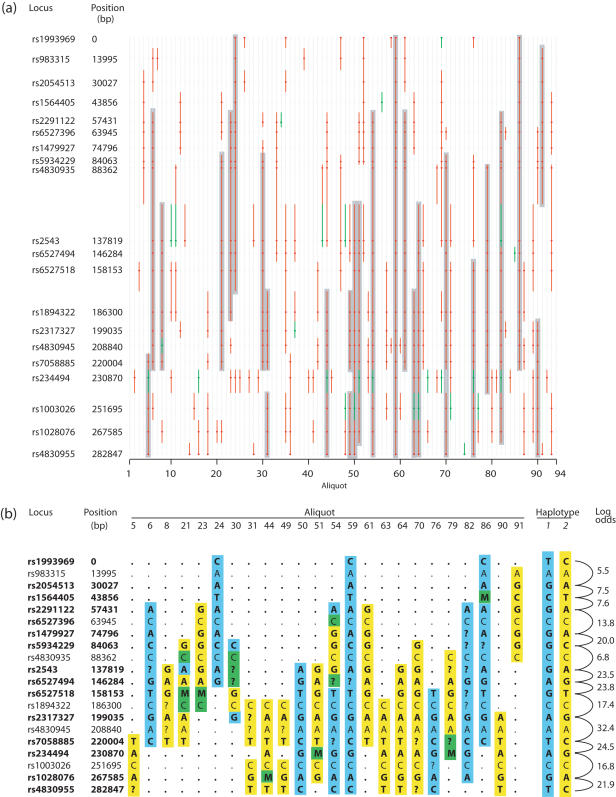
A panel of 94 aliquots of DNA (plus two negative controls) was prepared from a mixture of equal amounts of two unrelated male DNAs, and tested initially for the presence/absence in each aliquot of 20 SNP loci spanning a 280 kb region of the X-chromosome. As can be seen (Figure 2a), each locus is found to be present in only a minority of the aliquots, with an average of 23/94 aliquots being positive for any given locus. Assuming a random (Poisson) distribution of DNA fragments amongst the aliquots, this implies an average of 0.28 X-chromosome copies per aliquot (see Protocol S1). Many of the aliquots contain runs of several consecutive loci, implying that those aliquots contain DNA fragments which span long portions of the chromosome. We selected 24 aliquots that contained one or more such runs, and genotyped the corresponding PCR products; on average, each locus was genotyped in ∼12 of the aliquots (Figure 2b). Of the 20 loci, 14 proved to be ‘heterozygous’ (i.e. to be represented by two different alleles in the mixed DNA).
Figure 2.
Pre-screening, genotyping and haplotype reconstruction for X-chromosome markers. (a) Pre-screening results. The 94 sub-genomic aliquots are arranged from left to right, and the 20 X-chromosome loci are represented from top to bottom (spacing reflects genomic locations). Red dots indicate which loci are present in each aliquot (green dots = uncertain), and red/green lines represent the DNA fragments inferred to lie in each aliquot. Grey shading indicates the aliquots and loci which were selected for genotyping. (b) Genotyping results for the selected aliquots and loci. Letters show the allele found; M = mixed genotyping result; ? = no genotype (failed reaction). Two distinct haplotypes (blue and yellow) can be reconstructed. Each genotyping result is coloured to indicate which haplotype it is inferred to originate. Green indicates that the haplotype origin for that data point cannot be inferred, either because the locus is not heterozygous or because it gives a mixed (‘M’) genotype result. The two reconstructed haplotypes (denoted 1 and 2) are indicated; heterozygous loci in bold. On the right are shown the calculated log odds supporting the haplotypes for each step between consecutive heterozygous loci.
Inspection of these results allows the two haplotypes to be resolved directly: two tiling paths can be reconstructed, each consisting of a series of overlapping partial haplotypes. As expected, a proportion of the aliquots contain fragments from both of the haplotypes, revealing mixed alleles upon genotyping; these typings do not contribute towards establishing the resolved haplotypes. The two haplotypes reconstructed from these results are indicated in Figure 2b, and were found to be identical to those determined by direct genotyping of the original, unmixed DNA samples.
Recovery of chromosome 21 haplotypes from a diploid sample
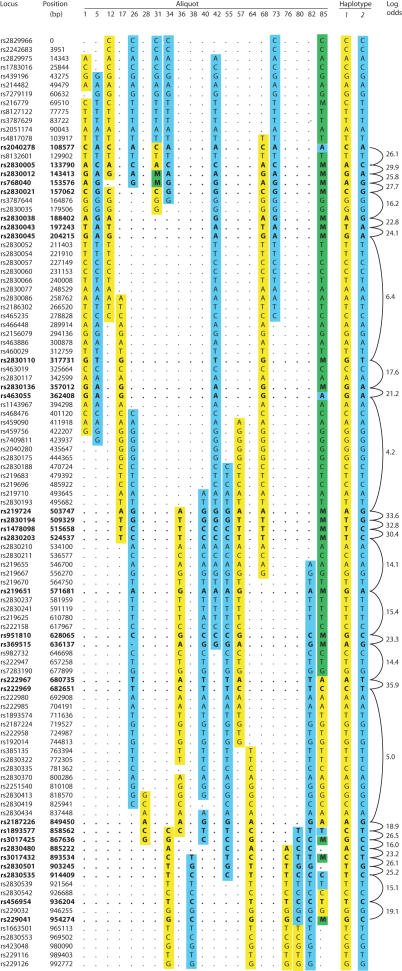
We next sought to recover true autosomal haplotypes from a normal diploid DNA sample. We selected 105 SNP loci from an arbitrarily chosen region of ∼1 Mbp on Chr21. DNA from a single individual was prepared, diluted and dispensed into a panel of 96 aliquots. The DNA concentration of the aliquots was lower than in the X-chromosome experiments, to reduce the incidence of aliquots containing mixed haplotypes. All aliquots were scored by PCR for the presence/absence of each of the 105 loci; on average, each locus was found in 13.3/96 aliquots, implying an average of 0.15 [autosomal] genomes per aliquot. As before, many of the aliquots contained one or more long runs of consecutive loci. Twenty-one of the aliquots were genotyped at selected loci (Figure 3); on average, each locus was genotyped in ∼7 aliquots. Of the 105 loci tested, 29 proved to be heterozygous in this individual. Only one of the aliquots tested proved to contain extensively overlapping molecules from different haplotypes, giving rise to mixed genotypes. Again, it is easy to reconstruct the two haplotypes by inspection of the data (Figure 3).
Figure 3.
Genotyping and haplotype reconstruction for chromosome 21 markers. Initial PCR pre-screening results for the 96 aliquots are not shown: only the genotyping results for the selected aliquots and loci are shown (format and notation as for Figure 2b).
Statistical analysis
In both the model (X chromosome) and real (chromosome 21) experiments, the haplotypes can be resolved unambiguously by inspection, without daunting statistical analysis. However, it is clearly reassuring to have a statistical measure of the confidence which can be placed in the reconstructed haplotypes. The confidence which can be assigned to the linkage phase across any two consecutive heterozygous loci depends only upon the number of informative genotyping results which agree or conflict with that phase, and on a statistical ‘power factor’ P which depends on the distance between loci and on the properties of the panel of aliquots. (See Methods section for an outline of the statistical procedure, and Protocol S1 for a detailed description; a simple look-up table for P is given in Table 1).
For the X-chromosome panel, we estimate the average size of the fragments at ∼90 kb from the initial pre-screening results (see Supplementary Data). Using this estimate of fragment size and the calculated DNA content of the panel as 0.28 X-chromosome copies per aliquot, we can calculate the confidence in the recovered haplotype over consecutive pairs of loci.
For example, taking loci rs2291122 and rs1479927 (consecutive heterozygous loci, since the intervening locus rs6527396 is monomorphic), nine aliquots were genotyped for both loci, of which eight were informative (the ninth failing to give a genotyping result for locus rs1479927). Of these eight, seven give result AA or GG, whilst one (from aliquot 54) gives result AG (Figure 2). The distance between the loci is 17.4 kb, or ∼0.2 of the average fragment size (90 kb), and we know the DNA content of the panel to be 0.28 copies per aliquot (see above). So, from Table 1, the value of P (see Materials and Methods section) is ∼2.3, and hence the LogL of the linkage phase being AA/GG:
or odds of ∼1014:1. The same analysis for all loci reveals that the linkage phase is weakest (least supported) between markers rs1993969 and rs2054513, where the haplotype structure (TG/CA) is supported by odds of 105.5:1. Given the statistical support for the linkage phase between successive pairs of markers along the haplotype, one can compute the confidence in the complete haplotype structures spanning all 20 loci (14 heterozygous loci): the probability of the complete haplotype being correct is the product of the probabilities that each consecutive step from one heterozygous locus to the next is correct. In this case, the log-odds supporting the complete haplotypes is >5 (odds > 100 000:1).
A similar analysis can be performed for the longer haplotypes reconstructed on chromosome 21. In this instance, the average DNA content of the panel is 0.15 copies (of chromosome 21) per aliquot, and the average fragment size is about 190 kbp (Protocol S1). Analysis shows that the weakest point in the reconstructed haplotypes is between loci rs463055 and rs219724 [supported at odds of 104.2:1], and support for the complete haplotypes across all 29 heterozygous loci is >10 000:1
DISCUSSION
Large-scale analyses have added greatly to our understanding of scope and significance of variation in the human genome. They have also drawn attention to the importance of haplotypes (the combination of alleles found on a single chromosome) as opposed to genotypes alone, in extracting useful information from this variation (1–5). The utility of haplotypes inferred from population studies [such as the HapMap (6)] has recently been challenged (7). In any case, it is more generally agreed that the prudent use of direct molecular haplotyping can add greatly to the power of association studies in identifying genes involved in complex diseases (36,37). Against this, however, must be weighed the extra difficulty or cost of obtaining haplotype data: the easier haplotyping becomes, the greater the number of studies in which it becomes advantageous to use haplotype data.
We have demonstrated, for the first time, the accurate reconstruction of haplotypes spanning both large numbers of loci and long distances, using diploid DNA. The method is technically simple, requires no family members for pedigree analysis, and does not depend on assumptions of non-recombination, which are valid only over short distances. The method is efficient, because each locus need be genotyped on only a handful of informative samples. In the present example, we reconstructed long haplotypes on chromosome 21 by genotyping only about seven samples per locus. Indeed, statistical analysis shows that strongly-supported haplotypes can be reconstructed with fewer genotypings than would be needed in conventional pedigree analysis.
Aside from the DNA concentration of the aliquots, the only factors which determine the statistical power of the method are the average size of the DNA fragments and the spacing between successive heterozygous loci along the chromosome. Clearly, haplotypes cannot be reconstructed when none of the fragments is long enough to span the distance from one heterozygous locus to the next. However, significant statistical power remains even when the distance between loci approaches the average fragment size (Table 1) since the occasional longer, informative fragments are identified in the initial pre-screening. We were able to build haplotypes which spanned >160 kb between successive heterozygous loci (chromosome 21, loci rs222969–rs2187226). Haplotype blocks in humans are rarely longer than this (38). If necessary, however, far longer DNA fragments can be isolated at extreme dilution directly from pulsed-field gels (33) and make it possible to reconstruct haplotypes between even more widely-spaced loci.
In this paper, we have focused on the common problem of reconstructing the two haplotypes present in a diploid DNA sample. However, there is no reason why the same approach cannot be applied to reconstruct multiple haplotypes from the mixed DNA of many individuals.
On a technical level (and returning to the analysis of diploid samples), we note that the initial DNA dilution and PEP need be done only once. This gives sufficient product for ∼30–50 multiplex pre-screening reactions, each of which can comprise many tens of markers [we have shown elsewhere (39) that several hundred loci can be multiplexed in this way and amplified from each fraction of the PEP]. So a single initial sample prep is sufficient to haplotype many hundreds or several thousands of SNP loci. In this respect, our method complements the ‘polony’ approach of Zhang et al. (32), which they claim can be used to analyse large numbers of individuals, but only for modest numbers of SNPs.
The dilution and multiplex amplification steps which we use are straightforward, and have been shown to be robust in other contexts inluding genome mapping (39–41), the analysis of ancient DNA (42) and the measurement of copy-number variation (43). Multiplexing the first phase of the hemi-nested PCR (in contrast to multiplexing conventional single-phase PCRs) is surprisingly easy and does not constrain primer design. We have evaluated a wide range of alternative whole-genome amplification protocols (data not shown), but still find PEP to be the most representative and reproducible when starting from truly sub-genomic quantities of template. Nevertheless, we continue to explore other techniques for the primary amplification step, which would ideally give both faithful representation of sub-genomic samples and, with a sufficient level of amplification, would permit the use of only a single-phase PCR afterwards.
In these experiments, the final genotyping step was done by sequencing, since this was the most convenient method for us in analyzing relatively small numbers of samples. However, the protocol gives conventional PCR products which should be amenable to genotyping by many (though not all) other available technologies, some suited to much higher throughput. Moreover, the PCR products of the initial pre-screening reactions can be ‘cherry-picked’ and used directly in the genotyping reactions, avoiding the need to repeat PCRs.
Finally, we note that this method requires only very small amounts of genomic DNA: a 10th of a nanogram or less. This should make it possible (though we have not tested this) to recover haplotype data from very small samples, such as biopsies of tumours in which chromosomal rearrangements may produce haplotypes not found in the normal DNA of the individual.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Martin E.R., Lai E.H., Gilbert J.R., Rogala A.R., Afshari A.J., Riley J., Finch K.L., Stevens J.F., Livak K.J., Slotterbeck B.D., et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am. J. Hum. Genet. 2000;67:383–394. doi: 10.1086/303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drysdale C.M., McGraw D.W., Stack C.B., Stephens J.C., Judson R.S., Nandabalan K., Arnold K., Ruano G., Liggett S.B. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris R.W., Kaplan N.L. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet. Epidemiol. 2002;23:221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- 4.Winkelmann B.R., Hoffmann M.M., Nauck M., Kumar A.M., Nandabalan K., Judson R.S., Boehm B.O., Tall A.R., Ruano G., Marz W. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharmacogenomics J. 2003;3:284–296. doi: 10.1038/sj.tpj.6500195. [DOI] [PubMed] [Google Scholar]
- 5.Clark A.G. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 2004;27:321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- 6.Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. International HapMap Consortium. (2005). A haplotype map of the human genome. Nature. 437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terwilliger J.D., Hiekkalinna T. An utter refutation of the ‘Fundamental Theorem of the HapMap’. Eur. J. Hum. Genet. 2006;14:426–437. doi: 10.1038/sj.ejhg.5201583. [DOI] [PubMed] [Google Scholar]
- 8.Yan H., Papadopoulos N., Marra G., Perrera C., Jiricny J., Boland C.R., Lynch H.T., Chadwick R.B., de la Chapelle A., Berg K., et al. Conversion of diploidy to haploidy. Nature. 2000;403:723–724. doi: 10.1038/35001659. [DOI] [PubMed] [Google Scholar]
- 9.Kim J.H., Leem S.H., Sunwoo Y., Kouprina N. Separation of long-range human TERT gene haplotypes by transformation-associated recombination cloning in yeast. Oncogene. 2003;22:2452–2456. doi: 10.1038/sj.onc.1206316. [DOI] [PubMed] [Google Scholar]
- 10.Burgtorf C., Kepper P., Hoehe M., Schmitt C., Reinhardt R., Lehrach H., Sauer S. Clone-based systematic haplotyping (CSH): a procedure for physical haplotyping of whole genomes. Genome Res. 2003;13:2717–2724. doi: 10.1101/gr.1442303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukita Y., Miyatake K., Stokowski R., Hinds D., Higasa K., Wake N., Hirakawa T., Kato H., Matsuda T., Pant K., et al. Genome-wide definitive haplotypes determined using a collection of complete hydatidiform moles. Genome Res. 2005;15:1511–1518. doi: 10.1101/gr.4371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruano G., Kidd K.K. Direct haplotyping of chromosomal segments from multiple heterozygotes via allele-specific PCR amplification. Nucleic Acids Res. 1989;17:8392. doi: 10.1093/nar/17.20.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalatos-Beloin S., Tishkoff S.A., Bentley K.L., Kidd K.K., Ruano G. Molecular haplotyping of genetic markers 10 kb apart by allele-specific long-range PCR. Nucleic Acids Res. 1996;24:4841–4843. doi: 10.1093/nar/24.23.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitan Y., Kashi Y. Direct micro-haplotyping by multiple double PCR amplifications of specific alleles (MD-PASA) Nucleic Acid Res. 2002;30:e62. doi: 10.1093/nar/gnf062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano M., Nakamura T., Ozawa S., Maekawa K., Saito Y., Jun-ichi S. Allele-specific long-range PCR/sequencing method for allelic assignment of multiple single nucleotide polymorphisms. J. Biochem. Biophys. Methods. 2003;55:1–9. doi: 10.1016/s0165-022x(02)00114-8. [DOI] [PubMed] [Google Scholar]
- 16.Greene C.N., Cordovado S.K., Mueller P.W. Polymorphism scan for differences between transmitted and nontransmitted drb1*030101 alleles outside of exon 2 for type 1 diabetes: the frequency of polymorphisms is similar. Hum. Immunol. 2004;65:737–744. doi: 10.1016/j.humimm.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Pont-Kingdon G., Jama M., Miller C., Millson A., Lyon E. Long-Range (17.7 kb) Allele-specific polymerase chain reaction method for direct haplotyping of R117H and IVS-8 mutations of the cystic fibrosis transmembrane regulator gene. J. Mol. Diagn. 2004;6:264–270. doi: 10.1016/S1525-1578(10)60520-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C., Devlin B., Galloway N., Loomisa E., Schellenberg G.D. ADLAPH: a molecular haplotyping method based on allele-discriminating long-range PCR. Genomics. 2004;84:600–612. doi: 10.1016/j.ygeno.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Kamio K., Matsushita I., Tanaka G., Ohashi J., Hijikata M., Nakata K., Tokunaga K., Azuma A., Kudoh S., Keicho N. Direct determination of MUC5B promoter haplotypes based on the method of single-strand conformation polymorphism and their statistical estimation. Genomics. 2004;84:613–622. doi: 10.1016/j.ygeno.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Pont-Kingdon G., Lyon E. Direct molecular haplotyping by melting curve analysis of hybridization probes: beta 2-adrenergic receptor haplotypes as an example. Nucleic Acids Res. 2005;33:e89. doi: 10.1093/nar/gni090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millson A., Pont-Kingdon G., Page S., Lyon E. Direct molecular haplotyping of the IVS-8 poly(TG) and polyT repeat tracts in the cystic fibrosis gene by melting curve analysis of hybridization probes. Clin. Chem. 2005;51:1619–1623. doi: 10.1373/clinchem.2005.052159. [DOI] [PubMed] [Google Scholar]
- 22.Hurley J.D., Engle L.J., Davis J.T., Welsh A.M., Landers J.E. A simple, bead-based approach for multi-SNP molecular haplotyping. Nucleic Acids Res. 2005;32:e186. doi: 10.1093/nar/gnh187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H.H., Gyllensten U.B., Cui X.F., Saiki R.K., Erlich H.A., Arnheim N. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature. 1988;335:414–417. doi: 10.1038/335414a0. [DOI] [PubMed] [Google Scholar]
- 24.Crouau-Roy B., Clayton J. Haplotypes without children: PCR applied to close loci on individual human sperm. Hum. Biol. 1995;67:171–178. [PubMed] [Google Scholar]
- 25.Ruano G., Kidd K.K., Stephens J.C. Haplotype of multiple polymorphisms resolved by enzymatic amplification of single DNA molecules. Proc. Natl Acad. Sci. USA. 1990;87:6296–6300. doi: 10.1073/pnas.87.16.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding C., Cantor C.R. Direct molecular haplotyping of long-range genomic DNA with M1-PCR. Proc. Natl Acad. Sci. USA. 2003;100:7449–7453. doi: 10.1073/pnas.1232475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra R.D., Butty V.L., Shendure J., Williams B.R., Housman D.E., Church G.M. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl Acad. Sci. USA. 2003;100:5926–5931. doi: 10.1073/pnas.0936399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul P., Apgar J. Single-molecule dilution and multiple displacement amplification for molecular haplotyping. Biotechniques. 2005;38:553–559. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- 29.Inbar E., Yakir B., Darvasi A. An efficient haplotyping method with DNA pools. Nucleic Acids Res. 2002;30:e76. doi: 10.1093/nar/gnf075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald O.G., Krynetski E.Y., Evans W.E. Molecular haplotyping of genomic DNA for multiple single-nucleotide polymorphisms located kilobases apart using long-range polymerase chain reaction and intramolecular ligation. Pharmacogenetics. 2002;12:93–99. doi: 10.1097/00008571-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Wu W.M., Tsai H.-J., Pang J.H., Wang T.-H., Wang H.-S., Hong H.-S., Lee Y.-S. Linear allele-specific long-range amplification: a novel method of long-range molecular haplotyping. Hum. Mutat. 2005;26:393–394. doi: 10.1002/humu.9369. [DOI] [PubMed] [Google Scholar]
- 32.Zhang K., Zhu J., Shendure J., Porreca G.J., Aach J.D., Mitra R.D., Church G.M. Long-range polony haplotyping of individual human chromosomal molecules. Nature Genet. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- 33.Dear P.H., Bankier A.T., Piper M.B. A high-resolution metric HAPPY map of human chromosome 14. Genomics. 1998;48:232–241. doi: 10.1006/geno.1997.5140. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., Cui X., Schmidt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA. 1992;89:5487–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens J.C., Rogers J., Ruano G. Theoretical underpinning of the single-molecule-dilution (SMD) method of direct haplotype resolution. Am. J. Hum. Genet. 1990;46:1149–1155. [PMC free article] [PubMed] [Google Scholar]
- 36.Gillanders E.M., Pearson J.V., Sorant A.J.M., Trent J.M., O'Connell R., Bailey-Wilson J.E. The value of molecular haplotypes in a family-based linkage study. Am. J. Hum. Genet. 2006;79:458–468. doi: 10.1086/506626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levenstein M.A., Ott J., Gordon D. Are molecular haplotypes worth the time and expense? A cost-effective method for applying molecular haplotypes. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020127. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenwood T.A., Rana B.K., Schork N.J. Human haplotype block sizes are negatively correlated with recombination rates. Genome Res. 2004;14:1358–1361. doi: 10.1101/gr.1540404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichinger L., Pachebat J.A., Glöckner G., Rajandream M.-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q., et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall N., Pain A., Berriman M., Churcher C., Harris B., Harris D., Mungall K., Bowman S., Atkin R., Baker S., et al. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- 41.Bankier A.T., Spriggs H.F., Fartmann B., Konfortov B.A., Madera M., Vogel C., Teichmann S.A., Ivens A., Dear P.H. Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genomics. 2003;1:1787–1799. doi: 10.1101/gr.1555203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause J., Dear P.H., Pollack J.L., Slatkin M., Spriggs H., Barnes I., Lister A.M., Ebersberger I., Pääbo S., Hofreiter M. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 43.Daser A., Thangavelu M., Pannell R., Forster A., Chung G., Dear P.H., Rabbitts T.H. Interrogation of genomes by molecular copy-number counting (MCC) Nature Methods. 2006;3:447–453. doi: 10.1038/nmeth880. [DOI] [PubMed] [Google Scholar]