Abstract
We use single-protein atomic force microscopy techniques to detect length phenotypes in an Ig module. To gain amino acid resolution, we amplify the mechanical features of a single module by engineering polyproteins composed of up to 12 identical repeats. We show that on mechanical unfolding, mutant polyproteins containing five extra glycine residues added to the folded core of the module extend 20 Å per module farther than the wild-type polyproteins. By contrast, similar insertions near the N or C termini have no effect. Hence, our atomic force microscopy measurements readily discriminate the location of the insert and measure its size with a resolution similar to that of NMR and x-ray crystallography.
Proteins are responsible for a wide variety of mechanical functions (1–7). Recent experiments have demonstrated that the Ig and fibronectin type III domains are extensible (8–13). The mechanical properties of these domains are thought to play an important physiological role when they are placed under mechanical stress (8–12, 14). A wide variety of mutations target these domains (15–18). Although some of the mutations and amino acid sequence variations observed in these domains alter their ligand-binding properties (15–18), it is likely that they also alter the mechanical properties of the domains, creating mechanical phenotypes. However, mechanical phenotypes in proteins have, to our knowledge, never been studied or observed before. Here we use single-protein atomic force microscopy (AFM) techniques (10–13) to examine the mechanical topology of an Ig module and to detect length phenotypes with single amino acid resolution. For our studies, we use a well characterized Ig-like module: the I27 module of human cardiac titin. The three-dimensional structure of the I27 module has been determined by NMR (19, 20) and shown to be 89 aa long, folding into a characteristic β-sandwich with seven strands (Fig. 1A). Molecular dynamics simulations of the mechanical unfolding of the I27 module (21) predict that a patch of hydrogen bonds between the A′ and G β-strands creates a point of high mechanical resistance that must be ruptured to trigger unfolding. The steered molecular dynamics observations predict that a folded I27 module will contain a mechanically isolated region that includes the amino acids that are placed behind the point of high mechanical resistance in the A′G overlap. While the protein is folded, these amino acids do not experience the applied force. However, these “hidden” amino acids are predicted to become exposed on module unfolding. To test these predictions with AFM, we have created a series of I27 mutants by inserting Gly residues in locations either inside the “hidden” region or outside this region near the N or C termini. The AFM techniques measure the length of a polymer with Ångstrom resolution (10–13). By measuring the protein length before and after unfolding, we could readily detect the relative position and the length of the Gly inserts and explicitly examine the mechanical topology of the I27 module. The approach demonstrated here can reveal mechanical phenotypes and may be used to study the effect of mutations in the mechanical properties of a wide range of proteins.
Figure 1.
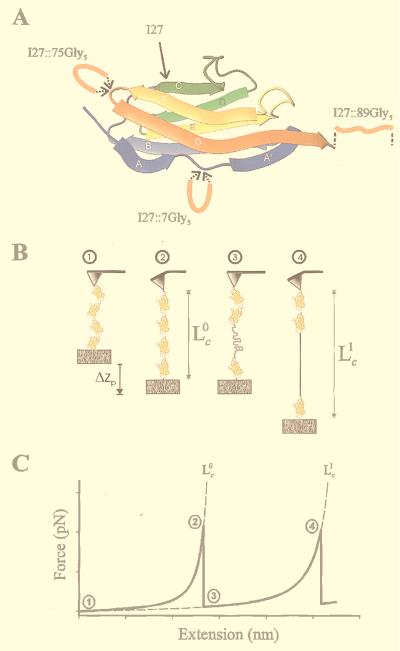
Single-molecule AFM measurements of the contour length of engineered polyproteins. (A) Structure of the I27 Ig-like module and location of the five Gly insertion sites. The N and the C termini of I27 are antiparallel and come in close apposition over the A′G strands. A mechanical linkage is thought to be present in the A′G overlap, providing continuity of force between the N and C termini of the folded module. Rupture of this linkage exposes the amino acids that are hidden in the fold, extending the contour length of the protein. We constructed polyproteins based on the wild-type (I27) and mutant forms of the I27 module with a five-Gly insert in the FG hairpin loop at position 75 (I27∷75Gly5), in the N terminus region at position 7 (I27∷7Gly5), and after the C terminus at position 89 (I27∷89Gly5). (B) Cartoon of the sequence of events during the stretching of a polyprotein engineered with identical repeats of an I27 module. Stretching the ends of the polyprotein with an atomic force microscope sequentially unfolds the protein modules, generating a saw-tooth pattern in the force-extension relationship that reveals the mechanical characteristics of the protein (C; 10–13). 1, an anchored polyprotein composed of four Ig domains. The protein is relaxed; 2, stretching this protein to near its folded contour length, Lc0, requires a force that is measured as a deflection of the cantilever; 3, the applied force triggers unfolding of a domain, increasing the contour length of the protein and relaxing the cantilever back to its resting position; 4, further stretching removes the slack and brings the protein to its new contour length Lc1. (C) Idealized force-extension curve that results from stretching a polyprotein. The numbers correspond to the stages marked in B. The dashed lines are fits of the WLC model of elasticity to the force-extension curves leading up to each force peak. The increase in contour length resulting from module unfolding is calculated from the fits as Δ = Lc1 − Lc0.
MATERIALS AND METHODS
Protein Engineering.
Recombinant DNA techniques were used to synthesize and express direct tandem repeats of the I27 monomer, as described before (13). I27 cDNA with flanking AvaI sites was amplified by PCR. Five Gly insertions were generated by PCR by using mutagenic primers, Pfu DNA polymerase (Stratagene), 5% DMSO as a cosolvent and a denaturing temperature of 98°C. The PCR products were verified by sequencing. Directional DNA concatemerization was done by self ligation of the sticky ends of the nonpalindromic CTCGGG AvaI restriction site. After self ligation, the concatemers were cloned into a custom-made expression vector by using Sure-2 Escherichia coli cells (Stratagene). The engineered polyproteins are: I2712 (12 repeats of the wild-type I27 module); I2710∷75Gly5 (10 repeats of a mutant I27 module containing a five-Gly insert in position 75); I275∷89Gly5 (five repeats of a mutant I27 module containing a five-Gly insert in position 89); and I279∷7Gly5 (nine repeats of a mutant I27 module containing a five-Gly insert in position 7). The polyproteins all have different numbers of repeats because we cannot control the extent of polymerization during the engineering of the cDNA concatemers by bulk ligation; however, we always take the longest possible clone. The polyproteins were expressed in the recombination-defective strain BLR(DE3) (Novagen), purified by Ni2+-affinity chromatography and kept in PBS/5 mM DTT.
Single-Protein AFM.
Our custom-made single-molecule AFM apparatus, as well as its mode of operation, is identical to those that we have recently described (11, 13, 22).
The AFM was constructed by using a Digital Instruments (Santa Barbara, CA) AFM detector head (AFM-689) mounted on top of a Physik Instrumente (Waldbronn, Germany) single-axis piezoelectric positioner. The positioner has a capacitative sensor resulting in a z-axis resolution of 0.1 nm (P-732.ZC). The data acquisition (force) and the voltage control of the movement of the piezoelectric positioner (Δzp; Fig. 1B) are done by means of a PC-mounted data-acquisition board (AT-MIO-16X; National Instruments, Austin, TX) and controlled by custom-made labview (National Instruments) software. The spring constant, kc, of each individual AFM tip (Si3N4 tips, Digital Instruments) is calibrated in solution before each experiment by using the equipartition theorem as described (23). kc varies between 30 and 120 mN/m. The force is measured by the deflection of the cantilever, and the extension is calculated from the travel of the piezoelectric actuator. To make single-protein measurements, the poly-I27 proteins were suspended in PBS buffer at a concentration of 10–100 μg ml−1 and allowed to adsorb onto freshly evaporated gold coverslips. Then, the coverslips were mounted onto the piezoelectric actuator of the AFM apparatus and brought into contact with the AFM tip to pick up a polyprotein.
Contour Length Measurements.
Fig. 1B shows a hypothetical sequence of events during the mechanical unfolding of a polyprotein. The first section shows an anchored but relaxed polyprotein composed of four Ig domains. Stretching first causes the protein to straighten to near its folded contour length, Lc0. Further stretching causes the unfolding of a domain and elongates the contour length to Lc1. This pulling sequence results in a characteristic saw-tooth pattern in the measured force-extension relationship (Fig. 1C). The force peaks observed in the force-extension curves obtained by AFM have been shown to correspond to true unfolding events of the individual protein domains (13). The increase in contour length observed on domain unfolding, Δ = Lc1 − Lc0, is a measure of the number of amino acids that were packed into the hidden core of the domain. However, contour lengths can be reached only at high force. Because unfolding is typically triggered at 100–300 pN of force, the actual contour lengths need to be estimated by using models of polymer elasticity.
The elasticity of a polypeptide chain is well described by the worm-like-chain (WLC) model that predicts the relationship between the extension of a polymer and the entropic restoring force generated (24, 25). The adjustable parameters of the WLC model are the persistence length, p, and the contour length of the polymer, Lc. The contour length counts only force-bearing amino acids, because those that are hidden within a protein fold do not contribute individually to generating the restoring force caused by the entropic drive to coil the polymer. Nonlinear fits of the WLC function to the force-extension curves that lead to each force peak showed that the WLC model adequately described the elasticity of the recombinant proteins (see Fig. 4A).
Figure 4.
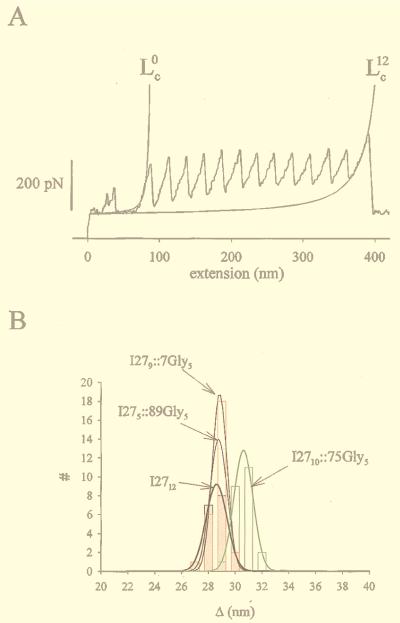
(A) Measurement of the contour length of an I2712 protein before (Lc0) and after (Lc12) unfolding of its 12 modules. The solid lines are fits of the WLC model to the data. We measured Lc0 = 92 nm (P = 0.57 nm) and Lc12= 436 nm (P = 0.29 nm) and calculate an elongation per module of Δ = (Lc12 − Lc0)/12 = 28.7 nm. The average values for the persistence length in the folded state were 0.87 ± 0.11 nm and 0.35 ± 0.02 nm in the unfolded state (n = 18). (B) Histogram of the contour length increment caused by the unfolding of a single module (Δ) measured for the wild-type and mutant polyproteins. Gaussian fits to each histogram are shown as solid lines.
Geometrical Errors in the Measurements of Contour Length.
Fig. 1B shows the simplest pulling geometry, where the polyprotein is pulled at a 90o angle with respect to the substrate. A more realistic pulling geometry is shown in Fig. 2. We assume that the polyproteins are initially relaxed and have a coiled end-to-end length l0 given by < l02 > ≈ 2pLc(1 − p/Lc), where Lc is the contour length and p is the persistence length (25). When picked up by the AFM tip, a polyprotein is stretched from its coiled length to near its folded contour length Lc at an angle θ1. Module unfolding extends the protein to a contour length Lc + Δ. The measured increase, Δm, is a function of θ1, according to:
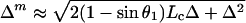 |
1 |
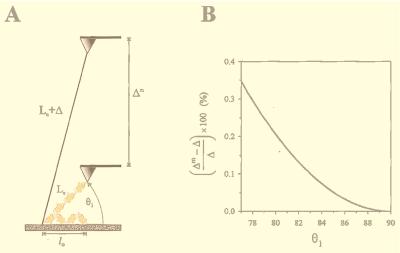
This equation is derived by using the law of cosines for an oblique triangle and assuming that the contour length increases because of unfolding and Δ is much larger than the initial folded length Lc (i.e., Δ ≫ Lc). When the protein is pulled at an angle of θ1 (θ1 < 90o), the measured increase in contour length, Δm, is always bigger than the actual change in contour length, Δ. This geometry introduces an error in the measurement of Δ. However, this error is small. For example, we calculate (Δm − Δ)/Δ as a function of θ1 for a typical polyprotein composed of 10 identical I27 domains. For a folded contour length Lc = 38 nm and a folded persistence length p = 1 nm, we estimate a coiled end-to-end length of l0 = 8.6 nm. This polyprotein expands by Δ = 280 nm because of the unfolding of all its modules. Under these conditions, the maximal error in the measurement of Δ is less than 1% (Fig. 2B) and occurs when the coiled protein starts out flat against the substrate (Fig. 2A). Under these conditions, the stretched but still folded protein will form an angle of at least θ1 = 77° with respect to the substrate. Unfolding of the modules always makes θ1 larger, approaching 90°. Because the proteins are randomly adsorbed within a layer that is 20- to 50-nm thick, it is very likely that their starting angles are bigger that the calculated worst case of θ1 = 77°. Polyproteins that are stretched from higher θ1 values have less error (Fig. 2B). When θ1 = 90°, the error is zero.
Figure 2.
Geometrical errors in the measurements of contour length. (A) A relaxed polyprotein with a length l0 is picked up by the AFM tip and stretched by force to a folded contour length Lc. The stretched polyprotein is pulled at an angle θ1. Module unfolding extends the protein to a contour length Lc + Δ. However, the measured increase, Δm, is a function of θ1. (B) We calculate this error as a function of θ1 for a typical polyprotein composed of 10 identical I27 domains with l0 = 8.6 nm; Lc = 38 nm and Δ = 280 nm. Under these conditions, the maximal error in the measurement of Δ is less than 1% and occurs at a θ1 = 77o. Molecules that are stretched from higher θ1 values have less error. When θ1 = 90°, the error is zero (see Materials and Methods).
RESULTS AND DISCUSSION
Length Mutations in the I27 Module.
Fig. 1A shows the structure of the I27 module and location of the five Gly insertion sites. The engineered insertions are placed either inside the folded domain (I27∷75Gly5, Fig. 1A), before the folded domain near the N terminus (I27∷7Gly5), or after the folded domain at the C terminus (I27∷89Gly5).
To examine the length phenotype of these insertions, we have constructed proteins made of identical tandem repeats of the wild-type I27 module (13) and of its mutant forms (see Materials and Methods). We use single-protein AFM techniques (10–13) to stretch these engineered polyproteins. Stretching a polyprotein with tandem repeats of an identical protein module generates force-extension curves with a characteristic saw-tooth pattern that amplifies the features of the module and allows for accurate measurements of its length (13).
Length Phenotypes of the I27 Module.
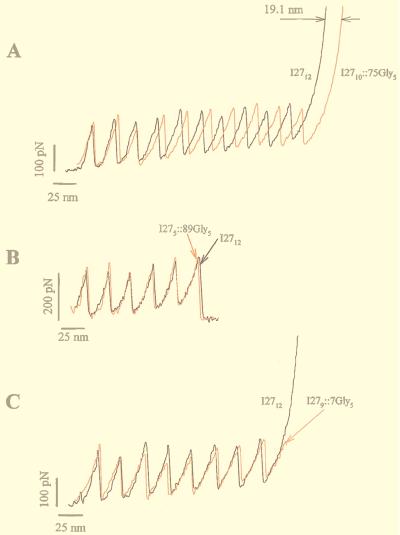
Fig. 3A shows a comparison between the force-extension curves of the wild-type (I2712) and an insertion mutant protein (I2710∷75Gly5). Stretching either polyprotein resulted in force-extension curves with equally spaced force peaks with an average value of 200 pN (Fig. 3A). The force-extension curves for both proteins are placed in register at the first unfolding peak. A difference in peak spacing is only slightly apparent after unfolding the first module; however, as more modules are unfolded, the force-extension curves of both proteins become increasingly out of register. Like in a moiré pattern, the two records are maximally out of phase after unfolding seven modules and should become in register again after unfolding 14 consecutive modules (not observed). After unfolding 10 modules, the I2710∷75Gly5 polyprotein is ≈19.1 nm longer than the I2712 polyprotein (measured at 600 pN; Fig. 3A). Hence, from this comparison it is evident that the folded core of the I2710∷75Gly5 protein is 19.1 Å per module longer than its wild-type form. The difference in length amounts to the five-Gly insert, because each amino acid contributes with 3.8 Å to the length of a polypeptide. By contrast, similar measurements made with the I279∷7Gly5 and I275∷89Gly5 mutants (i.e., with the five-Gly insert placed before or after the folded domains) showed force-extension curves displaying saw-tooth patterns that could be superimposed onto the wild-type I2712 polyprotein (Fig. 3 B and C). This means that these inserts contribute to the length of the folded polyprotein and do not contribute to the length of polypeptide exposed by unfolding domains.
Figure 3.
Amplification by polyprotein unfolding captures small changes in the length of a mutant I27 module. Insertion of five Gly residues into the FG loop of the I27 module increases the contour length of the unfolded module by 2 nm. (A) Comparison of the force-extension curves of a wild-type I27 polyprotein (I2712; black trace) and a mutant polyprotein, I2710∷75Gly5 (red trace). In both cases, domain unfolding increases the contour length of the proteins as they are extended. However, the I2710∷75Gly5 polyprotein extends more with each unfolding event. In the example shown, after 10 consecutive unfolding events the mutant polyprotein is 19.1 nm longer than the wild-type polyprotein (arrows), corresponding to a difference of 1.91 nm/module. (B and C) The saw-tooth pattern in the force-extension curves obtained from polyproteins engineered with five Gly inserts placed either in position 89 (I275∷89Gly5; red trace in B) or in position 7 (I279∷7Gly5; red trace in C) superimpose on the saw-tooth pattern obtained from the wild-type polyprotein (I2712; black trace in B and C).
These results demonstrate the remarkable precision attainable by combining amplification by polyproteins and AFM measurements. The advantages of polyprotein construction are evident here. By stretching a protein made of multiple repeats of an identical module, the mechanical features of the module are amplified linearly with the number of repeats. Higher-precision measurements will be possible with longer polyproteins.
Length of the Wild-Type and Mutant I27 Polyproteins Measured with the WLC Model of Elasticity.
The analysis shown in Fig. 3 is model independent and results directly from the measurements of distance obtained by the AFM. However, it is not always possible to compare pairs of force-extension curves with an equal number of unfolded modules, because the AFM tip picks up protein fragments of various lengths at random. Hence, to facilitate the analysis of these data, we also estimated the contour length of polyproteins by using the WLC model of polymer elasticity (24, 25). The force-extension curves of our polyproteins are well described by the WLC model, which predicts the entropic restoring force generated on the extension of a polymer. WLC fits to the force-extension curve that precede the first peak gave a measure of the contour length of the protein when all its modules are still folded, Lc0 (the upper index indicates the number of unfolded modules). Similarly, WLC fits to the force-extension curve before the last peak measures the contour length of the protein with all its modules in the unfolded state, Lcm, where m is the total number of modules in the protein segment. We applied this analysis to the force-extension curves obtained from the wild-type polyprotein I2712 (3 ≤ m ≤ 12; Fig. 4A). We calculate the increase in contour length caused by the unfolding of a wild-type I27 module as Δ = (Lcm − Lc0)/m = 28.1 ± 0.17 nm (3 ≤ m ≤ 12; n = 18; Fig. 4B). A similar analysis done on the I2710∷75Gly5 protein showed a larger Δ = 30.1 ± 0.14 nm (3 < m < 10; n = 23; Fig. 4B). The other mutant proteins, with the five-Gly insert before or after the folded domains, gave a value of Δ that was equal to that of the wild-type I27 (for I275∷89Gly5, Δ = 28.2 ± 0.13 nm, n = 23 and for I279∷7Gly5, Δ = 28.2 ± 0.12 nm, n = 27).
Hence, we observe that it is only the insertion of five Gly residues in the FG hairpin loop (Fig. 1A) that enlarges the size of the unfolded module by ΔΔ ≈ 2.0 nm, which corresponds to the length of the five inserted amino acids. By contrast, similar insertions near the N or at the C terminus leave Δ unchanged. Because these last two insertions do not change the value of Δ, they must be adding to the length of the linker region between modules and therefore would be expected to change the length of the folded polyprotein, Lc0. However, the measurements of Lc0 have a large error because these measurements are affected by the variability in the attachment point of the polyprotein to the gold substrate. By contrast, the differential measurement of Δ (Fig. 4A) is accurate to Ångstrom resolution.
Mechanical Topology of the I27 Module.
The measurements of the contour length changes observed during the extension of a polyprotein reveal the underlying mechanical topology of its repeating module. We group the amino acids of a folded I27 module in two categories: hidden amino acids that do not experience the applied force because they are mechanically isolated by the A′G patch, and force-bearing amino acids that support the applied force. The total length of the force-bearing amino acids corresponds to the contour length of the folded protein. By contrast, the hidden amino acids become exposed only after an unfolding event takes place and determine the contour length increment observed on unfolding, Δ. Our measurements readily distinguish between hidden and force-bearing amino acids and provide an accurate measurement of Δ, which reports the mutant length phenotype. Our results show that Δ = 28.1 nm, corresponding to ≈75 hidden amino acids. The I27 module is 89 aa long. In addition, we use a GLG linker, which makes the repeating unit of the polyprotein equal to 92 aa. Hence, in addition to the 75 “hidden” amino acids, there are 17 amino acids that are “force bearing” at all times. This distribution defines the mechanical topology of the folded modules of our I27 polyprotein.
CONCLUSIONS
It is likely that many mutations that target the Ig modules affect their mechanical properties. However, mechanical phenotypes in proteins have never been studied or observed before. Here we demonstrate a length phenotype that is sensitive to the precise location of the mutation with respect to the mechanical topology of an Ig module. Our results have a resolution comparable to that obtained by NMR and x-ray crystallography techniques. However, in contrast to these techniques, the AFM measurements are made on a single molecule, in real time, and in a standard saline solution. We anticipate that these techniques can be used to determine the mechanical topology of other protein modules and provide a framework to predict mechanical phenotypes that may play critical roles in understanding the molecular basis of many genetic diseases.
Acknowledgments
We thank H. Erickson for discussions and comments on the manuscript. We also thank W. J. Greenleaf and C. Badilla-Fernandez for their help in polyprotein engineering. This study was funded by National Institutes of Health and National Science Foundation grants to J.M.F., P.E.M. and A.F.O.
ABBREVIATIONS
- AFM
atomic force microscopy
- WLC
worm-like-chain
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Alon R, Chen S, Puri K D, Finger E B, Springer T A. J Cell Biol. 1997;138:1169–1180. doi: 10.1083/jcb.138.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon R, Hammer D A, Springer T A. Nature (London) 1995;374:539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 3.Lauffenburger D A, Linderman J J. Receptors. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 4.Erickson H P. Science. 1997;276:1090–1092. doi: 10.1126/science.276.5315.1090. [DOI] [PubMed] [Google Scholar]
- 5.Keller T C. Nature (London) 1997;387:233–235. doi: 10.1038/387233a0. [DOI] [PubMed] [Google Scholar]
- 6.Hynes R O. Proc Natl Acad Sci USA. 1999;96:2588–2590. doi: 10.1073/pnas.96.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher T A, Oberhauser A F, Carrion-Vazquez M, Marszalek P E, Fernandez J. Trends Biochem Sci. 1999;24:339–344. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- 8.Kellermayer M S, Smith S B, Granzier H L, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 9.Tskhovrebova L, Trinick J, Sleep J A, Simmons R M. Nature (London) 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 10.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 11.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 12.Rief M, Gautel M, Schemmel A, Gaub H E. Biophys J. 1998;75:3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrion-Vazquez M, Oberhauser A F, Fowler S B, Marszalek P E, Broedel S E, Clarke J, Fernandez J M. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohashi T, Kiehart D P, Erickson H P. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chothia C, Jones E Y. Annu Rev Biochem. 1997;66:823–862. doi: 10.1146/annurev.biochem.66.1.823. [DOI] [PubMed] [Google Scholar]
- 16.Bateman A, Jouet M, MacFarlane J, Du J S, Kenwrick S, Chothia C. EMBO J. 1996;15:6050–6059. [PMC free article] [PubMed] [Google Scholar]
- 17.Saffell J L, Walsh F S, Doherty P. J Cell Biol. 1994;125:427–436. doi: 10.1083/jcb.125.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenwrick S J. In: Ig Superfamily Molecules in the Nervous System. Sonderegger P, editor. Amsterdam: Harwood; 1998. pp. 287–303. [Google Scholar]
- 19.Improta S, Politou A S, Pastore A. Structure (London) 1996;4:323–337. doi: 10.1016/s0969-2126(96)00036-6. [DOI] [PubMed] [Google Scholar]
- 20.Improta S, Krueger J K, Gautel M, Atkinson R A, Lefevre J F, Moulton S, Trewhella J, Pastore A. J Mol Biol. 1998;284:761–777. doi: 10.1006/jmbi.1998.2028. [DOI] [PubMed] [Google Scholar]
- 21.Lu H, Isralewitz B, Krammer A, Vogel V, Schulten K. Biophys J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marszalek P E, Oberhauser A F, Pang Y-P, Fernandez J M. Nature (London) 1998;396:661–664. doi: 10.1038/25322. [DOI] [PubMed] [Google Scholar]
- 23.Florin E L, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy V T, Gaub H E. Biosens Biolelectron. 1995;10:895–901. [Google Scholar]
- 24.Marko J F, Siggia E D. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 25.Rivetti C, Walker C, Bustamante C. J Mol Biol. 1998;280:41–59. doi: 10.1006/jmbi.1998.1830. [DOI] [PubMed] [Google Scholar]