Abstract
Gene expression and processing during mouse male germ cell maturation (spermatogenesis) is highly specialized. Previous reports have suggested that there is a high incidence of alternative 3′-processing in male germ cell mRNAs, including reduced usage of the canonical polyadenylation signal, AAUAAA. We used EST libraries generated from mouse testicular cells to identify 3′-processing sites used at various stages of spermatogenesis (spermatogonia, spermatocytes and round spermatids) and testicular somatic Sertoli cells. We assessed differences in 3′-processing characteristics in the testicular samples, compared to control sets of widely used 3′-processing sites. Using a new method for comparison of degenerate regulatory elements between sequence samples, we identified significant changes in the use of putative 3′-processing regulatory sequence elements in all spermatogenic cell types. In addition, we observed a trend towards truncated 3′-untranslated regions (3′-UTRs), with the most significant differences apparent in round spermatids. In contrast, Sertoli cells displayed a much smaller trend towards 3′-UTR truncation and no significant difference in 3′-processing regulatory sequences. Finally, we identified a number of genes encoding mRNAs that were specifically subject to alternative 3′-processing during meiosis and postmeiotic development. Our results highlight developmental differences in polyadenylation site choice and in the elements that likely control them during spermatogenesis.
INTRODUCTION
Messenger RNA cleavage and polyadenylation is a necessary processing step for almost all eukaryotic mRNA transcripts. The mechanisms involved in 3′-processing have been studied in detail over the last 30 years, and the identities and roles of most of the proteins involved have been characterized (1,2). Comparative studies of the mechanisms and of the proteins involved in several organisms have shown a high level of primary sequence conservation between the homologous genes across a broad range of organisms (1,3).
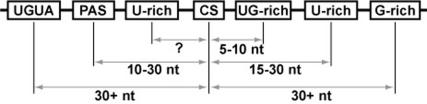
A number of cis-acting RNA sequences involved in the regulation, selection and processing of the 3′ ends of mammalian mRNA transcripts have been previously described [Figure 1, (1,4–6)]. By convention, these sequence elements are described as being (5′) or (3′) relative to the site of poly(A) addition. The most widely observed sequence, the polyadenylation signal (PAS), was identified 30 years ago (7), and is usually the canonical hexamer, AAUAAA, or its most common variant, AUUAAA; however, a number of recent studies have indicated that this element is more variable than originally thought (8,9). The PAS is usually 15–30 nt upstream of the site of poly(A) addition (1). Early studies of the downstream element indicated the presence of two distinct elements (10,11), though subsequent studies [e.g. (12–14)] resulted in an ambiguous picture, such that the downstream elements were jointly referred to as U-/UG-rich. We recently completed a computational study of large sets of 3′-processing sites in 10 different metazoans (5) that confirmed the presence of two separate elements, a proximal UG-rich element (typically 6–10 nt downstream of the 3′-processing site), and a more distal U-rich element (typically 15–30 nt downstream of the 3′-processing site). Our analysis also confirmed that these elements are highly degenerate, to the extent that functional 3′-processing sites often lack strong matches to one or both elements.
Figure 1.
A representation of the current model for the complete mouse 3′-processing regulatory sequences. The upstream PAS, most commonly AAUAAA or close variants, is the most prevalent feature (1). Recent studies have verified that the downstream U- and UG-rich elements, while degenerate, are distinct in both positioning and sequence content (5,6). The remaining elements are often referred to as auxiliary elements. While upstream U-rich elements have been postulated, no specific positioning requirements have been identified.
In addition to those commonly recognized sequence elements, ongoing studies from several laboratories have pointed to the existence of multiple auxiliary elements that can affect the efficiency or selection between alternative 3′-processing sites. These include upstream U-rich sequences [that can occur upstream or downstream of the PAS (15)] upstream UGUA-containing elements (16), and downstream G-rich elements (17–20). A recent study of human 3′-processing sites revealed potential systematic variations in the cis- and trans-acting elements that could be driving alternative processing (21).
The trans-acting factors responsible for 3′-processing include the core complexes cleavage-polyadenylation stimulation factor (CPSF), which interacts with the PAS element, cleavage stimulation factor (CstF), which interacts with the downstream U-rich [and possibly UG-rich elements (5)], and Papol, the poly(A) polymerase (1). A number of additional factors have been implicated in 3′-processing, especially in the context of alternative 3′-processing site selection (1,21,22).
Alternative 3′-processing, like alternative splicing, is a mechanism for altering the sequence and function of a mature mRNA transcript (15,16,21–28). While alternative polyadenylation can change the coding region of an mRNA, frequently only the 3′-untranslated region (3′-UTR) of the mRNA is affected. Variation of the 3′-UTR through alternate 3′-processing provides a means for gene regulation at the post-transcriptional stage, since 3′-UTRs frequently contain regulatory elements related to transcript localization, stability and translational control (29–42).
Spermatogenesis consists of a highly specialized program of development leading to a unique cell that requires unique protein products (43–45). Several studies have demonstrated unusual features in male germ cell mRNAs (43), both in the form of tissue-specific transcripts and tissue-specific isoforms of mRNAs with broader expression profiles. Several early studies noted a number of mRNAs that lacked the canonical AAUAAA PAS element upstream of the site of polyadenylation (8,46–49). Furthermore, alternative 3′-processing has been reported widely in male germ cells (22). More recent studies identified a mammalian germ cell-expressed variant of Cstf2 (50), a subunit of the CstF complex that binds to the precursor mRNA sequence downstream of the 3′-processing site (12). The variant, Cstf2t, is expressed predominantly in meiotic and postmeiotic male germ cells (pachytene spermatocytes through early elongating spermatids), and is thought to compensate for the lack of the somatic form of the X-linked Cstf2 in these cell types (50). CSTF2T protein is encoded by an autosomal intronless paralog of Cstf2 (51,52). Similarly, germ cell-specific variants of other factors, e.g. the cytoplasmic poly(A) polymerase (Papolb), have also been identified (53). These findings suggest that mechanisms of polyadenylation and 3′-processing in male germ cells might differ in significant ways from those in somatic cells, and that those differences could contribute to the specialized functions of germ cells. The discovery of germ cell-specific forms of proteins involved in 3′-processing provided a potential mechanism to account for some of this variation, yet the mechanism has remained elusive.
We initiated the present study to characterize differences in the 3′-processing regulatory sequences, and therein improve our understanding of the mechanism and regulatory implications of alternative 3′-processing during spermatogenesis. Our hypothesis was that mRNA transcripts expressed during male germ cell maturation would show differences in 3′-processing regulatory signals and/or 3′-processing sites that can be illuminated by sequence analysis of expressed sequence tags (ESTs). Using EST libraries, we identified and analyzed putative 3′-processing sites used at various stages (premeiotic, meiotic, and postmeiotic) of spermatogenesis. We compared 3′-processing characteristics represented in each library, looking initially at fidelity of putative cis-acting 3′-processing signals. We use the term fidelity to describe variation of a signal element among different genes in a single organism, reserving the term conservation for interspecies analysis. We also characterized the differences in 3′-UTR length distribution and use of tissue specific 3′-processing sites. We observed significant differences in 3′-processing characteristics (including both cis-element usage and 3′-UTR length distribution) in all three spermatogenic cell libraries, but not in the library derived from the testicular somatic Sertoli cells. Differences in 3′-processing can be correlated with changes in transcript abundances of known 3′-processing factors, as shown by a previously released set of microarray experiments generated from a spermatogenesis timecourse (54). We were further interested in regulatory implications of differences in 3′-processing, and therefore examined the portions of the 3′-UTR sequences that were differentially included in transcripts dependent on the choice of 3′-processing site, identifying evolutionarily conserved elements that represent putative functional portions of transcripts. Our findings are consistent with stage-specific variation in mRNA 3′-processing during spermatogenesis that can result in large-scale and systematic differences in posttranscriptional gene regulation.
MATERIALS AND METHODS
Cell type specific cDNA templates for sequencing
Primary, unamplified cDNA libraries from 18 day mouse Sertoli cells (10 pfu/μl), primitive Type A spermatogonia (0.116 pfu/μl), adult mouse pachytene spermatocytes (3.7 pfu/μl), and mouse round spermatids (5.5 pfu/μl ) were constructed in lambda Uni-Zap XR (Stratagene, CA) directionally cloned from oligo(dT)-primed-poly(A)+ RNA recovered from purified populations of each testicular cell type as described (55). Plasmid rescues were performed by mass excision on these libraries according to the manufacturer's protocol (Stratagene, CA). Briefly, XL-1 Blue MRF′ cells grown in Luria–Bertani (LB) supplemented with magnesium and maltose (2% w/v final) were resuspended to an A600 of 1.0 in 10 mM magnesium sulfate. A total of 300 μl of these XL-1 Blue MRF′ cells were co-infected with 100 μl of each primary cDNA library and 2 μl of ExAssist helper phage at 37°C for 15 minutes. Addition of 3.0 ml of NZY broth was followed by 3 hours of incubation at 37°C and 20 min at 70°C. Cultures were centrifuged and the supernatant was collected as a source of library phagemid. SOLR cells (300 μl) (Stratagene) at A600 = 1.0 was infected with 5 μl of each library phagemid supernatant at 37°C for 15 min. A total of 60 μl of 5 × NZY broth was added to each reaction and incubation was continued for an hour at 37°C. Appropriate volumes of each reaction were plated on LB ampicillin plates to obtain well-separated single colonies. Single colonies were used to inoculate LB medium in 96-well blocks to obtain plasmid DNA using the QIAprep 96 Turbo miniprep kit (Qiagen) and a BioMEK 2000 robot (Beckman). cDNA inserts in each plasmid were sequenced using a CEQ 2000XL automated sequencer (Beckman) with the T7 promoter primer. All EST libraries were supplemented by searching NCBI's dbEST for EST libraries that were generated from the same tissue type. In order to obtain adequate numbers of putative 3′-processing sites, we relaxed the tissue specificities of two libraries to any type of spermatogonia and any type of spermatocyte. While this relaxation means that the spermatogonia and spermatocyte sites come from a mixture of cell types, they are still premeiotic and meiotic, respectively, and the gain in number of sequences adds statistical confidence to our results. Furthermore, examination of the transcript abundances of common 3′-processing factors (e.g. components of CstF and CPSF) from a set of microarrays generated from a spermatogenesis timecourse [Figure 4, (54)] indicated that these genes were expressed at essentially constant transcript levels in all types of spermatogonia. Novel EST sequences have been entered in Genbank with accession numbers EG563088–EG565092.
EST analysis
EST sequences were used to identify putative 3′-processing sites, as described previously (56). Briefly, sequences were initially filtered for low complexity, contaminant and repetitive element sequences. The resulting masked sequences were stringently aligned to the genome using BLAT (57). Aligned sequences that included masked elements were unmasked and re-aligned with a Smith–Waterman approach (58). Putative 3′-processing sites were obtained from either 3′-ESTs or 5′-ESTs, with a restriction that the EST contains evidence of poly(A) tails in the form of at least five consecutive trailing As (or five consecutive leading Ts) that did not align to the genome. Gene assignments were made by determining the nearest correctly oriented annotated gene [according to Ensembl (59)] within 5000 bases, a distance that should include over 90% of all true assignments as determined by the distribution of 3′-UTR lengths in our database PACdb (56). All library-specific 3′-processing site collections were reduced to unique sites [including clustering of sites separated on the genome by <25 nt (26,56)], to prevent highly expressed genes from skewing pattern detection results.
EST library characterization
Complete EST libraries were characterized with regard to 3′-processing site selection and regulatory sequence usage. Of specific interest was the distribution of 3′-UTR lengths in each library; however, we have demonstrated that such distributions are subject to systematic bias depending on the length distribution of the sampled transcripts (60). The length distribution of the transcripts depends on the preparation of the library, including efficiency of reverse transcription and selection of specific clones for sequencing. We developed a simple method to identify EST libraries whose 3′-UTR length distributions could be compared safely. Briefly, we aligned all ESTs from a given library to a reference set of cDNA sequences downloaded from ENSEMBL (59), and kept only the best match that spanned at least 90% of the library-derived EST with at least 95% sequence identity. The collections of cDNA sequences matched by specific EST libraries were assumed to be representative of the transcripts from which the ESTs were derived (including any systematic biases), and were therefore used to generate estimated transcript length distributions. Finally, we restricted direct comparison of 3′-UTR length distributions to groups of EST libraries with sufficiently similar estimated transcript length distributions (60).
Regulatory sequence analysis
For regulatory signal characterization, we extracted genomic sequences extending 100 nt upstream and 100 nt downstream of all (unique) putative 3′-processing sites in each library. We performed an initial search for putative 3′-processing signals with the Gibbs Sampler (61), using the command line options -x -n -r -F -S 50. We also used our positional word counting (PWC) software that characterizes both count and specific positioning with respect to the 3′-processing site (5,62). PWC produces a 2D matrix, indexed by subsequences (words) of length k (k mers) in 1D and sequence position in the other dimension. Putative biologically functional sequences are inferred from significantly non-random positioning distributions. To infer degenerate motifs from the counts of exact sequence words, we developed a novel approach (L. N. Hutchins, P. Singh, J. M. Brockman, S. Murphy, J. Salisbury and J. H. Graber, manuscript in preparation) based on a Non-negative Matrix Factorization (NMF) (63) decomposition of the PWC matrix to identify sets of related sequence words. Briefly, NMF is a dimensionality reduction algorithm that identifies basis vectors of k mers that follow common positioning patterns. Each basis vector putatively represents a functional motif that has a specific positioning in the sequences. The output of NMF includes a weight vector that can be interpreted as a positioning probability for the motif, and a weighted list of k mers that contribute to the motif. Our studies indicate that the combined PWC/NMF analysis is robust with variation of word size k (manuscript in preparation). The results reported here were based on a tetramer (k = 4) analysis. The top five weighted tetramers for each putative motif (Figure 1) were used for the word set enrichment analysis (WSEA), described below. We further used the NMF weight vectors to restrict acceptable positioning of each motif in the word set enrichment analysis. A complete listing of the acceptable sequence words and positioning restrictions for each motif are shown in Table 2.
WSEA
For comparison of degenerate signals between sequence sets, we developed a method termed WSEA derived from the gene set enrichment analysis (GSEA) previously developed for analysis of differential expression of sets of related genes (64). GSEA was developed to identify statistically significant differences between phenotypically distinct microarray experiments in which a set or group of genes acts together in a complex fashion that reduces the likelihood that any single gene in the set will pass a multiple hypothesis test. In GSEA, all genes are ranked according to their correlation with the phenotype, and then sets are assessed within the ranking for non-random distribution (64). In WSEA, we characterize differences in complex regulatory sequences between different libraries in an analogous manner, in which sequence words replace genes, and library replaces phenotype assignment. Sets of words are chosen as the most probable manifestations of a putative degenerate regulatory sequence, (such as developed through the PWC/NMF analysis described above).
In brief, the method for comparing two sequence samples is as follows: all k mers are counted in each sample, and converted to either the represented fraction of all k mers counted, or to the fraction of the sampled sequences with at least one copy of the k mer. A Z-score is generated from a standard test of equal proportions, using the large-sample null hypothesis that the measured proportions of k mers will follow a normal distribution. All k mers are ranked based on the Z-scores, which by definition vary between plus and minus infinity. The GSEA methodology ranks genes via correlation values, which vary between plus and minus one, therefore we converted Z-scores to the same range via a logit transformation (the logit transform is implemented as an option, since the exact numerical nature of the ranking is not critical) (64).
The WSEA enrichment score (ES) for any set of words is assessed with the GSEA equation (http://www.broad.mit.edu/gsea/):
| 1 |
| 2 |
| 3 |
| 4 |
where W is the set of k mers under test, Nh is the number of k mers in W, N is the size of the total sorted list of words, R is the array of correlation values corresponding to the sorted list, I is an indicator function, equal to 1 if word wj is in set W and 0 otherwise, and i and j are index variables. Due to the normalization, ES is constrained to vary between −1 and +1. As with GSEA, we assess statistical significance in WSEA with two different permutation analyses: (i) permutation of the k mer labels, which compares the specific set of k mers under test to any random selection of k mers and (2) permutation of the assignment of individual sequences to sequence set, which compares the observed differences in k mer sets to differences that would arise through a random separation of the sequences into two groups. As with GSEA, we find the latter analysis to be more stringent, and report those values here. WSEA is a means of testing hypotheses (as is GSEA), rather than de novo identification. Putative word sets were generated from PWC/NMF analysis (described above) of the universe control set of sequences. The weighted word lists were used to provide word sets,and the positioning weight vector was used to restrict the portions of the sequences analyzed for each putative motif.
Gene Ontology (GO) analysis
To search for shared functionality of groups of genes that were alternatively 3′-processed in a specific tissue, we used the VLAD (VisuaL Annotation Display) tool (65). VLAD is based on the GO annotations (66), and assesses the statistical significance of a group of genes sharing a common annotation. The statistical significance is based on a hypergeometric distribution, and specifically assesses the probability of drawing x out of n genes with a given annotation, given that the superset from which they are drawn has known proportion X out of N. Each set was tested with two different supersets: (i) the tissue-specific set of all genes for which 3′-processing sites could be determined and (ii) the complete set of all annotated genes from the Mouse Genome Informatics (MGI) (67). The former is a more stringent analysis, assessing the alternatively processed genes as a subset of the genes present in the tissue, rather than as a subset of all genes in the genome.
Evolutionarily conserved sequences
We downloaded and constructed rudimentary databases for the conserved sequences from a multiple alignment of human, dog, mouse and rat genome sequences (42), and from a multiple alignment of human, rat, mouse, chicken and zebrafish genome sequences (38). Custom Perl scripts were developed to extract conserved elements from either source that overlap a user-provided genomic region. Putative elements were restricted to those identified through an alignment of mouse with at least two additional species in order to reduce false positive cases due to insufficient evolutionary divergence.
RESULTS
We characterized 3′-processing in specific testicular cell types (spermatogonia, spermatocytes, round spermatids and Sertoli cells) through genomic alignment of non-normalized EST libraries (see Materials and Methods). Because we were interested in characterizing subtle differences in 3′-processing regulatory sequences, we limited our analysis to putative 3′-processing sites with only the strongest support (56). Our EST libraries are a combination of new sequences from a non-normalized cDNA library (described in Materials and Methods) and sequences extracted from NCBI's dbEST that could be assigned to the same tissue type. Our final working sets (Table 1) included 305 sites from spermatogonia, 356 from spermatocytes, 190 from round spermatids and 322 from Sertoli cells. For comparative purposes, we generated two additional sets:
Universe: 8542 high-confidence, frequently used (at least five supporting ESTs) 3′-processing sites drawn with no restriction on the tissue or developmental stage. This set was generated as part of a recent multi-organism characterization of 3′-processing downstream regulatory elements (5).
Standard: 3674 high-confidence, moderately utilized (at least two supporting ESTs), drawn from non-normalized EST libraries that explicitly did not include samples from tissues that are known to utilize extensive alternate 3′-processing characteristics, e.g. testis, brain or cancerous tissues.
All datasets are available at http://harlequin.jax.org/spermatogenesis/.
Table 1.
A summary of the EST-genome analysis for identification of putative 3′-processing sites
| Sertoli cells | Spermatogonia | Spermatocytes | Round spermatids | |
|---|---|---|---|---|
| Starting ESTs | 11 014 | 7008 | 9941 | 4618 |
| ESTs after filtering | 10 070 | 6421 | 9662 | 4549 |
| ESTs after genomic alignment | 8739 | 5790 | 8588 | 4044 |
| Putative 3′-processing sites | 418 | 457 | 600 | 541 |
| Condensed 3′-processing sites | 359 | 346 | 417 | 243 |
| Unique 3′-processing sites | 322 | 305 | 355 | 190 |
| Sites with AAUAAA | 220 | 229 | 244 | 123 |
3′-Processing elements vary systematically in spermatogenic cell transcripts
Early studies indicated an unexpectedly low incidence of the canonical PAS hexamer (AAUAAA) in transcripts from mammalian testes (8). We measured the percentage of the unique sequences in each of our EST libraries that contained an exact copy of the canonical PAS signal (AAUAAA) between 10 and 40 nt upstream of the putative cleavage sites [this range was selected based on previous studies of AAUAAA positioning, (1,5,26)]. We found that AAUAAA is present in 64.7% of round spermatid, 68.5% of spermatocytes, 75.1% of spermatogonia and 68.3% of Sertoli cell mRNAs. In contrast, AAUAAA occurred in 70.4% of the standard mRNAs and 69.9% of the universe mRNAs. Differences between premeiotic spermatogonia and the postmeiotic round spermatids are statistically significant (P = 0.013, test of equal proportions), and, in general, the fraction of sequences containing AAUAAA is lower in meiotic and postmeiotic samples than in premeiotic spermatogonia, testicular somatic Sertoli cells or other somatic cells.
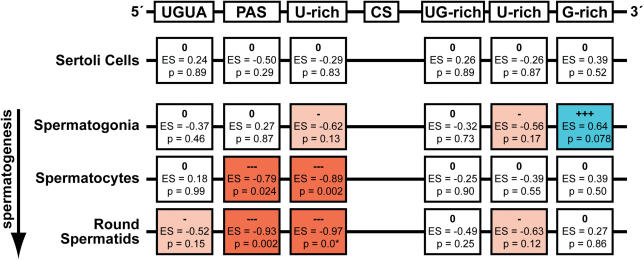
Characterizing differences in 3′-processing elements other than the PAS is challenging due to the well-characterized degeneracy of the other elements (1). We therefore developed a new method (WSEA, see Materials and Methods), explicitly to characterize variation in degenerate signal elements between sequence samples. We represent each element by its most probable set of sequence words. Sequence word sets and positioning restrictions (listed in Table 2) were generated to represent each of the elements shown in Figure 1 based on PWC/NMF analysis (Materials and Methods) of the universe set. Using WSEA, we tested all tissue-specific sequence sets against the standard set, and found significant differences in the fidelity and/or usage of 3′-processing elements in all three germ cell samples, but not in the Sertoli cells (Figure 2). A comparable analysis using the universe as a control produced similar results and is available in Supplementary Figure 1. The complete WSEA analysis, including comparisons between all pairs of samples, is available in Supplementary Table 1. We used a variety of other approaches to search for differences in the regulatory sequences, including positional word counting (5,62) and the Gibbs Sampler (61). The results of these analyses are consistent with the WSEA results (data not shown).
Table 2.
Groups of sequence words and positioning restrictions for mouse 3′-processing elements used in the WSEA
| 3′-Processing element | Accepted sequence words | Accepted start positions |
|---|---|---|
| Upstream UGUA | UGUA, CUGU, AUGU, UUGU, CCUG | −100 to −30 |
| PAS | AAUA, AUAA, UAAA, AAAA, AAAU | −40 to −10 |
| Upstream U-rich | UUUU, AUUU, UUUA, UUUG, CUUU | −100 to −5 |
| Downstream UG-rich | UGUG, GUGU, CUGU, UCUG, GUCU | −5 to +25 |
| Downstream U-rich | UUUU, AUUU, UUUA, UUUG, CUUU | 0 to +45 |
| Downstream G-rich | GGGG, GGGA, GGAG, GAGG, AGGG | +25 to +100 |
Accepted words and positioning for each element were approximations taken from PWC/NMF (manuscript in preparation) analysis of the universe sequence set.
Figure 2.
Variation in 3′-processing elements in various testis cell types. WSEA characterization of the variation in 3′-processing signals in spermatogenesis related tissues compared to the standard control set of 3′-processing sites. Over- and under-representation are signified by ‘+’ symbols and blue color, or ‘−’ symbols and red color, respectively. Significance values are shown reflecting the probability (P) of equal representation in the test and control sets according to permutation analysis (+/–: P < 0.2, ++/−−: P < 0.1, +++/−−−: P < 0.05, 0: no significant change). The reported P-values represent the probability of obtaining the measured difference when randomly selecting test sets of equivalent size from the standard control set. CS: cleavage site, PAS: canonical polyadenylation, ES: enrichment score (Equation 4).
Germ cell mRNAs have truncated 3′-UTRs compared with other cell types
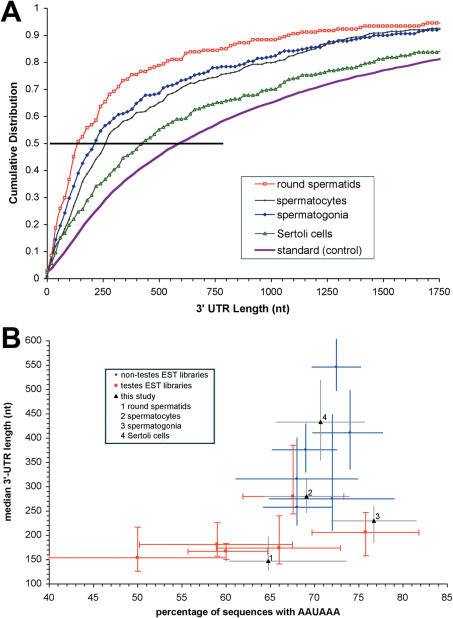
We used the genomic alignments of the ESTs to assign the putative 3′-processing sites to probable genes, and from these and the locations of the annotated stop codons, we determined projected 3′-UTR lengths. As shown in Figure 3A, the distribution of 3′-UTR sequence lengths in round spermatids was significantly shifted towards shorter values, with a median value of ∼150 nt, compared to 220 nt for spermatogonia, 260 nt for spermatocytes, 400 nt for Sertoli cells, 600 for the standard set and 800 for the universe set. Plotting the median 3′-UTR-length as a function of the fraction of sequences that contain the canonical PAS element (Figure 3B) revealed a positive correlation (r = 0.6). Note that the EST libraries represented in Figure 3B were restricted to those that we determined to be free of systematic biases related to sampled transcript lengths, as described in Materials and Methods. The libraries drawn from NCBI's dbEST were plotted separately based on whether or not they were derived from testis-generated tissues. Testis-derived sequences can be easily distinguished as a group from other sequences.
Figure 3.
Variation in 3′-UTR length distribution during spermatogenesis. (A) Cumulative length distribution of the 3′-UTR sequences implied by the EST libraries from various testis-related tissues. For comparison, the length distribution is also shown for the standard control set. (B) Median 3′-UTR length plotted as a function of sites with the canonical PAS hexamer (AAUAAA). Bars show the 95% confidence intervals. The libraries shown are a selection of dbEST (86) libraries with similar transcript sampling, as described previously (60).
Since many of the genes expressed in testis are known to be tissue specific (43), we investigated the possibility that the difference in 3′-UTR length distribution was due to differences in the genes being expressed rather than differences in 3′-processing. We searched for genes from each tissue sample for which we could identify 3′-processing sites in our standard set, and characterized genes as either:
Truncated: if the 3′-processing site in the tissue specific sample produced a 3′-UTR shorter than or equal to all sites for the same gene in the standard sample (with the further restriction that multiple sites be present in the standard set);
Elongated: if the 3′-processing site in the tissue specific sample produced a 3′-UTR longer than or equal to all sites for the same gene in the standard sample (with the further restriction that multiple sites be present in the standard set);
Invariant: if the 3′-processing site in the tissue specific sample was the same as in the standard sample; or
Indeterminate: if the sites in the standard set implied both longer and shorter 3′-UTRs than observed in the tissue specific sample.
We found (Table 3) that genes with 3′-processing specific to developing spermatogenic cells are twice as likely to result in 3′-UTR truncation as elongation, whereas the Sertoli sample showed roughly equal numbers of truncated and elongated 3′-UTRs. A complete list of all genes subject to alternative 3′-processing in germ cells is available in Supplementary Table 2.
Table 3.
A comparison of truncated and elongated 3′-UTR sequences in spermatogenesis related sequences (based on EST-genome analysis)
| 3′-UTR comparison | Sertoli cells | Spermatogonia | Spermatocytes | Round spermatids |
|---|---|---|---|---|
| Shorter | 15 | 11 | 15 | 18 |
| Shorter or equal | 13 | 15 | 24 | 9 |
| All truncated (x) | 28 | 27 | 39 | 27 |
| Longer | 10 | 7 | 13 | 2 |
| Longer or equal | 20 | 10 | 11 | 1 |
| All elongated (n − x) | 30 | 17 | 24 | 3 |
| Truncated fraction (f = x/n) | 0.483 | 0.614 | 0.619 | 0.900 |
| Probability (P = 0.5) | 0.896 | 0.174 | 0.077 | 8.68E-07 |
Statistical tests are for the null hypothesis that truncation and elongation are equally likely (P = 0.5). The two-sided probability was calculated as the sum of binomial probabilities for observing an equal or greater divergence of f from 0.5.
Variation in 3′-processing factor expression during spermatogenesis
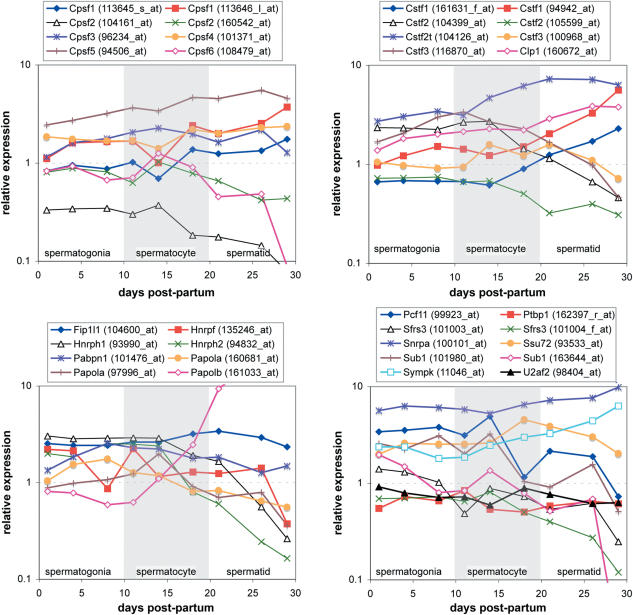
A recent report described a series of microarray experiments that were generated from a spermatogenesis timecourse (54). We extracted the data from these experiments from NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), and examined the measurements for a number of genes with known activity in 3′-processing (1,21,22), including Cpsf1-Cpsf6, Cstf1-Cstf3, Cstf2t, Papola, Papolb, Clp1, Fip1l1, Hnrpf, Hnrph1 (hnRNP H), Hnrph2 (hnRNP H′), Pabpn1, Pcf11, Ptbp1, Sfrs3, Snrpa, Ssu72, Sub1, Sympk and U2af2. Since we are interested in germ cell-specific processing, the expression levels were normalized to the average value observed in the two testicular somatic samples. In examining the transcript measurements, a few general patterns could be discerned, considering both absolute expression and variation throughout the timecourse. Cpsf5, Cstf2t, Clp1, Snrpa, Ssu72 and Sympk were significantly more abundant in all germ cell samples compared to the somatic cells. Conversely, Cpsf2 was downregulated in all germ cells, particularly so in the isoform with the longer 3′-UTR (probeset 104161_at).
Examination of the variation in transcript levels during the timecourse revealed additional features of interest. With only a few exceptions (e.g. Hnrpf), the transcript levels stayed constant throughout the spermatogonial stages, up to 10 days postpartum (dpp). Meiosis begins to be observed at ∼14 dpp (corresponding to the appearance of pachytene spermatocytes), with an accompanying change in the transcript levels of many genes in Figure 4. As expected from previous work (50–53), the transcript levels for Papolb and Cstf2t began to increase, while those for Papola and Cstf2 correspondingly decreased. Transcript measurements for Hnrph1 and Hnrph2 are much more highly expressed at the spermatogonia stages than either the spermatocyte or round spermatid stage(∼21 dpp). A number of additional transcripts appeared either to increase (Cpsf1, Cpsf5, Cstf1, Clp1 and Sympk) or decrease (Cpsf2, Cpsf6, Cstf3, Pcf11 and Sfrs3) continually from meiosis through spermatid formation.
Figure 4.
Variation in transcript measurements of known 3′-processing factors during spermatogenesis. Microarray-based transcript measurements of known 3′-processing factors extracted from a spermatogenesis timecourse (54). Transcripts with multiple probesets (e.g. Cpsf1) typically are designed to assay different isoforms. Accordingly, changes in the relative signal between such probesets across the timecourse potentially indicate alternative processing of the associated transcript at different stages of spermatogenesis (e.g. Cstf1). All probesets were normalized to the average measured value in the two somatic samples from the original data set (54).
Transcript levels can only provide indirect evidence of 3′-processing factor protein activity for two distinct reasons. First, 3′-processing factors can be subject to significant posttranscriptional control of translation, e.g. Cstf3 (68,69), potentially disrupting any correlation between transcript and protein levels. In addition, 3′-processing genes can also be subject to alternative processing resulting in variation in the 3′-UTR, e.g. Papola (1). Microarray measurement of isoforms with varied 3′-UTRs is dependent on the location of the hybridization probes, in as much as probes located within portions of the transcript that are excised in some isoforms can lead to misinterpretation of alternative processing events as changes in transcript abundance. At present, there is no systematic method to account for these differences using data from Affymetrix gene chips, however updated gene chips typically add additional probes to specifically assay alternate isoforms.
Despite these limitations, we were able to use the microarray data as a means of supporting alternative 3′-processing as indicated by ESTs. Using the annotation data made available by Affymetrix (http://www.affymetrix.com/), we identified specific locations on the genome and within transcripts for the U74 genechip probes. We identified several transcripts with probesets that were completely located within sequence that would be eliminated from the transcript when processed at the site indicated by ESTs. For example, at the round spermatid stage, ESTs indicate truncated forms of genes Gga2 (probeset 160812_at), Rpl21 (133729_at) and Lmnb2 (101414_at), whereas the microarrays indicated at least threefold reduction in expression, an expected result, since the truncated transcripts do not include the sequence complementary to the probes. A similar phenomenon is observed at the spermatocyte stage for genes Ubp1 (97304_at) and Bzw1 (94019_at).
Putative regulatory implications of alternative 3′-processing: evolutionarily conserved elements
It has been suggested that variation in 3′-UTR sequence is a potential mechanism for altering posttranscriptional regulation of the affected transcript, through specific inclusion or exclusion of functional sites [e.g. binding sites for RNA-binding proteins (29,33,39,40) or microRNAs (31,34,70–72)]. A mutational analysis revealed that cytoplasmic polyadenylation element (CPE) mediated control, based in the 3′-UTR is specifically needed for transcripts crucial for meiosis in males and females (73). As such regulatory sequences are often evolutionarily conserved (74), candidate functional elements can be identified through comparative analysis of orthologous genes in related organisms. We examined genes with evidence of altered 3′-processing in germ cells, searching for conserved sequence blocks (as described in Materials and Methods) in the portions of the 3′-UTR that are included or excluded based on the choice of 3′-processing site. We found that a majority of the differentially included 3′-UTR sequences overlapped conserved elements. A straight-forward interpretation is that the overlap between evolutionarily conserved sequence blocks and the differentially included portions of 3′-UTR reflects stage-specific changes in the regulation due to alternative 3′-processing. A complete listing of all putative conserved elements is available in Supplementary Table 2.
Recent reports identified a number of microRNA sequences that are active at various stages of spermatogenesis (75,76). From these reports, we extracted 32 distinct mature microRNA sequences for analysis with the 3′-UTR sequences implied by the 3′-processing sites in our datasets. For this analysis, we used the miRanda software package (77). Interestingly, we found that essentially all microRNAs we tested occurred at the same frequency (total matches divided by summed length of all target sequences considered) across all sequence samples. However, since the 3′-UTR sequences are significantly shorter in spermatocyte and round spermatid (Figure 3) samples, this resulted in proportionately fewer targeted transcripts in each of these two samples. The microRNA/miRanda analysis is available as Supplementary File 3.
Finally, we used the GO (66) to search for over-represented classifications among the genes that are subject to tissue-specific processing, but found no significant enrichment (data not shown). This result indicates that no specific processes, functions or cellular locations are systematically altered, but rather that the genes with germ-cell specific alternative 3′-processing are consistent with a random sampling of the rather specialized genes (43) that are expressed in these tissues.
DISCUSSION
3′-Processing control elements vary systematically during different stages of spermatogenesis
In this report, we have described differences in 3′-processing regulatory sequences during mammalian spermatogenesis. Developmental or tissue-specific variations in 3′-processing site selection are mediated by differences in expression of the trans-acting factors that interact with the precursor mRNA to select the site. Such changes can include expression of isoforms of common 3′-processing proteins, variation in the abundance of common 3′-processing proteins, and/or expression of tissue-specific auxiliary proteins. Our analysis of the spermatogenesis microarray timecourse [Figure 4; (54)] identified several known 3′-processing factors whose transcript abundances change significantly during spermatogenesis. Computational characterization of 3′-processing sites from a tissue- or developmental stage-specific sources, such as we have performed here, therefore can reveal the characteristics of the regulatory sequence patterns, and indirectly reflect differences in the expression of trans-acting protein factors.
It is important to keep in mind that the phenomena that we report here are statistical characterizations of the 3′-processing activities throughout spermatogenesis. Transcripts sampled at each stage represent a snapshot that is a mixture of current and previous transcriptional activity, e.g. transcripts generated and processed in early stages of spermatogenesis could last a significant length of time, and be sampled in EST libraries generated in later stages. Indeed, there are several documented examples of this phenomenon. For example, previous studies have shown that CPE mediated translational regulation is active and required for successful spermatogenesis (73). Transcripts regulated by CPEs are translationally inactive and stable for long periods (78). In addition, transcripts that are silenced by this mechanism are often deadenylated to a short (10–20 nt) poly(A) tail (78); nearly all EST library construction is initiated with a oligo(dT)-primed reverse transcription. A reduction or absence of the poly(A) tail has the potential to bias EST libraries against these transcripts.
Our results support at least two distinct types of variation in 3′-processing signals during spermatogenesis. We found that 3′-processing sites utilized in meiotic spermatocytes and postmeiotic round spermatids were characterized by a reduced incidence of most elements of the complete 3′-processing regulatory sequence, when compared to controls. Given the reduced usage of the typical 3′-processing elements, one might expect to find alternative elements. However, we were not able to identify any significant alternative pattern through standard characterizations [e.g. the Gibbs sampler (61)]. Instead, we found patterns that were similar to those from control sets, but with lower information content, signifying a weaker signal, or reduced fidelity, in the collected sites. Concurrent with the differences in the cis-elements, the transcript levels of several genes known to be involved in 3′-processing vary significantly starting at meiosis with continued variation through the spermatid stages (Figure 4). The reduction in strength of the computationally identified pattern supports a model in which changes in the 3′-processing complex result in a reduced specificity in selection of 3′-processing sites in meiotic and postmeiotic germ cells.
Our analysis of the 3′-processing sites used in premeiotic spermatogonia revealed a significant increase in transcripts with downstream G-rich elements, accompanied by a reduction in the fraction of transcripts with downstream U-rich elements (Figure 2). G-rich signals have been implicated as auxiliary elements in 3′-processing via several distinct mechanisms, including interactions with the HNRPH1 and HNRPH2 proteins (18,79), formation of secondary structure with the U-rich downstream elements (80), formation of an independent quadruplex secondary structure (19), and activity (acting as a DNA element) as a transcriptional pause site (20). Of these potential mechanisms,only the second can be eliminated as likely functioning in male germ cells, since we observe a concurrent decrease in U-rich sequences. Intriguingly, both Hnrph1 and Hnrph2 transcript levels were significantly increased in spermatogonia cells compared to either spermatocytes or round spermatids. Elevated HNRPH1 or HNRPH2 protein levels could provide a mechanism for increased use of 3′-processing sites with G-rich elements. Further experimental study will be required to test this possibility.
In contrast to the germ cells, the somatic Sertoli cells show no apparent difference in the usage of any of the 3′-processing elements, when compared with either of our controls. This result suggests that the differences observed in the spermatogonia, spermatocyte and round spermatid libraries are not systematic artifacts related to preparation or analysis, since the libraries were generated through common procedures. In addition, the similarity of 3′-processing of transcripts in Sertoli cells to our control sets indicates that spermatogenic cells utilize unique posttranscriptional regulatory activities that differ from those generally used in somatic cells, which might be expected given the different fates of these cells.
3′-Processing during spermatogenesis is characterized by truncated 3′-UTR sequences
The distribution of transcript and 3′-UTR lengths in a specific cell type is determined by which genes are expressed as well as how they are processed, including 3′-processing site selection. All cell types investigated here displayed a tendency towards shorter 3′-UTR sequences (Figure 3), although the effect was most pronounced in round spermatids. Truncated 3′-UTR sequences were typically associated with poor matches to the 3′-processing signal consensus (Figures 2 and 3). Computational surveys of somatic 3′-processing sites have previously indicated that short 3′-UTRs were associated with poor matches to the consensus PAS hexamer, AAUAAA (22,46,48). The correlation between signal and length of 3′-UTR sequences is also consistent with an association between the 3′-processing complex and the C-terminal domain of the largest subunit of RNA polymerase II (81,82). Under the assumption of 5′- to 3′-processing by RNA polymerase, a reduced specificity in recognition of 3′-processing elements would reduce the length of sequence that must be scanned to find an acceptable processing site. Our results are consistent with either less restrictive selection of 3′-processing sites, as we propose here, or alternatively with selective degradation of transcripts with longer 3′-UTRs. We favor the former mechanism in light of the combined evidence of systematic changes in 3′-processing) signals and correlated changes in expression of trans-acting factors (4), including paralogous copies of trans-acting 3′-processing factors in testes (50,53).
As noted above, the spermatogonial library revealed a relatively high percentage of 3′-processing sites with the canonical AAUAAA sequence concurrent with a relatively short 3′-UTR length distribution. This suggests that the mechanism(s) leading to fewer transcripts with the canonical AAUAAA hexamer in spermatogenic cells is independent of the mechanism(s) leading to reduced transcript length, or at least that there may be multiple means by which a shortened 3′-UTR is obtained. The relatively high incidence of downstream G-rich signals in spermatogonial transcripts indicate one potential mechanism by which these transcripts are truncated. Investigation of the spermatogonial 3′-processing sites revealed that transcripts with a reasonable match (in sequence and positioning) to the G-rich element displayed a truncated 3′-UTR length distribution when compared to the transcripts without a match (data not shown).
Systematic variation in 3′-processing may enable broad changes in post-transcriptional regulation
Significant changes in 3′-UTR length and composition can be accompanied by regulatory changes due to inclusion or exclusion of posttranscriptionally acting cis-elements. We found a large fraction of the differential sequences to be covered by evolutionarily conserved elements (Supplementary Table 2), supporting the regulatory relevance of the tissue-specific differences in mRNA 3′-processing. As noted, our analysis of microRNA targets and GO classification revealed no significant differences in either specific target sequences or functional classes of genes subject to alternative processing; rather these results indicate that altered 3′-processing is a general feature of the cell type.
The trend towards truncation of 3′-UTRs in late stage spermatogenic cells (Figure 3) and the evidence of evolutionary conservation in the cleaved sequence is intriguing in the context of previous studies of posttranscriptional regulation mediated by 3′-UTR sequences. These studies have reported effects on both transcript stability and translational control mediated by highly conserved sequences (83). While the functionality of any specific sequence could not be predicted solely from its evolutionary conservation, the general trend was that inclusion of such conserved elements in the transcript led to a decrease in stability. As spermatogenesis proceeds, the chromatin is remodeled, terminating all transcription until after fertilization and one cell division in the new embryo. Intriguingly, a recent study revealed evidence of mRNA translation in mature sperm (84). It is reasonable to assume that at least some transcripts expressed at this stage would benefit from an extended half-life, compared with somatic cells.
Variability of 3′-processing elements has complicated characterization efforts
Several previous studies found little or no consensus pattern among the downstream portions of the mammalian (mouse and human) 3′-processing regulatory sequences (15,19). Our results present a possible explanation for this: previous studies have been typically performed on sequences drawn from a broad range of tissues, cell types and growth conditions. In our detailed study of just four different and very specialized samples, we find that the exact nature of the regulatory sequences, specifically the balance between the different elements of a multi-partite signal, can vary significantly from one sample to another. [A recent study of human 3′-processing revealed similar variations among a different set of cell types (6)]. A training set drawn from multiple tissues, developmental stages or growth conditions would presumably include examples of many systematic variations of the full 3′-processing signal. A computational characterization of such a training set would be necessarily driven to an average pattern that only accurately reflects the strongest, common element(s), namely the PAS element and core downstream elements.
While significant evidence has accumulated for tissue-specific 3′-processing (6,15,25,85), our results indicate that the processing in any given tissue type or developmental stage is a mixture of condition-specific and condition-independent activities. Such a mixture complicates the identification of signal elements specific to the given conditions; we used our control sets as means of identifying the tissue-specific processing sites. As described above, we find evidence of at least two mechanisms of condition-specific 3′-processing activity. In addition, our results indicate that a significant fraction of the genes in any given sample are using a ‘typical’ 3′-processing site. A direct interpretation of these results is that, in general, only a small group of genes in a given cell/tissue type utilize specific differences in their posttranscriptional regulation as a mean to contribute to the overall pattern of differential gene expression required to establish a unique cellular phenotype.
In summary, we have identified and characterized systematic variations in the regulatory sequences that control 3′-processing site selection at different stages of mouse spermatogenesis. These systematic differences in posttranscriptional processing provide a means of altering the regulation of large groups of genes simultaneously. Our results provide a basis for further investigations into the role of variable 3′-processing (and corresponding 3′-UTRs) in the process of spermatogenesis.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Acknowledgments
J.H.G., L.N.H., P.S., DL and J.M.B. were partially supported by NIH contracts NCRR INBRE Maine 2 P20 RR16463, NIGMS GM072706 and NICHD HD037102. The authors thank Natalya Klueva for sequencing, Jose-Luis Redondo for early studies, and Carol Bult and Mary Ann Handel for critical review of the manuscript. Funding to pay the Open Access publication charges for this article was provided by contract NIGMS GM072706.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol. Biol. Rev. 1999;63:405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmonds M. A history of poly a sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 3.Keller W., Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell. Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 4.Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 5.Salisbury J., Hutchison K.W., Graber J.H. A multispecies comparison of the metazoan 3′-processing downstream elements and the CstF-64 RNA recognition motif. BMC Genomics. 2006;7(55):1471–2164. doi: 10.1186/1471-2164-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J., Lutz C.S., Wilusz J., Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proudfoot N.J., Brownlee G.G. 3′ Non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald C.C., Redondo J.L. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 9.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDevitt M.A., Hart R.P., Wong W.W., Nevins J.R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil A., Proudfoot N.J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald C.C., Wilusz J., Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagaki Y., Manley J.L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer K., Dandekar T., Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem. 1997;272:26769–36779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 15.Legendre M., Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkataraman K., Brown K.M., Gilmartin G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F., Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arhin G.K., Boots M., Bagga P.S., Milcarek C., Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonaha M., Proudfoot N.J. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Lee J.Y., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwalds-Gilbert G., Veraldi K.L., Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaudoing E., Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautheret D., Poirot O., Lopez F., Audic S., Claverie J.M. Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res. 1998;8:524–30. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- 25.Iseli C., Stevenson B.J., de Souza S.J., Samaia H.B., Camargo A.A., Buetow K.H., Strausberg R.L., Simpson A.J., Bucher P., Jongeneel C.V. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veraldi K.L., Arhin G.K., Martincic K., Chung-Ganster L.H., Wilusz J., Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse b cells. Mol. Cell Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J., Marr T.G. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakheet T., Williams B.R., Khabar K.S. Ared 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conne B., Stutz A., Vassalli J.D. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nature Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 31.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloc M., Zearfoss N.R., Etkin L.D. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 33.Kuersten S., Goodwin E.B. The power of the 3′ UTR: translational control and development. Nature Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 34.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 35.Mazumder B., Seshadri V., Fox P.L. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 36.Mendez R., Richter J.D. Translational control by CPEB: a means to the end. Nature Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 37.Pesole G., Liuni S., Grillo G., Licciulli F., Mignone F., Gissi C., Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickens M., Anderson P., Jackson R.J. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 40.Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 41.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 42.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRS by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy E.M. Male germ cell gene expression. Recent Prog. Horm. Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 44.Walker W.H., Delfino F.J., Habener J.F. RNA processing and the control of spermatogenesis. Front Horm. Res. 1999;25:34–58. doi: 10.1159/000060996. [DOI] [PubMed] [Google Scholar]
- 45.Venables J.P. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 2002;12:615–619. doi: 10.1016/s0959-437x(02)00347-7. [DOI] [PubMed] [Google Scholar]
- 46.Meijer D., Hermans A., von Lindern M., van Agthoven T., de Klein A., Mackenbach P., Grootegoed A., Talarico D., Della Valle G., Grosveld G. Molecular characterization of the testis specific c-abl mRNA in mouse. EMBO J. 1987;6:4041–4048. doi: 10.1002/j.1460-2075.1987.tb02749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millan J.L., Driscoll C.E., LeVan K.M., Goldberg E. Epitopes of human testis-specific lactate dehydrogenase deduced from a cDNA sequence. Proc. Natl Acad. Sci. USA. 1987;84:5311–5315. doi: 10.1073/pnas.84.15.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oppi C., Shore S.K., Reddy E.P. Nucleotide sequence of testis-derived c-abl cDNAs: implications for testis-specific transcription and abl oncogene activation. Proc. Natl Acad. Sci. USA. 1987;84:8200–8204. doi: 10.1073/pnas.84.23.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Øyen O., Myklebust F., Scott J.D., Cadd G.G., McKnight G.S., Hansson V., Jahnsen T. Subunits of cyclic adenosine 3′,5′-monophosphate-dependent protein kinase show differential and distinct expression patterns during germ cell differentiation: alternative polyadenylation in germ cells gives rise to unique smaller-sized mRNA species. Biol. Reprod. 1990;43:46–54. doi: 10.1095/biolreprod43.1.46. [DOI] [PubMed] [Google Scholar]
- 50.Wallace A.M., Dass B., Ravnik S.E., Tonk V., Jenkins N.A., Gilbert D.J., Copeland N.G., MacDonald C.C. Two distinct forms of the 64, 000 mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl Acad. Sci. USA. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dass B., McMahon K.W., Jenkins N.A., Gilbert D.J., Copeland N.G., MacDonald C.C. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J. Biol. Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- 52.Dass B., McDaniel L., Schultz R.A., Attaya E., MacDonald C.C. The gene CSTF2T, encoding the human variant CstF-64 polyadenylation protein τCstF-64, lacks introns and may be associated with male sterility. Genomics. 2002;80:509–514. [PubMed] [Google Scholar]
- 53.Lee Y.J., Lee Y., Chung J.H. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett. 2000;487:287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- 54.Schultz N., Hamra F., Garbers D. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl Acad Sci. USA. 2004;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarrey J.R., O'Brien D.A., Skinner M.K. Construction and preliminary characterization of a series of mouse and rat testis cDNA libraries. J. Androl. 1999;20:635–639. [PubMed] [Google Scholar]
- 56.Brockman J.M., Singh P., Liu D., Quinlan S., Salisbury J., Graber J.H. PACdb: PolyA cleavage site and 3′-UTR database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- 57.Kent W.J. Blat—the blast-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 59.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu D., Graber J.H. Quantitative comparison of EST libraries requires compensation for systematic biases in cDNA generation. BMC Bioinformatics. 2006;7:77. doi: 10.1186/1471-2105-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrence C.E., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C. Detecting subtle sequence signals: a gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 62.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee D.D., Seung H.S. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Proc. Natl. Acad. Sci. USA. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drabkin H.J., Hollenbeck C., Hill D.P., Blake J.A. Ontological visualization of protein-protein interactions. BMC Bioinformatics. 2005;6:29. doi: 10.1186/1471-2105-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology the gene ontology consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blake J.A., Eppig J.T., Bult C.J., Kadin J.A., Richardson J.E. The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res. 2006;34:D562–D567. doi: 10.1093/nar/gkj085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Audibert A., Simonelig M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc. Natl. Acad Sci. USA. 1998;95:14302–14307. doi: 10.1073/pnas.95.24.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan Z., Zhang H., Hague L.K., Lee J.Y., Lutz C.S., Tian B. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene. 2006;366:325–334. doi: 10.1016/j.gene.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 70.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 71.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 72.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tay J., Richter J.D. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev. Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 74.Lenhard B., Sandelin A., Mendoza L., Engstrom P., Jareborg N., Wasserman W.W. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu Z., Raabe T., Hecht N.B. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol. Reprod. 2005;73:427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 76.Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Richter J.D. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bagga P.S., Ford L.P., Chen F., Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3( end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu C., Alwine J.C. Secondary structure as a functional feature in the downstream region of mammalian polyadenylation signals. Mol. Cell Biol. 2004;24:2789–2796. doi: 10.1128/MCB.24.7.2789-2796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 82.Hirose Y., Manley J.L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–6. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 83.Spicher A., Guicherit O.M., Duret L., Aslanian A., Sanjines E.M., Denko N.C., Giaccia A.J., Blau H.M. Highly conserved RNA sequences that are sensors of environmental stress. Mol. Cell Biol. 1998;18:7371–7382. doi: 10.1128/mcb.18.12.7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gur Y., Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–416. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boguski M.S., Lowe T.M., Tolstoshev C.M. dbest—database for expressed sequence tags. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.