Abstract
The human thrombopoietin (THPO) gene displays a series of alternative splicing events that provide valuable models for studying splicing mechanisms. The THPO region spanning exon 1–4 presents both alternative splicing of exon 2 and partial intron 2 (IVS2) retention following the activation of a cryptic 3′ splice site 85 nt upstream of the authentic acceptor site. IVS2 is particularly rich in stretches of 3–5 guanosines (namely, G1–G10) and we have characterized the role of these elements in the processing of this intron. In vivo studies show that runs G7–G10 work in a combinatorial way to control the selection of the proper 3′ splice site. In particular, the G7 element behaves as the splicing hub of intron 2 and its interaction with hnRNP H1 is critical for the splicing process. Removal of hnRNP H1 by RNA interference promoted the usage of the cryptic 3′ splice site so providing functional evidence that this factor is involved in the selection of the authentic 3′ splice site of THPO IVS2.
INTRODUCTION
Several studies have shown that exonic and intronic sequences in addition to the well-conserved donor and acceptor sites may play a role in nuclear pre-mRNA splicing (1). One of these motifs consists in runs of Gs placed within introns immediately downstream of 5′ splice sites as well as upstream of 3′ splice sites (2–4). Functional studies have supported the hypothesis that small introns contain intronic cis-acting elements based on G runs that drive efficient selection of particular splice sites. The relevance of G-rich sequences in the control of splicing has been shown in many studies dealing with constitutive and alternative splicing both in physiological (5–8) and pathological (9–13) events. Investigations on the trans-acting factors potentially involved in splicing models carrying intronic G runs have identified at least one main factor, hnRNP H, as one of the proteins that bind to G-rich sequences (11,13,14) and can regulate splicing of different genes with variable effects (8,13). These studies have also proposed that mechanisms of splicing control by G runs can be further regulated by the interplay with different sequences and by the recruitment of other factors (8,13). Therefore, it is of interest to characterize the mechanism of action of G runs-dependent splicing modulation in different models in order to establish if they function through the same biochemical language and their possible link with physiological or pathological events.
Human thrombopoietin (THPO) stimulates the proliferation and maturation of megakaryocytes and controls the production of platelets (15). The gene coding for this cytokine maps to chromosome 3q27–28 and displays a complex pattern of alternative splicing (16–18). Our molecular studies on the expression of THPO gene have identified an alternative spliced isoform corresponding to a mRNA carrying an insertion of 85 nt generated by the activation of a cryptic 3′ splice site within intron 2. We have shown that the 3′ end of intron 2 contains G-rich elements that work coordinatively mainly for the selection of the authentic 3′ splice site of IVS2. In this work, the heterogeneous ribonucleoprotein H1 (hnRNP H1) has been identified as the trans-acting factor that interacts specifically with G runs in the THPO context. In particular, siRNA-mediated down regulation of hnRNP H indicates that this factor is important to secure the constitutive splicing of intron 2.
MATERIALS AND METHODS
Constructs
The THPO genomic region spanning from exon 1 to exon 4 was amplified from human genomic DNA in two rounds to shorten intron 1. In the first round, two overlapping PCR fragments were generated. Fragment A was amplified using primer sense Ex1-EcoRV (5′-attggatatctgcagaattcgccctttta-3′) and primer antisense (5′-cagggtgaacagatgcattctgggaaggctacactcttctcgaca-3′), whereas fragment B was amplified using primer IVS1-s (5′-tgtcgagaagagtgtagccttcccagaatgcatctgttcaccctg-3′) and Ex4–XhoI (5′-acgtctcgagcatctgggttttccattctc-3′). In the second round, fragment A and B were used as templates in the same PCR with the external oligos Ex1-EcoRV and Ex4–XhoI in order to generate the THPO minigene. The PCR product was EcoRV-XhoI cloned in pcDNA expression vector (Invitrogen) so generating the construct IVS2-wt (Figure 1c). Subsequently, the constructs carrying disruption of single or multiple G runs were obtained by PCR site directed mutagenesis. All constructs were verified by sequence analysis using the CEQ 2000 sequencer machine (Beckman Coulter) to exclude the presence of mutations.
Cell culture and transfections
Human cell lines (HEK-293T, Hep3B and HeLa) were cultured in Dulbecco's modified Eagle's medium with glutamax (Invitrogen) supplemented with 10% fetal bovine serum and gentamicin (100 μg/ml). The DNA used for transfections was purified with JetStar columns (Genomed). In HEK293 cells, 2 μg of each constructs were transfected with the calcium phosphate coprecipitation method (19). The total RNA was extracted using RNAwiz solution (Ambion) and retrotranscribed with poly(dT) primer. To amplify only the messengers derived from the transfections, RT–PCR were carried out using as sense primer, T7 promoter (annealing to the plasmid) and the antisense primer specific for THPO, Ex4–XhoI. The conditions used for the PCRs were the following: 94°C for 3 min for the initial denaturation, 94°C for 30 s, 54°C for 30 s, 72°C for 60 s for 35 cycles and 72°C for 7 min for the final extension. The PCRs were optimized to be in the exponential phase of amplification. The results of all the transfections are the representative of at least three independent experiments. The variability among different experiments was always <20%.
UV-Cross-linking of proteins and RNA
THPO G789 and THPO G789m plasmids were generated by annealing the oligos THPO G789s (5′-ctgaggggtggattccctgggtttcaggtctgggtccta-3′) and THPO G789as (5′-catggaggacccagacctgaaacccagggaatccacccctcattcga-3′) for the wild type construct as well as of THPO G789mut-s (5′-ctgacgtgtggattccctgagtttcaggtctgagtccta-3′) and THPO G789mut-as (5′-catggtaggactcagacctgaaactcagggaatccacacgtcagttcga-3′) for the mutant construct, followed by direct cloning under the T7 promoter control into KpnI-HindIII digested pBluescript SK. Plasmids to be transcribed into RNA were first linearized by HindIII digestion. The UV-cross-linking assay was performed by incubation of the [α−32P]UTP-labeled RNA probes with 60 μg of HeLa nuclear extracts in 20 μl of final volume at 30°C for 15 min. Final binding conditions were 20 mM HEPES, pH 7.9, 72 mM KCl, 1.5 mM MgCl2, 0.78 mM magnesium acetate, 0.52 mM DTT, 3.8% glycerol, 0.75 mM ATP, 1 mM GTP and Heparin at a 5 μg/μl final concentration as a nonspecific competitor. In the competition experiments increasing amounts of cold RNA (the molar ratios of cold/labeled RNA were 3 and 6) was also added as a competitor 5 min before addition of the labeled RNAs. The samples were then transferred in the wells of an HLA plate (Nunc) and irradiated with UV light on ice (800 000 kJ for 5 min). Unbound RNA was then digested with 30 μg of RNase A (Sigma) by incubation at 37°C for 30 min. The samples were then analyzed by 10% SDS-polyacrylamide gel electrophoresis followed by autoradiography.
RNA oligos (Proligo) were used to test the binding ability of G7 run to hnRNP H1. 200 ng of G7wt (5′-ucugagggguggau-3′) and G7pyr (5′-ucucugggguccau-3′) G7m (5′-ucugacguguggau-3′) RNAs were labeled by phosphorylation with [γ−32P]ATP and T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C and 10 ng of each oligo were used for UV- cross-linking. The gels were dried and exposed to X-OMAT AR films for 1–3 h.
Affinity purification of G789 runs-binding proteins
One nmole (∼8 μg) of cold THPO G789 or G789m RNA was placed in a 400 μl reaction mixture containing fresh 0.1 M Sodium Acetate (NaOAc) pH 5.0 and 5 mM sodium m-periodate (Sigma). Reaction mixtures were incubated for 1 h in the dark at RT. The RNA was ethanol precipitated and resuspended in 500 μl of 0.1 M NaOAc, pH 5.0. Then, 400 μl of adipic acid dehydrazide agarose bead 50% slurry (Sigma) were washed four times in 10 ml of 0.1 M NaOAc pH 5.0 and pelleted after each wash at 3000 r.p.m. for 3 min in a Eppendorf minifuge. After the final wash, 300 μl of 0.1 M NaOAc pH 5 were added to the beads. The slurry was then divided in two aliquots that were mixed with each periodate-treated RNA sample and incubated over night at 4°C on a rotator. The beads with the bound RNA were then pelleted and washed three times in 2 ml of 2 M NaCl and three times in 3 ml of RNA wash buffer (52 mM HEPES-KOH,pH 7.5, 10 mM MgCl2, 8 mM Mg(CH3COO)2, 5.2 mM DTT, 38% v/v glycerol). They were incubated in 1x RNA binding buffer (i.e. RNA wash buffer added with 7.5 mM ATP, 10 mM GTP and 5 mg/ml Heparin) with 0.3 mg of HeLa cell nuclear extract for 30 min at RT in 500 μl final volume, pelleted by centrifugation at 1000 r.p.m. for 3 min and washed four times with 1.5 ml of RNA wash buffer. After the final centrifugation 60 μl of SDS–PAGE sample buffer were added to the beads and heated for 5 min at 90°C before loading on a 10% SDS–PAGE gel. Internal sequence analysis from the Coomassie stained bands excised from the SDS–PAGE gel was performed using an electrospray ionization mass spectrometer (LCQ DECA XP-ThermoFinnigam). The bands were digested by trypsin and the resulting peptides were extracted with water and 60% acetonitrile/1% trifluoroacetic acid. The fragments were then analyzed by mass spectrometry and the proteins were identified by analysis of the peptide MS/MS data with Turbo SEQUEST (ThermoFinnigam) and MASCOT (Matrix Science).
Small interfering RNA (siRNA) transfections
siRNA transfections were performed in HeLa cells by using Oligofectamine Reagent (Invitrogen). One day before siRNA transfection cells were trypsinized to achieve 40–50% confluency. For a 35 mm-well plate, 2.5 μl of two different siRNA duplexes (10 μM, Ambion) were mixed with 175 μl of Opti-MEM medium and, separately, 5 μl Oligofectamine with 15 μl of Opti-MEM medium. The 19 nt-target sequences in hnRNP H1 were: 5′-GGAAATAGCTGAAAAGGCT-3′ and 5′-CCACGAAAGCTTATGGCCA-3′. The samples were incubated for 7 min at room temperature. Then the Oligofectamine mix was added to precomplexed siRNA mix and incubated for 20 min at room temperature. At the end of this incubation, the mix was dropped onto the cells maintained in 800 μl of Opti-MEM without antibiotics and FCS. After 24 h, a second siRNA transfection was performed as described above, followed by the transfection with the THPO minigenes. The following day, cells were harvested for RNA extractions, RT–PCR analyses and western blots. Each transfection experiment was repeated at least three times. The proportion of hnRNP H1 silencing was quantified by optical densitometry using Imagej 1.37 software (http://rsb.info.nih.gov/ij/).
RESULTS
G runs are involved in the control of THPO intron 2 3′ splice site selection and exon 2 definition
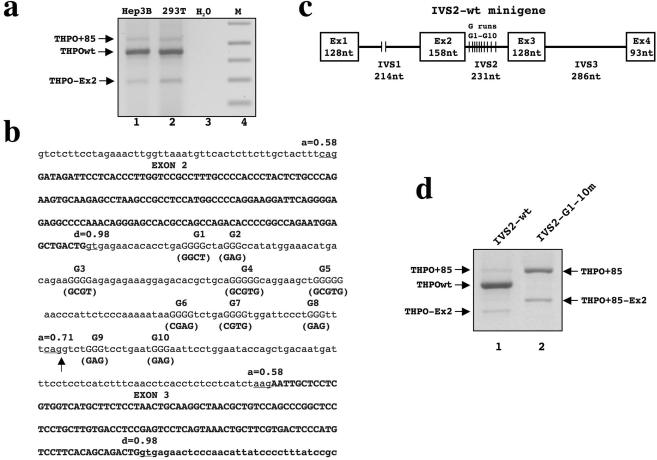
RT–PCR analysis of the region spanning exons 1–4 of the human THPO gene in cell lines that express this cytokine (human hepatocarcinoma Hep3B and human embryonic kidney HEK-293T) showed the presence of three PCR products (Figure 1a). Cloning and sequencing of these bands established that: (i) The band labeled THPO wt, by far the most abundant PCR product, corresponds to the full length cDNA encompassing exons 1–4; (ii) the shorter amplicon labeled THPO-Ex2 represents an alternative splicing isoform of THPO lacking exon 2; and (iii) the band labeled THPO + 85, the longer PCR product, is another splice variant corresponding to THPO cDNA carrying the inclusion of the 3′ end of intron 2. This isoform is the results of the selection of a cryptic acceptor site placed 85 nt upstream of the authentic 3′ splice site.
Figure 1.
Comparison of splicing pattern of endogenous THPO and IVS2-wt construct. (a) Analysis of the exon 1-exon 4 splicing pattern by RT–PCR of endogenous THPO gene constitutively expressed in Hep3B and HEK293 cell lines. THPOwt is the full length transcript (exon 1+2+3+4). THPO+85 is the full length transcript including 85 nt from IVS2. THPO-Ex2 is the mRNA excluding exon 2. H2O indicates the PCR control. M indicates the lane with 1 Kb plus DNA marker. (b) Nucleotide sequence encompassing intron 1-exon 2- intron 2-exon 3 of human THPO gene. Introns are shown in lower case, exons are in bold upper case. The score of acceptor (a=) and donor (d=) splice sites calculated using the Splice Site Prediction by Neural Network program (SSPNN, http://www.fruitfly.org/seq_tools/splice.html) are indicated. The ten G runs (in uppercase) within intron 2 are numbered 1–10 and the mutagenesis carried out for each G runs is shown in brackets. An arrow indicates the cryptic 3′ splice site within IVS2. (c) Scheme of the human THPO minigene used for transfection experiments. White boxes correspond to the human THPO exons (Ex) 1–4 and thin lines represent introns (IVS). Sizes of exons and introns are shown. (d) Analysis of pre-mRNA splicing of IVS2-wt and IVS2-G1-G10m constructs. Amplicons were separated on a 1.3% (w/v) agarose gel.
A search in human expressed sequence tag databases confirmed the presence of +85 THPO mRNA species (Genebank accession number AB102892). We tried to correlate these findings with calculations of the strength of potential splice sites of exon 2 and 3 in human THPO gene. The analysis carried out according to the Splice Site Prediction by Neural Network program (SSPNN, http://www.fruitfly.org/seq_tools/splice.html) shows that the authentic acceptor splice site of IVS2 is weak and its score (0.58) is even lower than that of the potential acceptor site of IVS2 (0.71) corresponding to the cryptic 3′ splice site activated in the THPO + 85 splice variant (Figure 1b, arrow). Therefore, the splicing behavior of exon 2 and 3 is not consistent with the in silico analysis. It is therefore plausible that accessory cis-acting elements might be involved in an accurate selection of the authentic acceptor sites of intron 1 and 2. We have observed that the human THPO intron 2 contains a high number of guanosine motifs of lengths ranging from three to five (Figure 1b, G1–G10). In order to understand whether the guanosine stretches within this intron play a role in the control of splicing, we developed an in vivo minigene system by cloning the genomic region of the human THPO gene spanning exon 1 to exon 4. (Figure 1c, IVS2-wt). We then constructed a second minigene where, through site directed mutagenesis, all the G runs were interrupted by point mutations (IVS-2-G1-10m). Following transfection and expression of the constructs in HEK293 cells, RT–PCR analysis was performed to determine the splicing patterns produced by these constructs. Specific primers were used for the minigene sequence to differentiate its product from one derived from the endogenous gene. IVS2-wt construct predominantly expresses the THPO wt transcript together with two additional less abundant mRNAs (Figure 1d, lane 1). Direct sequencing of these purified PCR products showed that they correspond to the THPO-Ex2 and THPO + 85 splice variants previously detected in vivo. As shown in (Figure 1a and d) the minigene reproduces the splicing pattern of the endogenous gene and hence contains all the necessary information for correct THPO splicing. The transfection of IVS2-G1-10m minigene strongly impairs the pre-mRNA processing and changed the relative proportion of the THPO splicing variants (Figure 1d, lane 2). The THPO wt isoform was no longer present and the predominant PCR products were shown by direct sequencing to correspond to the THPO+85 splice variant. Additionally, a novel splicing isoform containing the 85 nt inclusion and excluding totally exon 2 was also detected (THPO+85-Ex 2 variant). Therefore, it can be concluded that the guanosine rich elements within intron 2 can control both the selection of its 3′ splice site and exon 2 definition.
Mapping of the G motifs-splicing control elements within THPO intron 2
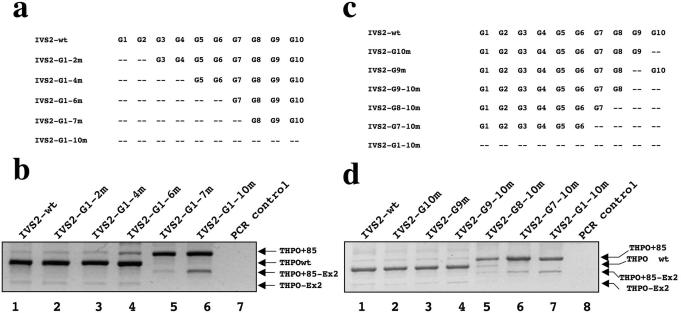
To determine the contribution of different G runs in the control of intron 2 splicing, we generated a series of IVS2-wt minigene mutants where G runs were progressively disrupted by point mutation starting from the 5′ end of intron 2 (Figure 2a). After transfection in HEK293 cells, we observed that the THPO+85 splice variant was present in a low but increasing amount, following the disruption of the first six G runs (Figure 2b, lanes 1–4). The strongest effect was observed when we extended the mutagenesis to the G7 run (Figure 2b, lane 5), which resulted in a similar splicing pattern to that obtained with the minigene carrying all G runs mutated (Figure 2b, lane 6) Thus, the G7 run represents a critical element for the control of intron 2 splicing.
Figure 2.
Mapping of the G runs splicing control elements within THPO IVS2. (a) and (c) Schemes of the constructs used for transient transfections. The presence of G runs is indicated by its position-number. (b) and (d) Splicing pattern analyses of the RT–PCR products derived from cellular RNA, separated on a 1.3% agarose gel and stained by ethidium bromide. THPOwt is the full length transcript (exon 1+2+3+4). THPO+85 is the full length transcript including 85 nt from IVS2. THPO-Ex2 is the mRNA excluding exon 2. THPO+85-Ex2 indicates the mRNA species carrying the 85 nt inclusion along with exon 2 skipping.
Other five variants of the IVS2 construct were generated where G runs were disrupted by point mutation starting from the 3′ end of intron 2 (Figure 2c). The THPO+85 splicing isoform was predominant over the wild type species only when G8+G9+G10 elements were mutated at one time (Figure 2d, lane 5). Moreover, a progressively increasing amount of THPO+85-Ex2 isoform was also evident following transfections of IVS2-G8-10m, IVS2-G7-10m and IVS2-G1-10m constructs (Figure 2d, lanes 5–7, respectively). These results imply that multiple G runs must be present to support the correct intron 2 removal and an efficient exon 2 splicing.
Coordinate action of different G runs influences the selection of the 3′ splice site within IVS2
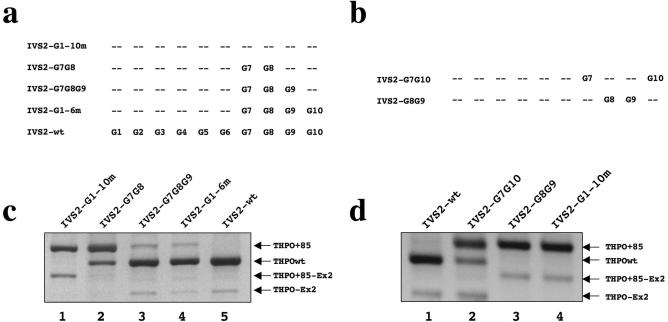
The contribution of the different G runs in intron 2 splicing was then examined by generating a series of minigenes, in which through site directed mutagenesis we reintroduced multiple G elements (G7-G8, G7-G8-G9, G7-G8-G9-G10, G7-G10, G8-G9, Figure 3a and b). It is of interest to note that multiple G runs such as G7-G8, G7-G8-G9 and G7-G8-G9-G10 elements caused a significant and progressive increase in the selection of the authentic 3′ acceptor site as compared with the effects of IVS2-G1-10m transfections (Figure 3c, see lanes 2 to 4). On the other hand, the minigene carrying only G8 and G9 was not sufficient to shift splicing towards the authentic 3′ acceptor site whereas the construct with only G7 and G10 was able to rescue the wild type 3′ splice site selection just partially (Figure 3d). These results suggest that G runs in the range G7-G10 work coordinatively to influence the splicing of intron 2. The presence of at least G7,G8 and G9 is therefore necessary for the recognition of the proper 3′ splice site of THPO intron 2 and in the efficient inclusion of exon 2.
Figure 3.
Effects of combined G7, G8 and G9 runs on THPO splicing. (a) and (b) Schemes of the constructs used for transient transfections. The presence of G runs is indicated by its position-number. (c) and (d) Splicing pattern analyses of the RT–PCR products derived from cellular RNA, separated on a 1.3% agarose gel and stained by ethidium bromide.
Role of G7 run in the 3′ splice site selection within intron 2
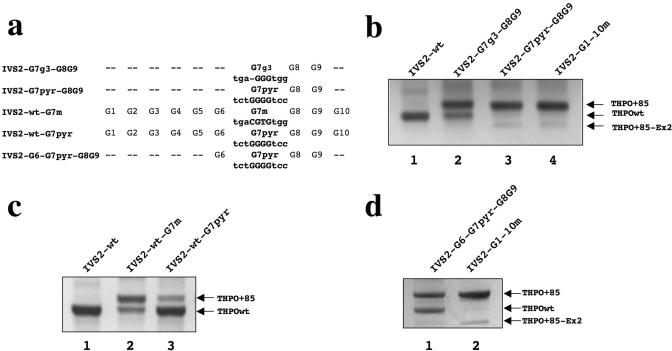
Taking into consideration the apparently central role of the G7 run in intron 2 splicing, we were interested to test whether the nucleotides flanking that G stretch could also be involved in the modulation of the IVS2 splicing. The G7 run is made of four Gs and surrounded by purine rich nucleotide sequences, as opposed to G8 and G9 runs, that contain three Gs and are flanked by pyrimidine rich sequences. This prompted us to construct a first mutant where using the IVS2-G7-G8-G9 template, G7 run was reduced from four to three Gs (IVS2-G7g3-G8G9) and a second variant where the purines immediately adjacent to G7 were replaced by pyrimidines (IVS2-G7pyr-G8G9, Figure 4a). We found that the deletion of one G in the G7 run strongly increased the THPO+85 isoform (Figure 4b, lane 2) and the transfection of the IVS2-G7pyr-G8G9 minigene shifted the splicing pattern toward the complete selection of the isoform that retain the last 85 nt of the intron 2 (Figure 4b, lane 3). Overall, these experiments indicate that the number of guanosines within G7 as well as the purines immediately adjacent to this G run are critical for the correct processing of this intron.
Figure 4.
Role of G7 run and its flanked nucleotides in the 3′ splice site selection within IVS2. (a) Scheme of the constructs used for transient transfections. (b) RT–PCR of the constructs carrying G7-G8-G9 carrying the reduction from 4 to 3 G in the G7 element and with the flanked nucleotides mutated are indicated with G7g3 and G7pyr, respectively. (c) RT–PCR products derived from the constructs where the G7 was disrupted at the level of the G-core (IVS-wt-G7m) or in its flanking nucleotides (IVSwt-G7pyr). (d) The construct IVS2-G6-G7pyr-G8G9 shows a significant rescue of the THPOwt isoform (lane 1).
We then confirmed the relevance of G7 for IVS2 splicing also when the other nine G runs are intact. In fact, we generated and transfected two mutants of IVS2-wt construct where the G7 was disrupted at the level of the G-core (IVS2-wt-G7m, Figure 4a) or of its flanking nucleotides (IVS2-wt-G7pyr, Figure 4a). The relevance of the G-core was confirmed since its mutation resulted in the expression of equal amounts of the THPO+85 as well as of the THPOwt variants (Figure 4c, lane 2). Surprisingly, the mutation of the nucleotides flanking G7 did not produce a dramatic change in the IVS2 splicing pattern (Figure 4c, lane 3). The latter result suggested that the nucleotides adjacent to G7 can modulate the 3′ splice site selection within IVS2 only when the surrounding G runs context is weakened. In fact, the reintroduction of an intact G6 run in the IVS2-G7pyr-G8G9 template (IVS2-G6-G7pyr-G8G9, Figure 4a) promoted the significant rescue of the THPOwt isoform (Figure 4d, lane 1).
hnRNP H1 binds the G runs critical for THPO intron 2 splicing
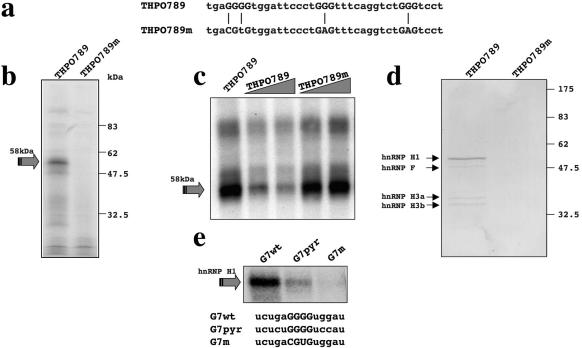
In order to identify the trans-acting factors able to bind the G runs in the THPO context, UV-cross-linking assays were performed with in vitro transcribed RNAs containing either the G7-G8-G9 region of the THPO intron 2 (Figure 5a, THPO789 construct) or the same sequence carrying point mutations within the G runs (Figure 5a, THPO789m construct). Figure 5b shows that a 58 kDa protein interacts specifically with THPO789 RNA and was no longer present when THPO789m RNA was used. Competition experiments then confirmed the binding specificity. As expected, the 58 kDa protein was rapidly competed away from labeled THPO789 RNA sequence by the addition of an increasing amount of cold THPO789 RNA but not by the addition of an increasing amount of cold THPO789m RNA (Figure 5c). Next, we used an RNA-affinity purification procedure followed by mass spectrometry sequencing to identify the G runs-binding proteins. After incubation of RNA-beads preparations with HeLa nuclear extract, the RNA protein complexes formed were pulled down and the proteins present in the complexes fractionated on an SDS–PAGE gel and then visualized by Coomassie Blue staining (Figure 5d). The THPO789 RNA specifically pulled down a 58 kDa protein, whose molecular weight was similar to that of the band observed in UV-cross-linking assays. Moreover, three less abundant proteins of apparent 53 kDa, 38 kDa and 34 kDa molecular weight were also specifically pulled down. Internal sequence analysis by mass spectrometry of the excised 58 kDa band, yielded seven peptides whose sequences belong to hnRNP H1. The less abundant proteins were also sequenced and were found to correspond to hnRNP F (53 kDa) and to two hnRNP H3 isoforms (38 kDa and 34 kDa). These results are consistent with previous studies showing that members of the hnRNP H family have binding specificity for RNA sequences enriched in guanosine (20).
Figure 5.
Interaction of nuclear proteins with G runs within THPO IVS2. (a) Sequence of oligos KpnI/Hind III cloned in pBS KS under T7 promoter control for in vitro transcription. G7, G8 and G9 runs are in uppercase and the position of G mutagenized by point mutations are indicated with a vertical line. (b) UV-cross-linking assay using HeLa nuclear extract with in vitro transcribed THPO789 and THPO789m RNAs. The arrow shows the position of the 58 kDa complex. (c) Competition analysis confirms the binding specificity of the 58 kDa complex following addition of cold THPO789 and THPO789m RNAs to labeled THPO789 RNA in the presence of HeLa nuclear extract. The molar ratio of cold/labeled RNA was three and six. The arrows indicate the 58 kDa complex. (d) Coomassie stained SDS–PAGE of a pull-down assay using adipic dehydrazide beads derivatized with the RNAs following incubation with HeLa nuclear extract. In the lane from the wild type RNA-derivatized beads (THPO789), the arrows indicate the proteins that are present only in the wild type THPO789 RNA-derivatized beads. (e) UV-cross-linking assay using HeLa nuclear extract with RNA oligos spanning only wild type G7 run (G7wt), G7 with its flanking nucleotides mutagenized (G7pyr) and of G7 with G-core disrupted (G7m). The arrow shows the position of the hnRNP H1 proteins.
In consideration of the peculiar functional role of the G7 run, we then asked which is the specific contribution of this G run to the interaction with hnRNP H1. We evaluated the ability of RNA oligos spanning only G7 run to bind hnRNP H1. Therefore, we compared the binding capacity of G7 wild type (G7wt) sequence to those of G7 with G-core disrupted (G7m) and of G7 with its flanking purines mutagenized to pyrimidines (G7pyr). We found a consistent reduction in hnRNP H1-binding with G7pyr RNA, whereas it is completely abolished with G7m oligo (Figure 5e). Thus, there is a good correlation between the ability of G7 element to bind hnRNP H1 and the in vivo effects of its mutagenesis.
RNAi against hnRNP H promotes activation of the cryptic acceptor site within THPO intron 2
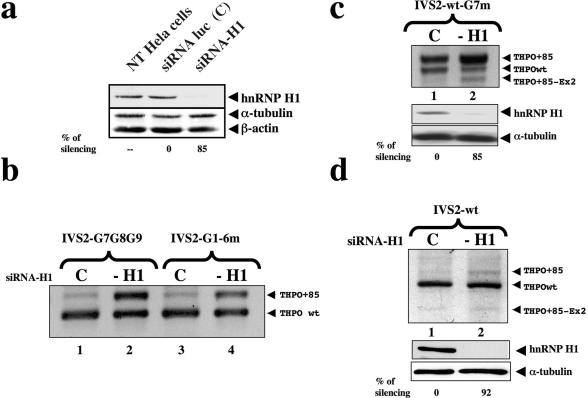
To obtain more direct evidence for the role of hnRNP H in splicing of human THPO mRNA, we performed siRNA experiments against endogenous hnRNP H1 protein. HeLa cells were used because they were transfected more efficiently with siRNA. The THPO constructs IVS2 G7G8G9 and IVS2-G1-6m were co-transfected after the last round of siRNA. Cell lysates were collected and analyzed for levels of endogenous hnRNP H1 by western blot analysis using anti-H1 rabbit serum. We observed a strong reduction in the hnRNP H expression in cells transfected with hnRNP H1-siRNA but not in untransfected cells or in cells treated with an unrelated siRNA. The expression level of two housekeeping genes (β-actin and α-tubulin) was unaffected (Figure 6a). To determine whether the knock-down of hnRNP H1 has a function in the splicing of the second intron of THPO gene, RT–PCR assays for THPO mRNA obtained from the constructs were carried out. SiRNA-mediated down regulation of hnRNP H promoted a strong increase of the THPO+85 transcript when the constructs IVS2 G7G8G9 and IVS2-G1-6m were used (Figure 6b). These results indicate that the reduction in the expression of cellular hnRNP H1 promotes a significant but not yet complete usage of the −85 nt cryptic acceptor site of THPO intron 2. To confirm the critical role of G7 element also in the IVS2 wild type context, RNAi experiments against hnRNP H1 were repeated with cotransfection of the IVS2-wt-G7m minigene. The THPO+85 isoform became predominant following transient knock-down of hnRNP H1. This experiment confirmed the key role of the interaction of hnRNP H1 to G runs probably primed by the G7 element.
Figure 6.
Knock-down of hnRNPH1 causes the activation of the cryptic acceptor site within IVS2. (a) Western blot carried out with polyclonal anti-hnRNP H1 antiserum on HeLa cell non transfected (NT HeLa cells) or transfected with siRNA against hnRNP H1 (siRNA-H1) or luciferase (siRNA-luc). β-tubulin and α-actin were probed with polyclonal antibodies as a control of total protein loading. Relative amounts of hnRNP H1 silencing are indicated below each lane. Standard deviations were always ≤20%. (b) The panel shows splicing pattern analysis of RT–PCR products after cotransfection of IVS2 G7G8G9 and IVS2-G1-6m along with either the siRNA-H1 (lanes 2 and 4) or a control siRNA-luc oligonucleotides (lanes 1 and 3). (c) RT–PCR experiments after RNAi against hnRNP H1 and cotransfection with the IVS2-wt-G7m minigene. Lane 1 shows the cotransfection of IVS-wt-G7m with a control siRNA. After siRNA anti-hnRNP H the THPO+85 isoform became predominant (lane 2). Lower panels show the related anti-hnRNP H1 western blot analyses. β-tubulin was used as a control of total protein loading. Relative amount of hnRNP H1 silencing is indicated below each lane. Standard deviation were ≤20%. (d) RT–PCR experiments in siRNA-H1-treated in HeLa cells. Lanes 1 and 2 show the results of PCR after cotransfecting the IVS2-wt minigene. After the reduction in the expression of hnRNP H there is a small increase of the THPO+85 isoform. The control transfection was loaded in lane 1. Lower panels show the related anti-hnRNP H1 western blot analyses with indication of relative amounts of hnRNP H1 silencing. Standard deviations were ≤20%.
Transfections of the IVS2-wt minigene in siRNA-H1-treated HeLa cells and RT–PCR assays specific for the plasmid confirmed the increment of the THPO+85 transcript, although to a lesser extent in comparison with that obtained with IVS2 G7G8G9 and IVS2-G1-6m minigenes (Figure 6d, compare lanes 1 and 2). Altogether these results indicate that G7-G8-G9-G10 runs represent the core elements whose interaction with hnRNP H1 control the selection of the authentic acceptor splice site of THPO IVS2.
DISCUSSION
In this work, we have characterized the molecular mechanisms underlying the alternative splicing of THPO intron 2 that arises from the activation of a cryptic 3′ splice site 85 nt upstream of the authentic 3′ splice site. Our interest was aimed at identifying possible cis-acting elements in THPO intron 2 able to influence its processing. We focused on the peculiar G run cluster present in this short intron as intronic G runs have been shown to be involved in modulating splicing of several genes in different species (7,11,12,21,22). Our results overlap only partially with previous studies that demonstrated how G runs can enforce weak 3′ splice sites. In fact, the mechanism by which G runs control the splicing and the processing efficiency of THPO IVS2 seems to be different from that found in other models. In α-globin intron 2, G triplets have been shown to act additively both to enhance splicing and to facilitate exon-intron border recognition (7). Consistently with this model, in IVSB7 of chicken β-tropomyosin, six (A/UGGG) repeats had an additive effect and were essential for spliceosome assembly (5). In the apoptotic Bcl-x gene, a cis-acting element with two G runs was shown to enhance the 5′ splice site of Bcl-xs (13). Conversely, in the CI cassette exon (exon 19) of the glutamate NMDA R1 receptor, a 5′ splice site proximal GGGG motif was shown to work in combination and cooperatively with a different element (an exonic UAGG sequence) to silence CI exon (8). Finally, in the 844ins68 allele of the CBS gene there is a duplication of the 3′ splice site at the CBS intron7/exon8 junction and G runs in between the two identical splice sites were shown to promote the selection of the distal one (11). In the THPO gene, in vivo experiments show that intronic G motifs affect splicing through a complex combinatorial mechanism derived by different actions of the G7, G8, G9 and G10 motifs. Splice site recognition depends on coordinate action of different cis-acting elements and our in vivo results show that G runs in the G7-G10 range are not simply a redundant back-up system for splicing fidelity.
In silico and in vivo results suggest that there is a competition among the two ‘weak’ authentic 3′ splice sites of exon 2-exon 3 and the ‘strong’ cryptic 3′ splice site within intron 2. In particular, the observation that G8+G9 runs alone are not able to inhibit the cryptic 3′ splice site is compatible with an enhancing role of these elements of the authentic 3′ splice site. On the other hand, the data showing that an increasing number of G runs in IVS2 is associated with a higher proportion of wild type transcripts lets us speculate that the higher number of G runs might stabilize the interaction with hnRNP H1 and consequently block the access to the cryptic 3′ splice site by steric hindrance.
The G7 run is the splicing hub of intron 2: its presence is determinant to secure the constitutive processing of intron 2 as demonstrated by the results of siRNA-H1 assays with IVS2-wt-G7m and by the fact that the control the 3′ splice site selection is determined by the G-core as well as the flanking nucleotides of this element. This latter observation represents an important corollary of these data because it demonstrates that the effects of G runs on splicing can be further modulated by the nucleotides surrounding the G cores. Investigations into the proteins interacting with G runs in THPO IVS2 context have shown that RNA containing G7-G8-G9 motifs bind mainly hnRNP H1, and other members of hnRNP H family (hnRNP F and hnRNP H3), but to a lesser extent. The co-purification by RNA-affinity of hnRNP F together with hnRNP H1 is consistent with previous work showing that both hnRNP H and hnRNP F bind to poly(G) sequences (23) and can form heterodimers (24). Other studies have reported the involvement of hnRNP H1 in the splicing of different genes with silencing or enhancing effects depending on its interaction with exonic (11,25–28) or intronic (24) sequences, respectively. Apparently, the G7 run might have more affinity for hnRNP H1 because it is the only element (among the last four) that contains four Gs instead of three. Therefore, it is possible that the interaction of hnRNP H1 with the G7 run might strengthen in vivo the interaction of this trans-acting factor onto the downstream G elements. These data support the hypothesis that the stable interaction of hnRNP H1 with the last four G runs favors the constitutive splicing of intron 2.
On the other hand, the finding that the IVS2-wt-G7m construct is still responsive to siRNA-H1 assays whereas the IVS2-wt construct is less sensitive gives a hint onto the possible involvement of further elements in the THPO IVS2 splicing. From our experiments, it is apparent that accessory components are involved in the modulation of THPO IVS2. Intriguingly, intronic G runs within THPO IVS2 might play a structural role, e.g. as previously shown for G quartets in fragile X retardation (29). Alternatively, they might enroll additional proteins such as other members of the hnRNP H family that could exhibit redundant activity with hnRNP H1. The recruitment of these factors might be favored by the RNAi-mediated depletion of hnRNP H1 and seems to be effective only in the presence of intact G runs. In fact, it is sufficient for the mutagenization of G7 to weaken the G-runs-framework and to make inefficient the complementation of hnRNP H1. This finding is consistent with the ability of G runs to work cooperatively (7) similarly to what already documented for hnRNP A1 (30–32). Therefore, we have demonstrated that the G7 run has a pivotal role over the last four G runs for the control of IVS2 splicing through interaction with hnRNP H1, but more complex mechanisms may be involved. Future studies will be required to establish whether accessory elements are involved.
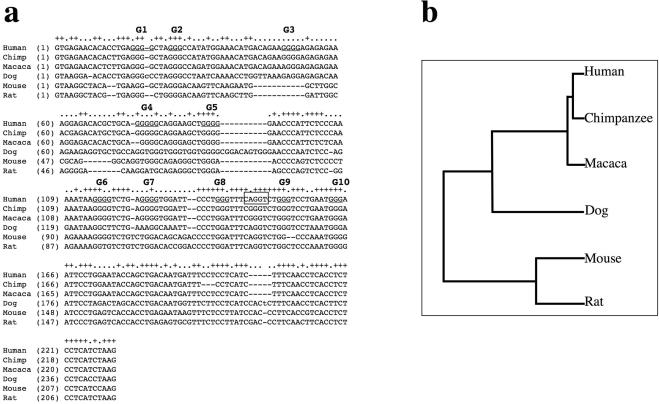
Finally, we should consider what could be the reason for such a complex control of the alternative splicing of both THPO intron 2 and exon 2 other than a bad evolutionary ‘joke’. Several studies have proven the relevance of alternative splicing in the regulation of translational efficiency of different protein involved both in physiological (18,33–35) and pathological events (36–39). In particular, a recent report has shown that a mRNA isoform generated by alternative splicing affects the composition of the 5′ end of THPO and may be an additional control mechanism for its production (18). The insertion of 85 nt in the novel THPO+85 splicing variant should produce a shift in the reading frame after codon 4 with the generation of a THPO precursor with a completely subverted sequence of the final 52 amino acids and a premature stop codon after codon 56. Interestingly, a phylogenetic analysis of THPO IVS2 shows that, on one hand, the composition of −85 cryptic acceptor site and of its flanking nucleotides is substantially conserved in different species such as dog, mouse and rat but not in other primates. On the other hand, the distribution of G runs and, in particular, the presence of those corresponding to human G7, G8 and G9 is not likewise conserved (Figure 7). Intriguingly, the comparison of THPO IVS2 in primates outlines that the −85 cryptic acceptor site is present neither in Pan troglodytes or in Macaca mulatta because it is disrupted by a A->G substitution that creates a new G run. Therefore, if the alternative splicing of IVS2 controlled by G runs is involved in regulation of THPO expression, its modulation in human might occur in different way in comparison not only with distant but also with closely related species. In conclusion, the peculiar role of the IVS2 G runs in human might be part of the circuit that control the expression of the THPO protein.
Figure 7.
Evolutionary comparison of the splicing control elements found in THPO intron 2 in human, macaca, chimpanzee, dog, mouse and rat. (a) The alignment of intron 2 from different species was generated using the GeneBee method (http://www.genebee.msu.ru/genebee.html). All G runs in human intron 2 are underlined. The human cryptic 3′ splice site between G8 and G9 runs is boxed. (b) Graphical phylogenetic tree generated by the GeneBee server that depicts the distances among different species for THPO IVS2.
Acknowledgments
The authors wish to thank Dr Emanuele Buratti for helpful suggestions and comments, and Ms Anna Meneghello for technical support. This work was supported by grants from Telethon Onlus Foundation–Italy (n. GGP02453) and EU FPVI (LSHG-CT-2005-518238) to F.E.B. Funding to pay the Open Access publication charges for this article was provided by ICGEB Institutional funding.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Nussinov R. (A)GGG(A), (A)CCC(A) and other potential 3′ splice signals in primate nuclear pre-mRNA sequences. Biochim. Biophys. Acta. 1987;910:261–270. doi: 10.1016/0167-4781(87)90119-9. [DOI] [PubMed] [Google Scholar]
- 3.Nussinov R. Conserved quartets near 5′ intron junctions in primate nuclear pre-mRNA. J. Theor. Biol. 1988;133:73–84. doi: 10.1016/s0022-5193(88)80025-0. [DOI] [PubMed] [Google Scholar]
- 4.Nussinov R. Conserved signals around the 5′ splice sites in eukaryotic nuclear precursor mRNAs: G-runs are frequent in the introns and C in the exons near both 5′ and 3′ splice sites. J. Biomol. Struct. Dyn. 1989;6:985–1000. doi: 10.1080/07391102.1989.10506526. [DOI] [PubMed] [Google Scholar]
- 5.Sirand-Pugnet P., Durosay P., Brody E., Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlo T., Sterner D.A., Berget S.M. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. Rna. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 7.McCullough A.J., Berget S.M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogan J.D., Prince M.A., Lekhakula S., Bundey S., Futrakul A., McCarthy E.M., Phillips J.A., 3rd A novel mechanism of aberrant pre-mRNA splicing in humans. Hum. Mol. Genet. 1997;6:909–912. doi: 10.1093/hmg/6.6.909. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy E.M., Phillips J.A., 3rd Characterization of an intron splice enhancer that regulates alternative splicing of human GH pre-mRNA. Hum. Mol. Genet. 1998;7:1491–1496. doi: 10.1093/hmg/7.9.1491. [DOI] [PubMed] [Google Scholar]
- 11.Romano M., Marcucci R., Buratti E., Ayala Y.M., Sebastio G., Baralle F.E. Regulation of 3′ splice site selection in the 844ins68 polymorphism of the cystathionine b-synthase gene. J. Biol. Chem. 2002;12:12. doi: 10.1074/jbc.M208107200. [DOI] [PubMed] [Google Scholar]
- 12.Mine M., Brivet M., Touati G., Grabowski P., Abitbol M., Marsac C. Splicing error in E1alpha pyruvate dehydrogenase mRNA caused by novel intronic mutation responsible for lactic acidosis and mental retardation. J. Biol. Chem. 2003;278:11768–11772. doi: 10.1074/jbc.M211106200. [DOI] [PubMed] [Google Scholar]
- 13.Garneau D., Revil T., Fisette J.F., Chabot B. Heterogeneous Nuclear Ribonucleoprotein F/H Proteins Modulate the Alternative Splicing of the Apoptotic Mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 14.Grabowski P.J. A molecular code for splicing silencing: configurations of guanosine-rich motifs. Biochem. Soc. Trans. 2004;32:924–927. doi: 10.1042/BST0320924. [DOI] [PubMed] [Google Scholar]
- 15.Kaushansky K. Thrombopoietin: understanding and manipulating platelet production. Annu. Rev. Med. 1997;48:1–11. doi: 10.1146/annurev.med.48.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Gurney A.L., Kuang W.J., Xie M.H., Malloy B.E., Eaton D.L., de Sauvage F.J. Genomic structure, chromosomal localization, and conserved alternative splice forms of thrombopoietin. Blood. 1995;85:981–988. [PubMed] [Google Scholar]
- 17.Li B., Pan H., Winkelmann J.C., Dai W. Thrombopoietin and its alternatively spliced form are expressed in human amygdala and hippocampus. Blood. 1996;87:5382–5384. [PubMed] [Google Scholar]
- 18.Ghilardi N., Wiestner A., Skoda R.C. Thrombopoietin production is inhibited by a translational mechanism. Blood. 1998;92:4023–4030. [PubMed] [Google Scholar]
- 19.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Short Protocols in Molecular Biology. New York: Greene Publishing Associates; 1992. [Google Scholar]
- 20.Caputi M., Zahler A.M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 21.Expert-Bezancon A., Sureau A., Durosay P., Salesse R., Groeneveld H., Lecaer J.P., Marie J. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B. J. Biol. Chem. 2004;279:38249–38259. doi: 10.1074/jbc.M405377200. [DOI] [PubMed] [Google Scholar]
- 22.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad Sci. USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matunis M.J., Xing J., Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C.D., Kobayashi R., Helfman D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquenet S., Mereau A., Bilodeau P.S., Damier L., Stoltzfus C.M., Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 2001;276:40464–40475. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- 27.Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 28.Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 30.Damgaard C.K., Tange T.O., Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. Rna. 2002;8:1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoltzfus C.M., Madsen J.M. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J., Mayeda A., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura H., Washizu J., Nakamura N., Enomoto A., Yoshikai Y. Translational efficiency is up-regulated by alternative exon in murine IL-15 mRNA. J. Immunol. 1998;160:936–942. [PubMed] [Google Scholar]
- 34.Arora S., Chauhan S.S. Identification and characterization of a novel human cathepsin L splice variant. Gene. 2002;293:123–131. doi: 10.1016/s0378-1119(02)00700-x. [DOI] [PubMed] [Google Scholar]
- 35.Shalev A., Blair P.J., Hoffmann S.C., Hirshberg B., Peculis B.A., Harlan D.M. A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology. 2002;143:2541–2547. doi: 10.1210/endo.143.7.8920. [DOI] [PubMed] [Google Scholar]
- 36.Konig H., Ponta H., Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. Embo J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwicky R., Muntener K., Csucs G., Goldring M.B., Baici A. Exploring the role of 5′ alternative splicing and of the 3′-untranslated region of cathepsin B mRNA. Biol. Chem. 2003;384:1007–1018. doi: 10.1515/BC.2003.113. [DOI] [PubMed] [Google Scholar]
- 38.Minn A.H., Kayton M., Lorang D., Hoffmann S.C., Harlan D.M., Libutti S.K., Shalev A. Insulinomas and expression of an insulin splice variant. Lancet. 2004;363:363–367. doi: 10.1016/S0140-6736(04)15438-X. [DOI] [PubMed] [Google Scholar]
- 39.Boissel J.P., Zelenka M., Godtel-Armbrust U., Feuerstein T.J., Forstermann U. Transcription of different exons 1 of the human neuronal nitric oxide synthase gene is dynamically regulated in a cell- and stimulus-specific manner. Biol. Chem. 2003;384:351–362. doi: 10.1515/BC.2003.041. [DOI] [PubMed] [Google Scholar]