Abstract
Regulation of endogenous genes by DNA-binding polyamides requires effective nuclear localization. Previous work employing confocal microscopy to study uptake of fluorophore-labeled polyamides has demonstrated the difficulty of predicting a priori the nuclear uptake of a given polyamide. The data suggest that dye identity influences uptake sufficiently such that a dye-conjugate cannot be used as a proxy for unlabeled analogs. Polyamides capable of nuclear localization unaided by fluorescent dyes are desirable due to size and other limitations of fluorophores. Recently, a polyamide-fluorescein conjugate targeted to the hypoxia response element (HRE) was found to inhibit vascular endothelial growth factor (VEGF) expression in cultured HeLa cells. The current study uses inhibition of VEGF expression as a biological read-out for effective nuclear localization of HRE-targeted polyamides. We synthesized a focused library of non-fluorescent, HRE-targeted polyamides in which the C-terminus ‘tail’ has been systematically varied. Members of this library bind the HRE with affinities comparable or superior to that of the fluorescein-labeled analog. Although most library members demonstrate modest or no biological activity, two non-fluorescent polyamides are reported with activity rivaling that of the previously reported fluorescein-labeled polyamide. We also show evidence that promoter occupancy by HIF-1, the transcription factor that binds the HRE, is inhibited by HRE-targeted polyamides.
INTRODUCTION
Polyamides containing N-methylimidazole (Im) and N-methylpyrrole (Py) comprise a class of programmable DNA-binding ligands capable of binding to a broad repertoire of DNA sequences with affinities and specificities comparable to those of natural DNA-binding proteins (1,2). Sequence specificity is programmed by side-by-side pairings of the heterocyclic amino acids in the minor groove of DNA: Im/Py distinguishes G•C from C•G; Py/Py binds both A•T and T•A; and 3-chlorothiophene/N-methylpyrrole (Ct/Py) prefers T•A over A•T at the N-terminus position (3–5). The use of polyamides to modulate the expression of selected genes through interaction with transcriptional machinery could have applications in biology and human medicine (1,2). Regulation of endogenous genes by DNA-binding small molecules requires cellular uptake and nuclear localization (6–9), chromatin accessibility (10,11) and site-specific interactions with gene promoters sufficient to interfere with specific transcription factor–DNA interfaces (12–15).
The use of confocal microscopy to visualize subcellular localization of fluorophore-labeled molecules is a convenient method to study uptake and trafficking of polyamides in living cells (6–9). Nuclear localization of more than 100 hairpin polyamide-fluorophore conjugates in several human cell lines has been examined using this method (8,9). Py/Im content, number and location of positive and negative charges, presence of a β-alanine residue at the C-terminus, choice of fluorophore, linker composition and attachment point have been shown to influence nuclear localization of polyamide-fluorophore conjugates (8,9). Although a significant number of eight-ring hairpin-polyamides with fluorescein at the C-terminus were shown to have good nuclear uptake in 10 cell lines, this complexity makes it difficult to predict a priori the subcellular distribution of a particular polyamide-fluorophore conjugate. Efficient nuclear localization of a fluorophore conjugate is not necessarily predictive for non-fluorescent analogs. Screening polyamides for a biological effect dependent on nuclear localization is a viable approach for the assessment of nuclear uptake and does not require incorporation of an optical tag. As polyamides are studied in model systems of greater biological complexity, molecules that can access the nuclei of cells without the use of fluorescent dyes are desirable.
Polyamide-fluorescein conjugate 1 (Figure 1) was designed to bind the sequence 5′-ATACGT-3′ within the hypoxia response element (HRE) in the vascular endothelial growth factor (VEGF) enhancer. It was found to bind its target site with an affinity of 6.3 × 109 M−1, disrupt hypoxia-inducible factor 1 (HIF-1) binding to the HRE in vitro, enter the nuclei of HeLa cells, and inhibit hypoxia-induced VEGF mRNA and protein production in cultured HeLa cells (Figure 2A–C) (16). A control polyamide 2 designed to bind the DNA sequence 5′-WGGWCW-3′ (where W denotes an A or T) also exhibited nuclear localization, but had a modest effect on VEGF expression. Independently, the DNA-binding antibiotic echinomycin, which binds 5′-ACGT-3′, was found to inhibit VEGF expression in U251 cells (17,18). The finding that two molecules of different structure bind the same HRE sequence 5′-TACGT-3′ and exhibit similar biological effects on hypoxia-driven gene expression validates the concept of using small molecules to interfere with protein–DNA interfaces.
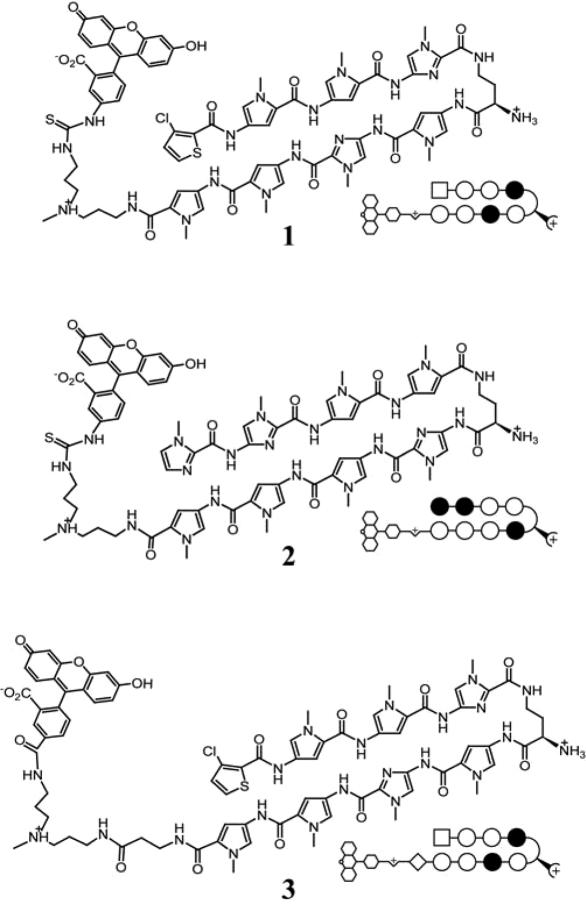
Figure 1.
Structures of polyamide-fluorescein conjugates 1–3. Polyamides 1 and 3 target the sequence 5′-WTWCGW-3′, while polyamide 2 targets the sequence 5′-WGGWCW-3′. Polyamide 3 differs from 1 and 2 in the linker region between the polyamide core sequence and the fluorescein moiety. Open circles designate N-methylpyrrole, closed circles designate N-methylimidazole, squares designate 3-chlorothiophene and diamonds designate β-alanine.
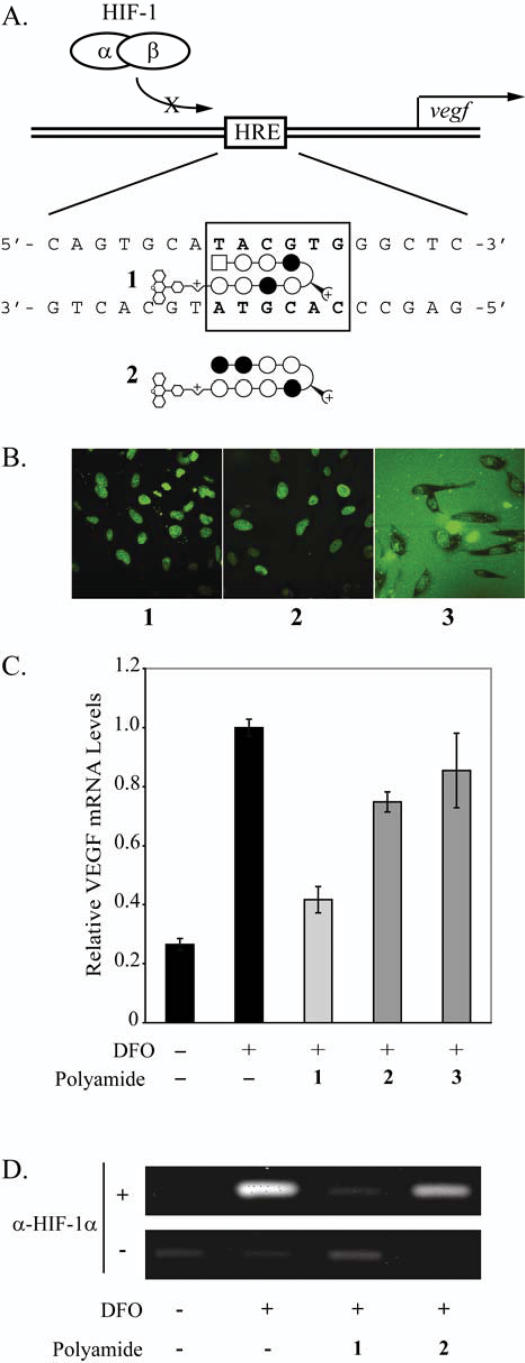
Figure 2.
(A) Schematic illustration of polyamide 1 binding to the VEGF HRE. (B) Uptake of polyamides 1–3 in HeLa cells. Polyamides 1 (left) and 2 (center) localize in the nucleus while polyamide 3 (right) localizes in the nucleus to a limited extent but is largely extracellular. (C) Induction of VEGF mRNA by the hypoxia mimetic deferoxamine (DFO) in the presence of polyamides 1–3 measured by quantitative real-time PCR. Polyamide 1 inhibits the expression of DFO-induced VEGF expression. Polyamides 2 and 3 have a more modest effect. (D) Chromatin immunoprecipitation assays using HeLa cells with anti-HIF1α or mock antibody treatment are consistent with inhibition of HIF-1 binding to the VEGF HRE in the presence of polyamide 1, and, to a more modest degree, 2. Concentrations of polyamides are 1 μM.
The current study utilizes the inhibition of hypoxia-induced VEGF expression as a biological read-out for nuclear localization of polyamides targeted to the HRE. As a control, we show that a polyamide-fluorescein conjugate 3 that targets the HRE but exhibits poor nuclear localization in HeLa cells due to a non-optimal linker (8) has a limited effect on hypoxia-induced VEGF expression. We have synthesized a focused library of polyamides with an identical core sequence that targets the HRE while varying C-terminus tail moieties. The C-terminus was chosen as the site of chemical variation in our library, as subtle modifications to this area were previously found to influence nuclear localization of polyamide-fluorophore conjugates (8,9). The polyamides generated in this study retain high-binding affinity and specificity to the target site. Synthesized as a control group was a library of ‘mismatch’ polyamides with identical C-terminus modifications but a different core targeted to the DNA sequence 5′-WGGWCW-3′, which is not found at the VEGF HRE. The majority of polyamides from both libraries failed to inhibit hypoxia-induced VEGF expression. We identified, however, two non-fluorescent polyamides with biological activity and binding affinities and specificities comparable to those of polyamide-fluorescein conjugate 1.
MATERIALS AND METHODS
Synthesis of polyamides
Polyamides were synthesized by solid phase methods on Kaiser oxime resin (Nova Biochem) except polyamide 3, which was synthesized on Boc-β-alanine-PAM-resin (Peptides International) (19,20). Fluorescein-labeled polyamides 1–3 were prepared as described previously (16). Polyamides 4–8 and 16–20 were cleaved from resin using the appropriate amine and purified by reverse-phase HPLC. For the synthesis of 9–15 and 21–27, the polyamide was first cleaved with 3,3′-diamino-N-methyl-dipropylamine and purified by reverse-phase HPLC. The designated acid was activated with PyBOP (Nova Biochem) and coupled to the polyamide in dimethylformamide and N,N-diisopropylethylamine to yield the final product after reverse-phase HPLC purification. The purity and identity of all polyamides were verified by analytical HPLC and MALDI-TOF MS.
Confocal microscopy
Microscopy of live, unfixed cells was performed as described previously (8,9). Briefly, cells were plated on glass-bottom culture dishes for 24 h prior to overnight incubation with 2 μM polyamide. Imaging was performed on a Zeiss LSM 5 Pascal inverted laser scanning microscope.
Determination of DNA-binding affinities and sequence specificities
Quantitative DNase I footprint titration experiments were used to determine polyamide-binding affinities (Ka) to the 5′-ATACGT-3′ sequence within the HRE of the VEGF promoter. These experiments were performed using the 5′-32P-labeled 197 bp PCR amplification product of the plasmid pGL2-VEGF-Luc isolated by non-denaturing gel electrophoresis (16,21). Quantitative DNase I footprint titration experiments were conducted as reported previously (21).
Measurement of hypoxia-induced relative VEGF mRNA
Cells were plated in 24-well plates at a density of 15–20 × 103 cells per well (30–40 × 103 cells/ml). Polyamides were added to adhered cells in solutions of cell media at the appropriate concentration and allowed to incubate with the cells for 48 h. Then, hypoxic induction of gene expression was chemically induced by adding deferoxamine (DFO) to 300 μM for an additional 16 h (22,23). Cells were harvested, RNA isolated, cDNA synthesized and quantitative real-time RT–PCR conducted as described previously (16). Briefly, RNA was isolated using the RNeasy kit (Qiagen), cDNA synthesized using Powerscript (BD Biosciences) and quantitative real-time RT–PCR performed using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7300 instrument. VEGF expression was measured relative to β-glucuronidase (GUSB) as an endogenous control. Statistical analyses of results were determined using three independent biological replicates. The primer sequences used for VEGF and GUSB are available upon request. HeLa cells were purchased from ATCC and U251 cells were received as a gift from Dr Giovanni Melillo of the National Cancer Institute.
Chromatin immunoprecipitation
Cells were plated on 15 cm diameter culture dishes and left to attach overnight. Polyamides were incubated with the cells for 48 h, and the cells were incubated for an additional 16 h after the addition of DFO to a final concentration of 300 μM. Cells were then treated with 1% formaldehyde for 10 min. Chromatin was isolated and sheared. HIF-1α antibodies (Novus Biologicals) were used to immunoprecipitate HIF-1 bound DNA fragments. After crosslink reversal, PCRs using primers flanking the HRE of VEGF were used to assess enrichment of bound fragments as compared to mock-precipitated (no antibody) controls. PCRs were monitored either using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7300 instrument, or directly visualized using gel electrophoresis.
RESULTS
Confocal microscopy of polyamide-fluorophore conjugates 1–3
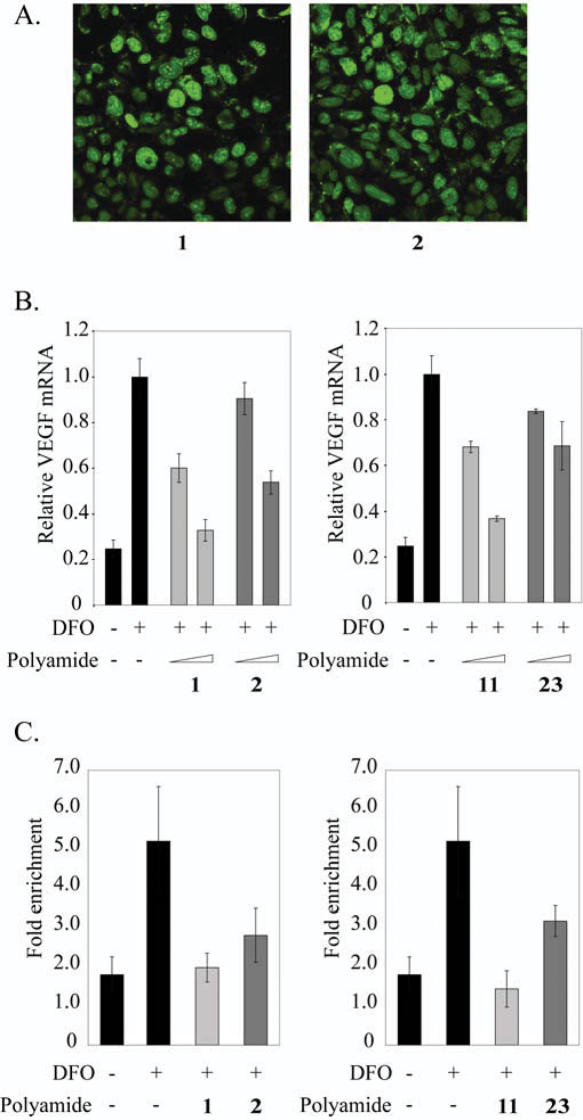
Cellular uptake of polyamide-fluorescein conjugates 1–3 (Figure 1) was examined by confocal microscopy in live, unfixed HeLa cells (Figure 2B). These data demonstrate that polyamide-FITC conjugates 1 and 2, targeted to 5′-WTWCGW-3′ and 5′-WGGWCW-3′, respectively, both localize in the nucleus of HeLa cells. In contrast, polyamide conjugate 3, also targeted to 5′-WTWCGW-3′ but with a β-alanine residue at the C-terminus, is largely extracellular.
Suppression of hypoxia-inducible transcription in cell culture by polyamide-fluorophore conjugates 1–3
Induction of VEGF mRNA by the hypoxia mimetic DFO in the presence of polyamides 1–3 in HeLa cells was measured by quantitative real-time RT–PCR (Figure 2C). Polyamide 1 inhibits the expression of DFO-induced VEGF mRNA production by ∼50%. Polyamides 2 and 3 show diminished biological activity compared to 1. Together, the uptake and gene expression data for 1–3 suggest that a polyamide targeted to the HRE of VEGF must have efficient nuclear uptake in order to inhibit hypoxia-induced expression of VEGF. Chromatin immunoprecipitation experiments in cultured HeLa cells were performed to assess whether changes in VEGF promoter occupancy by HIF-1α were consistent with displacement of this protein by polyamide 1. Chromatin immunoprecipitation assays with anti-HIF1α or mock antibody treatment are consistent with inhibition of HIF-1 binding to the HRE in the presence of polyamide 1 and polyamide 2 to a modest degree (Figure 2D). In the absence of either polyamide 1 or 2, HIF-1 occupies the VEGF HRE following DFO-induction. HIF-1 occupancy at the VEGF HRE is reduced in cells treated with 1 prior to DFO-induction. Polyamide 2 has a more limited effect.
Non-fluorescent HRE-targeted polyamides
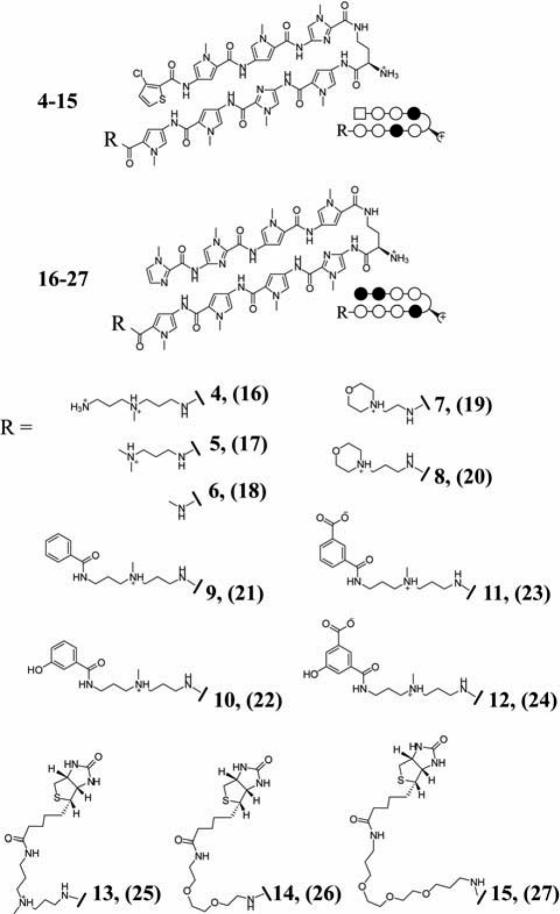
We set out to synthesize a series of non-fluorescent HRE-targeted polyamides (4–15) and their mismatch analogs (16–27; Figure 3). Polyamides 4–15 can be considered in three groups: 4–8 were designed with minimized tail motifs; 9–12 were designed to probe the functional groups present in fluorescein dyes potentially responsible for the favorable nuclear uptake profile of 1; and 13–15 were biotin conjugates. Polyamides 4 and 5 were designed to have tail moieties resembling the linker region of polyamide-fluorescein conjugate 1. The dimethylaminopropylamine tail present in 5 is often placed at the C-terminus of polyamides used in biophysical studies (1,2,24). Polyamide 6 was designed to have a tail group of minimal size. Polyamides 7 and 8 have 2-morpholinoethanamine and 3-morpholinopropan-1-amine tail groups, respectively. A four ring polyamide with a morpholino tail was previously shown to have antibacterial activity in a mouse peritonitis model (25). The next group, polyamides 9–12, was designed to reduce molecular weight while retaining functional groups present in fluorescein and maintaining the linker moiety found in polyamide 1. Polyamides 9–12 are derived from benzoic acid; 11 and 12 each contain a carboxylic acid meta- to the attachment site, and 10 and 12 each have a hydroxyl group meta- to the attachment site. Polyamides 13–15 are polyamide-biotin conjugates. The high affinity of the biotin moiety to streptavidin may be useful for biochemical pull-down experiments. Polyamide 13 retains the linker found in 1–3 while 14 and 15 contain 2- and 3-oxygen PEG linkers, respectively. Polyamides 16–27 were synthesized as mismatch control polyamides for 4–15, respectively.
Figure 3.
Structures of polyamides 4–27. Polyamides 4–15 target 5′-WTWCGW-3′ DNA sequences; 16–27 target 5′-WGGWCW-3′.
Binding affinities and specificities
Polyamides 4–15, designed to bind the 5′-ATACGT-3′ HRE-binding site, share the same hairpin polyamide core as polyamides 1 and 3 but have different C-terminus tail groups. The DNA-binding affinities of 4–15 for the HRE of VEGF were measured by quantitative DNase I footprint titrations using a 5′-32P-labeled PCR amplification product of the plasmid pGL2-VEGF-Luc, which contains the VEGF HRE. Polyamide conjugate 1 was previously found to have Ka = 6.3 × 109 M−1 at this site (16). Polyamides 4–15 were footprinted (Figure 4, Supplementary Figure S1), and the Ka values obtained range from 4.4 (±1.7) × 109 for 15 to 7.7 (±0.9) × 1010 for 4 (Table 1). With the exception of 15, the values obtained are all higher than that measured for 1. Polyamides 4–15 retain good specificity for the 5′-ATACGT-3′ HRE-binding site, although a modest degree of non-specific binding is observed at high concentrations for polyamides 4, 5, 7, 8 and 10. Modifications to the tail moiety did not significantly abrogate high-binding affinity and specificity.
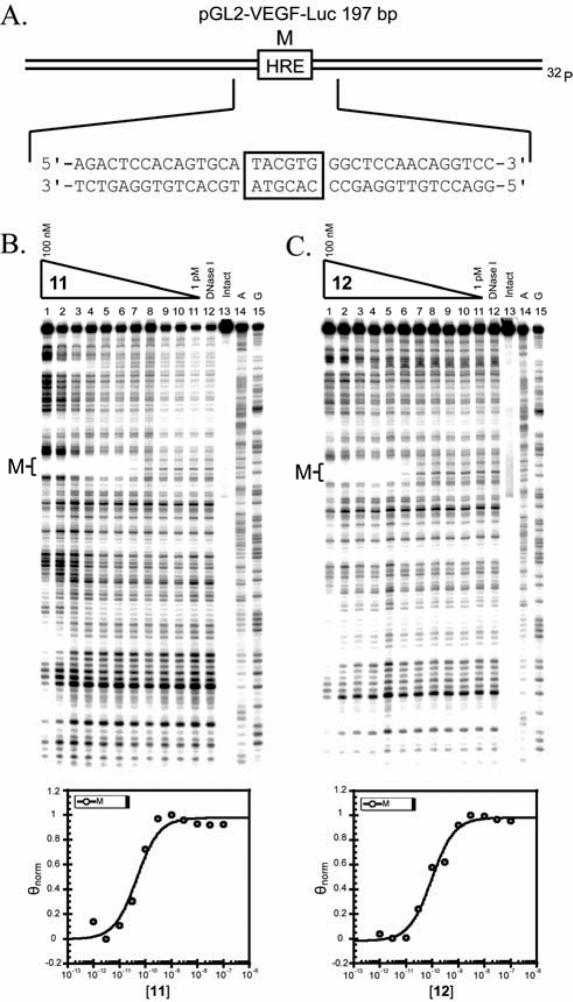
Figure 4.
Plasmid sequence and DNA-binding properties of polyamides 11 and 12. (A) Illustration of plasmid pGL2-VEGF-Luc. The HRE match binding site M is indicated by the box. (B and C) Quantitative DNase I footprint titration experiments for polyamides 11 (B) and 12 (C) on the 197 bp, 5′-end-labeled PCR product of plasmid pGL2-VEGF-Luc: lanes 1–11, 100 nM, 30 nM, 10 nM, 3 nM, 1 nM, 300 pM, 100 pM, 30 pM, 10 pM, 3 pM and 1 pM polyamide, respectively; lane 12, DNase I standard; lane 13, intact DNA; lane 14, A reaction; lane 15, G reaction. Each footprinting gel is accompanied by a binding isotherm for polyamides binding the match site M (below).
Table 1.
Binding affinities (M−1) for polyamides 4–15
| Polyamide | 5′-TACGTG-3′ |
|---|---|
| 4 | 7.7 (±0.9) × 1010 |
| 5 | 6.2 (±1.2) × 1010 |
| 6 | 2.5 (±0.3) × 1010 |
| 7 | 4.0 (±0.9) × 1010 |
| 8 | 3.8 (±0.5) × 1010 |
| 9 | 1.5 (±0.6) × 1010 |
| 10 | 2.1 (±0.4) × 1010 |
| 11 | 2.6 (±0.4) × 1010 |
| 12 | 1.1 (±0.2) × 1010 |
| 13 | 7.2 (±1.1) × 109 |
| 14 | 4.8 (±1.9) × 109 |
| 15 | 4.4 (±1.7) × 109 |
Equilibrium association constants reported are mean values from three DNase I footprint titration experiments; standard deviations are shown in parentheses.
Screening for suppression of hypoxia-inducible transcription by polyamides 4–27 in HeLa cells
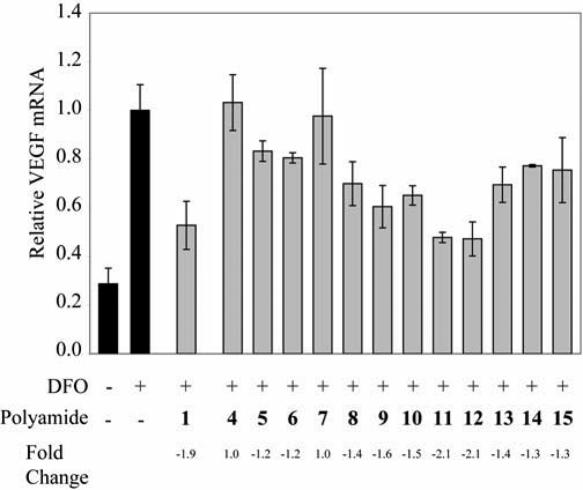
Induction of VEGF mRNA by the hypoxia mimetic DFO in HeLa cells in the presence of polyamides 4–15 was measured by quantitative real-time RT–PCR and compared to that of polyamide 1 (Figure 5). Polyamides 5, 6 and 8 showed modest inhibition of induced VEGF expression and polyamides 4 and 7 showed no difference relative to the untreated, induced control. Mismatch polyamides 16–19 showed no difference in VEGF mRNA relative to the untreated, induced control. Polyamide 20 showed a modest effect (Supplementary Figure S2).
Figure 5.
Induction of VEGF mRNA by the hypoxia mimetic deferoxamine (DFO) in the presence of polyamides 1 and 4–15 measured by quantitative real-time PCR. Concentrations of polyamides are 1 μM.
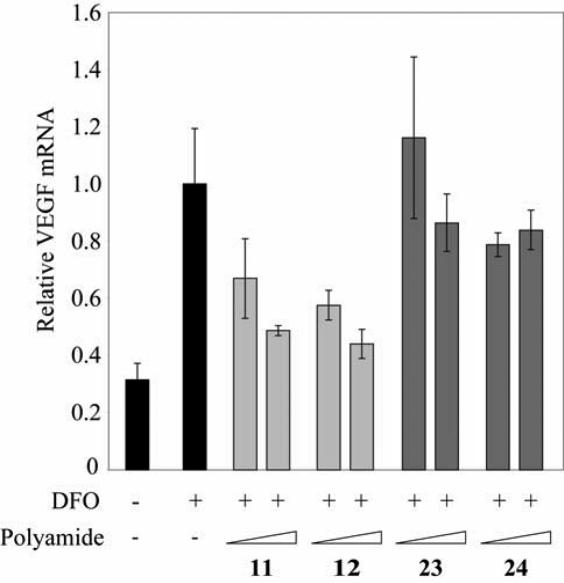
Polyamides 9 and 10 had a modest effect on induced VEGF expression, whereas 11 and 12 inhibited induced VEGF expression comparably to polyamide 1. Control polyamides 23 and 24 had a reduced effect as compared with their match congeners 11 and 12 (Figure 6). HeLa cell growth was not inhibited by 11, 12, 23 or 24 at 1 μM (Supplementary Figure S3).
Figure 6.
Induction of VEGF mRNA by the hypoxia mimetic deferoxamine (DFO) in the presence of polyamides 11, 12, 23 and 24 measured by quantitative real-time PCR. Polyamides 11 and 12, which bind the 5′-ATACGT-3′ sequence found in the VEGF HRE, show inhibition of DFO-induced VEGF expression. Polyamides 23 and 24, which target 5′-WGGWCW-3′, have a more modest effect. Concentrations of polyamides are 0.2 and 1 μM.
Polyamides 13–15 had a modest effect on induced VEGF expression. An appreciable difference between relative VEGF mRNA levels was seen in cells treated with biotin conjugates 13 and 14 and their mismatch congeners 25 and 26 (Supplementary Figure 2C). It is noted that cells treated with polyamides 25 and 26 had VEGF expression slightly higher than that of the untreated, induced control.
Suppression of hypoxia-inducible transcription by polyamides in U251 cells
The effects of HRE targeted polyamides 1 and 11 and control polyamides 2 and 23 were tested in U251 cells, which express a higher level of VEGF than HeLa cells (Figure 7). Uptake of optically tagged polyamides 1 and 2 was examined by confocal microscopy in live, unfixed U251 cells dosed with 2 μM of 1 or 2 (Figure 7A). Induction of VEGF mRNA in the presence of polyamides 1 and 11 was inhibited dose dependently (Figure 7B). Polyamide 2 had minimal effect on VEGF expression at 0.2 μM. However, at 1 μM, polyamide 2 had a moderate effect, though still less than that of its match congener 1. Polyamide 23 had a more modest effect than its match congener 11 at both 0.2 and 1 μM. We also undertook chromatin immunoprecipitation experiments in U251 cells to assess changes in VEGF promoter occupancy by HIF-1α in the presence of 1, 2, 11 and 23 (Figure 7C). Chromatin immunoprecipitation assays with anti-HIF1α or mock antibody treatment are consistent with decreased occupancy of HIF-1α at the VEGF HRE in the presence of polyamides 1 and 11 at 1 μM. Polyamide 23 has a more modest effect compared with that of its match congener 11. At the concentration tested (1 μM), polyamide 2 has an effect statistically similar, though slightly less than that of 1. This is not unexpected; 2 also showed some inhibition of VEGF mRNA at this concentration. PCR amplification at an irrelevant locus in the coding region of GUSB showed minimal enrichment (∼1- to 1.5-fold over background) for polyamide-treated and control samples (data not shown).
Figure 7.
(A) Uptake of polyamides 1 (left) and 2 (right) in U251 cells. (B) Induction of VEGF mRNA by the hypoxia mimetic deferoxamine (DFO) in the presence of polyamides 1, 2, 11 and 23 in U251 cells measured by quantitative real-time PCR. Concentrations of polyamides are 0.2 and 1 μM. (C) Chromatin immunoprecipitation assays with anti-HIF1α or mock antibody treatment expressed as fold-enrichment (specific/mock) of a 120 bp sequence at the VEGF HRE measured by real-time PCR. Concentrations of polyamides are 1 μM.
DISCUSSION
Previous studies on fluorophore-labeled polyamides have revealed several parameters that affect cellular trafficking of these molecules, including number of heterocyclic ring pairs, relative Py/Im content and sequence, and choice of fluorophore and attachment site (8,9). Among fluorophores considered, the protonation state of fluorescein is pH sensitive, while those of tetramethylrhodamine and Bodipy are not. In previous work, nuclear localization of polyamide 2 in HeLa cells was ablated when the cells were grown in glucose- and sodium pyruvate-free media, but subsequently restored when normal growth media were added (8). This suggests that for some polyamides, nuclear localization is energy dependent. Many fluorophore-polyamide conjugates that exhibit a punctate, cytoplasmic staining pattern co-localize with LysoTracker, a lysosomal stain. Verapamil, a calcium channel and P-glycoprotein efflux pump inhibitor, was shown to facilitate nuclear uptake of a Bodipy-polyamide conjugate otherwise sequestered within cytoplasmic vesicles (7). Some polyamides with minimal nuclear localization, e.g. 3, localize mostly in the extracellular space. Nevertheless, the fact that many eight-ring hairpin polyamide-fluorophore conjugates do localize in the nuclei of live cells is encouraging.
The inhibition of induced VEGF expression in the presence of polyamide 1, which binds the VEGF HRE, is an important proof-of-principle experiment for the use of designed DNA-binding ligands to modulate the expression of specific sets of genes. Moving ahead, anticipated experiments include studies in mammalian model systems to account for biodistribution, availability and metabolism of polyamides. Toward this end, we have attempted to identify polyamides capable of nuclear localization but of simpler structures and lower molecular weights than dye conjugates. The inhibition of induced VEGF expression with HRE-targeted polyamide 1, but not 2, which targets a different DNA sequence or 3, which exhibits considerably less nuclear localization, demonstrates the viability of VEGF inhibition as a proxy for nuclear localization of polyamides that bind the HRE.
The ability to interrogate polyamide cellular uptake and nuclear localization by affecting expression of an endogenous inducible gene enables exploration of the nuclear uptake profiles of non-fluorescent tail moieties. The quantitative real-time RT–PCR data presented show that removal of the fluorescein dye (polyamide 4) and step-wise ablation of the linker moiety (polyamides 5 and 6) diminish the biological activity of these compounds as compared with polyamide 1 in this system. The Ka values measured for 4–6 are between 4- and 12-fold higher than that measured for the parent polyamide 1 on the same DNA sequence. Similarly, the binding affinities of N-alkylaminemorpholino tail polyamides 7 and 8 are ∼6-fold higher than that of the parent polyamide 1 for the same binding site, but only modest biological activities are observed.
Data generated from cultured HeLa cells treated with polyamides 9–12 were more encouraging. These compounds were designed to probe contributions of the alcohol and carboxylic acid groups of fluorescein to the cellular uptake and nuclear localization observed for 1. This was done by synthesizing a focused series of polyamides composed of an amide-bond linked benzene ring containing four possible permutations of alcohol and/or carboxylic acid substituents at the meta- positions. Polyamides 9–12 demonstrated good sequence specificities and binding affinities greater than that of 1. Polyamide conjugates of benzoic acid and 3-hydroxy benzoic acid, 9 and 10, respectively, show a modest degree of biological activity, but those conjugated to either isophthalic acid or 5-hydroxy isophthalic acid, 11 and 12, respectively, demonstrate a biological effect similar to that observed for the parent polyamide 1. These results suggest that the presence of the carboxylic acid group on the benzene ring is a positive determinant of nuclear localization in this system.
Biotin conjugates 13–15 differ in the identity of the linker moiety and were of interest due to the potential utility of these conjugates for pull-down experiments. The binding affinities measured by quantitative DNase I footprint titration experiments for these conjugates were similar to that of the parent polyamide 1. By quantitative real-time RT–PCR, polyamides 13–15 all demonstrated modest biological activities and the results do not suggest strong differences in activities based on linker identity. The modest activities of the biotin conjugates, which might reflect a moderate degree of nuclear uptake, are neither encouraging nor dismissive of potential applications of these conjugates.
Our working hypothesis regarding VEGF inhibition by polyamides is through direct interactions with the DNA–HIF-1 interface at the HRE that prevents HIF-1 binding, most likely by an allosteric mechanism. To explore this model further, we used chromatin immunoprecipitation to assess HIF-1 occupancy at the VEGF HRE under hypoxia-induced conditions in HeLa cells in the presence and absence of match (and mismatch) polyamides with variable C-terminus tails. Antibodies against HIF-1α were found to enrich fragments of DNA containing the VEGF HRE after DFO-induction. This enrichment was reduced in samples treated with polyamide 1 and, to a lesser extent, polyamide 2. We also undertook experiments in U251 cells, which express VEGF more robustly and have previously been used as a model cell line for studying hypoxia-induced VEGF expression (17).
Polyamide-fluorescein conjugates 1 and 2 localize in the nucleus of U251 cells. Induction of VEGF mRNA in the presence of polyamides 1, 2, 11 and 23 at 0.2 and 1 μM in U251 cells was measured by quantitative real-time RT–PCR. Polyamide 1 showed significant inhibition at both concentrations. Interestingly, at 1 μM polyamide 2 showed inhibition greater than that measured in HeLa cells but still significantly less than 1. Polyamide 11 exhibits dose-dependent inhibition of VEGF similar to that measured in HeLa cells, while 23 showed modest activity at both concentrations comparable to that measured in HeLa cells. Chromatin immunoprecipitation of HIF-1α from U251 cells treated with HRE-targeted polyamides 1 and 11 and mismatch polyamides 2 and 23 shows that enrichment of HIF-1α bound DNA fragments containing the VEGF HRE after DFO induction is inhibited by pre-treatment with polyamides 1 and 11, and to a lesser extent 2 and 23. This is generally consistent with a mechanism whereby polyamides 1 and 11 exert their effect by preventing HIF-1 binding to the cognate HRE. This is also consistent with DNase I footprint titrations and gel shift assays in vitro (16).
This report identifies two polyamides, 11 and 12, with non-fluorescent moieties at the C-terminus that bind the VEGF HRE that have biological activity similar to that of the fluorescein-polyamide conjugate 1 known to localize in the nucleus. Both polyamides retain an amide-linked benzene ring with a carboxylic acid meta- to the attachment point and are similar in structure to the proximal portion of fluorescein. Work is ongoing to synthesize a second series of polyamides designed to examine the role of the aromatic ring and the carboxylic acid functional group in aiding nuclear localization. As the mechanism of polyamide uptake and nuclear localization continues to be elucidated, it is currently unclear whether the biological activity of 11 and 12 is due to improved ability to buffer the wide range of pH values that may be encountered in the cell (e.g. lysosomes), improved nuclear localization resulting in an increased effective polyamide concentration at the DNA, or decreased efflux of polyamide from the cell perhaps through one or several ATP-dependent drug efflux pumps (26,27).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering at the California Institute of Technology, supported in part by the National Science Foundation Materials and Research Science and Engineering Program. The authors gratefully acknowledge the gift of U251 cells from Dr G. Melillo of the National Cancer Institute, and helpful discussions regarding chromatin immunoprecipitation with Dr E. Soragni and Prof. J. M. Gottesfeld of the Scripps Research Institute. We thank the National Institutes of Health for support (GM51747) and predoctoral training fellowships (C.S.J., M.E.F.). We also thank the Ralph M. Parsons Foundation for support (N.G.N.). Funding to pay the Open Access publication charges for this article was provided by NIH GM51747.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dervan P.B., Edelson B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 2.Dervan P.B., Poulin-Kerstien A.T., Fechter E.J., Edelson B.S. Regulation of gene expression by synthetic DNA-binding ligands. Top. Curr. Chem. 2005;253:1–31. [Google Scholar]
- 3.White S., Szewczyk J.W., Turner J.M., Baird E.E., Dervan P.B. Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 4.Kielkopf C.L., Baird E.E., Dervan P.D., Rees D.C. Structural basis for G•C recognition in the DNA minor groove. Nature Struct. Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 5.Foister S., Marques M.A., Doss R.M., Dervan P.B. Shape selective recognition of T•A base pairs by hairpin polyamides containing N-terminal 3-methoxy (and 3-chloro) thiophene residues. Bioorg. Med. Chem. 2003;11:4333–4340. doi: 10.1016/s0968-0896(03)00502-9. [DOI] [PubMed] [Google Scholar]
- 6.Belitsky J.M., Leslie S.J., Arora P.S., Beerman T.A., Dervan P.B. Cellular uptake of N-methylpyrrole/N-methylimidazole polyamide-dye conjugates. Bioorg. Med. Chem. 2002;10:3313–3318. doi: 10.1016/s0968-0896(02)00204-3. [DOI] [PubMed] [Google Scholar]
- 7.Crowley K.S., Phillion D.P., Woodard S.S., Schweitzer B.A., Singh M., Shabany H., Burnette B., Hippenmeyer P., Heitmeier M., Bashkin J.K. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg. Med. Chem. Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 8.Best T.P., Edelson B.S., Nickols N.G., Dervan P.B. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl Acad. Sci. USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelson B.S., Best T.P., Olenyuk B., Nickols N.G., Doss R.M., Foister S., Heckel A., Dervan P.B. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suto R.K., Edayathumangalam R.S., White C.L., Melander C., Gottesfeld J.M., Dervan P.B., Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 11.Dudouet B., Burnett R., Dickinson L.A., Wood M.R., Melander C., Belitsky J.M., Edelson B., Wurtz N., Briehn C., Dervan P.B., et al. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 2003;10:859–867. doi: 10.1016/j.chembiol.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Gottesfeld J.M., Neely L., Trauger J.W., Baird E.E., Dervan P.B. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson L.A., Gulizia R.J., Trauger J.W., Baird E.E., Mosier D.E., Gottesfeld J.M., Dervan P.B. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl Acad. Sci. USA. 1998;95:12890–12895. doi: 10.1073/pnas.95.22.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen-Hackley D.H., Ramm E., Taylor C.M., Joung J.K., Dervan P.B., Pabo C.O. Allosteric inhibition of zinc-finger binding in the major groove of DNA by minor-groove binding ligands. Biochemistry. 2004;43:3880–3890. doi: 10.1021/bi030223c. [DOI] [PubMed] [Google Scholar]
- 15.Wurtz N.R., Pomerantz J.L., Baltimore D., Dervan P.B. Inhibition of DNA binding by NF-kappa B with pyrrole-imidazole polyamides. Biochemistry. 2002;41:7604–7609. doi: 10.1021/bi020114i. [DOI] [PubMed] [Google Scholar]
- 16.Olenyuk B.Z., Zhang G.J., Klco J.M., Nickols N.G., Kaelin W.G., Dervan P.B. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl Acad. Sci. USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong D.H., Park E.J., Stephen A.G., Calvani M., Cardellina J.H., Monks A., Fisher R.J., Shoemaker R.H., Melillo G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 18.Vandyke M.M., Dervan P.B. Echinomycin binding-sites on DNA. Science. 1984;225:1122–1127. doi: 10.1126/science.6089341. [DOI] [PubMed] [Google Scholar]
- 19.Baird E.E., Dervan P.B. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 20.Belitsky J.M., Nguyen D.H., Wurtz N.R., Dervan P.B. Solid-phase synthesis of DNA binding polyamides on oxime resin. Bioorg. Med. Chem. 2002;10:2767–2774. doi: 10.1016/s0968-0896(02)00133-5. [DOI] [PubMed] [Google Scholar]
- 21.Trauger J.W., Dervan P.B. Footprinting methods for analysis of pyrrole-imidazole polyamide/DNA complexes. Methods Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 22.Wood S.M., Ratcliffe P.J. Mammalian oxygen sensing and hypoxia inducible factor-1. Int. J. Biochem. Cell Biol. 1997;29:1419–1432. doi: 10.1016/s1357-2725(97)00129-5. [DOI] [PubMed] [Google Scholar]
- 23.Bianchi L., Tacchini L., Cairo G. HIF-1-mediated activation of transferrin receptor gene transcription by iron chelation. Nucleic Acids Res. 1999;27:4223–4227. doi: 10.1093/nar/27.21.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dervan P.B. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 25.Kaizerman J.A., Gross M.L., Ge Y.G., White S., Hu W.H., Duan J.X., Baird E.E., Johnson K.W., Tanaka R.D., Moser H.E., et al. DNA binding ligands targeting drug-resistant bacteria: structure, activity, and pharmacology. J. Med. Chem. 2003;46:3914–3929. doi: 10.1021/jm030097a. [DOI] [PubMed] [Google Scholar]
- 26.Juliano R.L., Ling V. Surface glycoprotein modulating drug permeability in Chinese-hamster ovary cell mutants. Biochim. Biophys. Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 27.Ueda K., Cardarelli C., Gottesman M.M., Pastan I. Expression of a full-length cDNA for the human Mdr1 gene confers resistance to colchicine, doxorubicin, and vinblastine. Proc. Natl Acad. Sci. USA. 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]