Abstract
Certain G-rich DNA sequences readily form four-stranded structures called G-quadruplexes. These sequence motifs are located in telomeres as a repeated unit, and elsewhere in the genome, where their function is currently unknown. It has been proposed that G-quadruplexes may be directly involved in gene regulation at the level of transcription. In support of this hypothesis, we show that the promoter regions (1 kb upstream of the transcription start site TSS) of genes are significantly enriched in quadruplex motifs relative to the rest of the genome, with >40% of human gene promoters containing one or more quadruplex motif. Furthermore, these promoter quadruplexes strongly associate with nuclease hypersensitive sites identified throughout the genome via biochemical measurement. Regions of the human genome that are both nuclease hypersensitive and within promoters show a remarkable (230-fold) enrichment of quadruplex elements, compared to the rest of the genome. These quadruplex motifs identified in promoter regions also show an interesting structural bias towards more stable forms. These observations support the proposal that promoter G-quadruplexes are directly involved in the regulation of gene expression.
INTRODUCTION
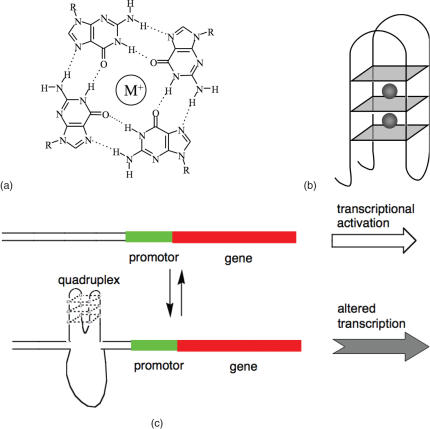
The nucleobase guanine (G) is capable of self-assembling to form hydrogen bonded motifs called G-tetrads [Figure 1a, (1–4)]. G-rich nucleic acid sequences that contain stretches of tandem Gs can form four stranded structures called G-quadruplexes that comprise stacked G-tetrads (Figure 1b). While the formation of G-quadruplexes in vitro has been known for several decades, (1) there has been considerable recent interest in the potential formation and function of such non-classical nucleic acid structures in biology. It is known that telomeres in many species can form G-quadruplexes (5–7) and it was recently found that telomeric quadruplex formation in vivo is regulated by telomere binding proteins, under the control of a cell-cycle dependent phosphorylation event (8). However, the role of quadruplexes outside telomeric regions is still unclear, although the quadruplex motif is prevalent in the human genome, (9,10) with an average incidence of ∼1 quadruplex every 10 000 bases.
Figure 1.
(a) Structure of a G-tetrad, showing hydrogen bonds and monovalent cation. (b) Schematic of an intramolecular G-quadruplex. G-tetrads are shown as blue squares, and monovalent cations as grey spheres. The structure shown is folded in an antiparallel conformation, with the strands of the G-tetrads alternately running up and down. (c) Model for transcription modulation via formation of a quadruplex in a promoter region.
Dynamic behaviour of the quadruplex motif in duplex DNA could be directly involved in gene regulation at the level of transcription (Figure 1c). This dynamic behaviour of G-quadruplex DNA has been studied in vitro for a single-stranded system (11,12). The possibility that this dynamism regulates gene activity was supported by the discovery of a G-quadruplex in the promoter of the chicken β-globin gene, (13,14) and the proposal has been directly investigated for the case of the c-myc protooncogene (15–17). Recently, a small number of quadruplexes have been located in promoter regions of other genes (13,15,16,18–23).
In the absence of general evidence for this hypothesis, our goal has been to analyse the relationship between promoters and G-quadruplex elements throughout the human genome. Using computational methods, we mapped out quadruplex motifs in gene promoters that may be involved in transcription regulation. Quadruplexes were found to be highly prevalent in human gene promoters. The analysis revealed evidence of evolutionary selection pressure to concentrate quadruplex elements in gene promoters and proximal to the transcription start sites (TSSs) of genes. Promoter quadruplexes were also found to be strongly associated with the open form of chromatinised DNA as judged by experimental genome-wide nuclease hypersensitivity data. Quadruplex loops are critical to the stability and folded structure, and promoter quadruplexes show an enrichment of stabilizing loops that promote a defined fold. We propose that quadruplex elements may be cis-acting regulatory elements for a large proportion (40%) of the genes in the human genome.
MATERIALS AND METHODS
Bioinformatic data
Promoter regions were defined using the ENSmart module of ENSEMBL, collecting the 5′ ends of every gene in the ENSEMBL database marked as ‘known’, using NCBI build 34. PQS positions were generated using the program quadparser (9), which searches for sequences of the form G3+N1−7G3+N1−7G3+N1−7G3+ (or the corresponding C-rich pattern). Loop-lengths were provided from quadparser. Where loop lengths could not be attributed to a PQS because it had an extremely long run of G, it was discarded for loop study purposes. Where PQS had more than three loops (because of having more than 4 runs of GGG), the 3′ end set of loops was used to avoid over-counting. All coordinates used NCBI build 34 of the human genome. NHS data were taken from the published work of Crawford et al. (24,25) and were generated from quiescent human CD4+ T cells. A total of 161 715 data points were provided, and 5158 clusters derived, each containing at least three data points within 500 bp. Sites where multiple nuclease cleavage points were identified in such close proximity were argued by Crawford et al. to have high reliability, and where such a cluster contained three or more cleavage points, they were over 80% likely to be valid.
Oligonucleotides
Oligonucleotides were purchased from Invitrogen (Paisley, UK). They were synthesized on a 50 nmol scale and supplied lyophilized. Stock solutions (50 μM) of the oligonucleotides were made with Milli-Q water. The sequences of the oligonucleotides used in these studies are shown in Table 2.
UV melting
UV melting curves were collected using a Varian Cary 1E UV-vis spectrophotometer, measuring the spectral absorbance at 295 nm, which has been previously identified as a key absorbance region for quadruplex formation (26). Samples were prepared at 4 μM in a buffer containing 100 mM KCl and 10 mM Tris–HCl (pH 7.4). These samples were then heat-annealed to 90°C and allowed to slowly cool to 4°C over a period of several hours. In a typical experiment 150 μl of a sample was degassed in a Speedvac for 3 min, transferred to a 1 cm path length quartz cuvette, and then covered with a layer of mineral oil (Sigma–Aldrich). It was then transferred to the spectrometer and equilibrated at 5°C for 10 min. It was then heated to 90°C and cooled to 5°C at 0.2°C/min, with data collection occurring every 0.5°C on both the annealing and melting steps. Tm values for the sequences were determined from alpha plots of the melting profiles following the method of Mergny (26). This analysis assumes simple two-state equilibrium between the folded and unfolded forms, with linear variations of absorbance with temperature for both species.
CD
CD experiments were performed on a Jasco J-810 spectropolarimeter using inbuilt software. Samples were prepared at 4 μM in a buffer containing 100 mM KCl and 10 mM Tris–HCl (pH 7.4). These samples were then heat-annealed to 90°C and allowed to slowly cool to 4°C over a period of several hours. In a typical experiment 400 μl of a sample was transferred to a 1 cm path length quartz cuvette, and scans were performed over the range 220–320 nm. Each trace is the result of the average of five scans at 50 nm/min, with a 2 s response time, 1 nm pitch and 1 nm bandwidth. The samples were left to equilibrate at 4°C for 10 min before the scans were performed. A constant flow of dry nitrogen ensured there was no condensation. A blank sample containing only buffer was treated in the same manner and subtracted from the collected data. For graphing purposes, the data were smoothed slightly, with each data point being graphed as the average of those around it and a zero correction being applied at 320 nm.
RESULTS AND DISCUSSION
We have previously mapped out putative quadruplex sequences (PQS) in the human genome by a computational methodology, which identifies motifs of the form G3+N1−7G3+N1−7G3+N1−7G3+ that we have shown are predisposed to the formation of intramolecular quadruplexes (9). As we show later in this paper, such in silico predicted motifs do form stable G-quadruplexes. To investigate the relationship between quadruplex formation and transcriptional regulation, we have generally defined putative promoter regions as being 1 kb upstream of the 5′ end of the TSS for a given gene, unless otherwise stated. Our study was carried out on the 19 268 known human genes in ENSEMBL (NCBI 34), so this definition covers ∼19.3 Mb of the genome. Within all these gene promoters, we identified 14 769 PQS, at an average density of 0.77 PQS/kb. Thus PQS are enriched by a factor of 6.4 in gene promoters as compared to the average throughout the whole genome (0.13 PQS/kbase) (Table 1). This is indicative of an evolutionary selection pressure for quadruplexes to occur within gene promoters.
Table 1.
PQS densities in promoters and near nuclease hypersensitive sites
| Location | PQS density (PQS/kbase) | Enrichment compared to genome average |
|---|---|---|
| Genomic DNA | 0.13 | 1× |
| 1 kb upstream of TSS | 0.77 | 6.1× |
| Near NHS cluster (500 bp) | 1.07 | 8.6× |
| Near cluster and in promoter | 28.6 | 230× |
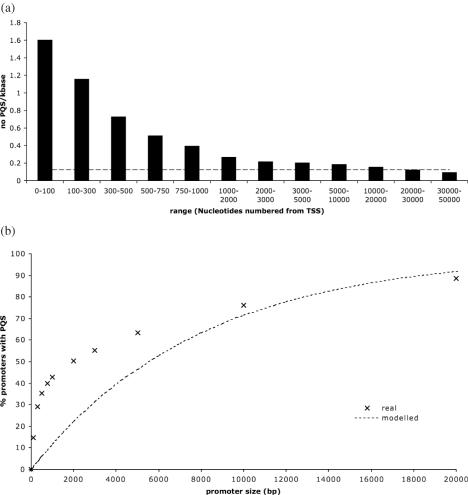
A second observation arising from this analysis is that 42.7% of the gene promoters contained at least 1 PQS. Within the promoters the probability of finding a PQS is directly related to its proximity with the TSS (Figure 2a). The highest concentration of PQS occurs very near the TSS, with the PQS density in the first 100 upstream bases being over 12 times higher that the genome average. There is a progressive reduction in PQS density away from the TSS until it decreases to below the genome average at distances remote from the TSS (>20 000 bases). Figure 2b shows the proportion of gene promoters that contain at least one quadruplex as the size of the promoter is varied, by increasing the distance from the TSS. The most rapid increase in the distribution occurs at much smaller promoter sizes, and for 29% of promoters at least one PQS exists within the first 300 bases upstream from the TSS. That PQS density is highest proximal to the TSS is consistent with quadruplex elements functioning as a topological switch mechanistically connected with transcription.
Figure 2.
(a) Density of PQS with distance upstream from the TSS. The genome as a whole has a density of 0.13 PQS/kb, shown by the dashed line. (b) The percentage of promoter regions containing at least one PQS increases as the size of the promoter increases. This increase is extremely fast over the first 1000 bases. The dashed line shows what percentage would be predicted if the density of PQS were equal to that across the genome as a whole.
We have also investigated which types of genes have such promoter PQS sequences. Investigating the promoters of 95 proto-oncogenes (27) showed that the have a significantly elevated frequency of having PQS, with 66 (69%) having at least one promoter PQS. We also examined the Gene Ontology (GO) codes of all promoters, to investigate, which categories showed significantly greater or lower frequencies of PQS promoters. GO categories which were significantly more likely to contain promoter PQS included those for transcription factor activity, development, neurogenesis and kinase activity. GO categories significantly less likely to have promoter PQS include olfaction, G-protein signalling, immune response, nucleic acid binding and protein biosynthesis. A full list of genes with significant deviations from chance (P < 1.25 × 10−4) is included in Supplementary Data.
We also considered the accessibility of DNA in these locations, and so looked at nuclease hypersensitivity data. Measurement of nuclease hypersensitivity is an experimental approach to identify regions within genomic DNA that have undergone a structural transition from the double helix to an unwound form that exposes single stranded DNA (28). Such a transition is required in gene promoters during transcription activation. Crawford et al. (24,25) (http://research.nhgri.nih.gov/DNaseHS/May2005/) have generated a genome wide map of DNase I hypersensitive sites (NHS) within nuclear chromatin, generated from quiescent human CD4+ T cells. We analysed this dataset to evaluate associations with quadruplex motifs, and also with promoter quadruplexes. Following Crawford et al. (24) we considered regions of the genome characterized by having three or more nuclease cleavage points, each separated by no more than 500 bp, which generated a dataset of 5158 NHS clusters. We computationally analysed the incidence of PQS within 500 bp of NHS clusters. Two-thirds (66.0%) of these NHS clusters were found to have at least one PQS, on either the plus or minus strand, within this range resulting in 8038 NHS-PQS motifs (a number of NHS clusters has more than one PQS). The mean density of PQS in NHS clusters was 1.07 PQS/kb, which is 8.6 times higher than the genome average (0.13 PQS/kb) (Table 1). Tightening the definition used for NHSs by reducing the separation between experimentally determined cleavage points tended to increase the density of PQS found. For example, using a 200 bp range for the NHE cluster gives 3204 clusters with 4271 PQS within 500 bp, an average density of 4.5 PQS/kbase. The same effect was observed when the maximum separation of PQS from NHS was reduced. Consideration of only the areas within an NHS cluster gives 3135 PQS at a mean density of 1.35 PQS/kb, greater than 10 times the genome average. Thus there is a strong association between nuclease hypersensitivity and the incidence of quadruplexes genome-wide.
By considering PQS that occur both within NHS clusters and 1 kb promoters, an even more pronounced PQS density might be expected if there is a functional role for G-quadruplexes in regulating transcription (Figure 1). A total of 94.5 kb of genomic DNA were found to be both within 500 bp of a NHS and within a 1 kb promoter. There were 2702 PQS elements within the promoter-NHS cluster regions giving a mean density of 28.6 PQS/kb. Thus, the density of PQS elements is 230 times higher within promoter-NHS cluster regions as compared to the average PQS incidence throughout the genome. This is a very high enrichment, greater than two orders of magnitude and also much greater than the product of the independent enrichments. This density of PQS (1 every 35 bases) is very high considering that a PQS must be at least 15 bases long. (see Table 1).
We have considered that NHS regions are rich in the normally rare CpG dinucleotide (29,30). We have shown that our observation of PQS enrichment was not simply caused by CpG enrichment. Theoretically, the presence of PQS is determined by the frequency of the ‘strong’ homodiads GpG and CpC (for the other strand), (9) and enrichment in competing homodiads would tend, if anything, to reduce the frequency of PQS formation. This was confirmed by analysis of the genomic data (see Supplementary Data).
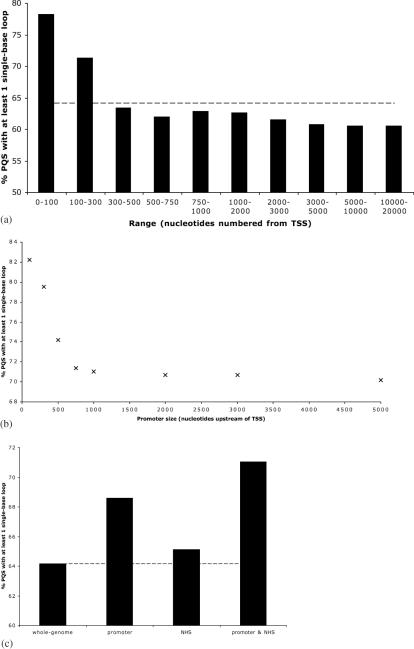
The length of the intervening loops between G tracts directly control the folded geometry and thermodynamic stability of quadruplexes. In particular, biophysical experiments (31,32) suggest that the presence of single-nucleotide loops is likely to increase the thermal stability of the folded quadruplex structure, a feature likely to be linked to biological function. Furthermore, a single nucleotide quadruplex loop must adopt a well defined geometry, called a double chain reversal that pre-disposes the quadruplex to adopt a parallel fold. (31) A number of structurally characterized promoter quadruplexes that occur within gene promoters have been found to contain one or more single-base loop (15,16,19–23). In the genome as a whole, 64% of PQS have at least one single nucleotide loop. However, in promoters the occurrence of single nucleotide loops in PQS is substantially higher. Of the 3087 PQS in the first 100 bases upstream of the TSS, 78% of them have at least one single-base loop (Figure 3a). This proportion gradually declines towards the genome average (64%) on moving away from the TSS. This suggests that proximal to the TSS, evolutionary selective pressure has favoured PQS with stabilizing loop lengths and a predisposition towards a parallel folded structure. Folding topology is important in the recognition of quadruplexes, as has been demonstrated for both protein (33) and small molecule (34) quadruplex ligands, because different surfaces are presented for recognition.
Figure 3.
(a) Percent of PQS identified in given ranges 5′ of a TSS (‘promoter regions’), which have at least one single-base loop. The genome wide average figure is 64% (dashed line). (b) Proportion of PQS upstream of a TSS and within 500 bp of an NHS cluster which have at least one single-base loop decreases rapidly with increasing promoter size. (c) Proportion of PQS with at least one single-base loop in bulk genomic DNA, within 1000 bp upstream of a TSS (promoter), within 500 bp of an NHS cluster, and in promoter/NHS cluster overlap regions. The whole-genome figure is shown as a dashed line.
A comparable analysis of single base loops for NHS cluster, and also for promoter regions that coincided with NHS clusters, revealed a higher incidence of single-base loops in these PQS (see Figure 3b and c). For the PQS that were both near NHS clusters and also within promoter regions, high-proportions were observed to have single-nucleotide loops. Using our standard definitions of NHS clusters (within 500 bp) and promoters (1000 bp upstream of the TSS), 71% of these PQS have single-nucleotide loops. If these definitions are tightened, by reducing the ranges in each case, then the proportions of single-base loops go up, so for example if NHS clusters are defined as being within 200 bp, and promoters only the first 300 bp, then 81% of the PQS identified have single-nucleotide loops. This suggests that selection pressures have favoured these highly-filtered PQS to be stable, and hence more likely to be folded into a quadruplex under physiological conditions.
In our analysis we have assumed that all PQS can form G-quadruplexes. To test this assumption, 10 sample PQS sequences were synthesized and studied using ultraviolet (UV) melting (26) and circular dichroism (CD) spectroscopy, (6,35,36), which are biophysical techniques widely employed for the validation of quadruplex folded structures. Two sets of PQS were characterized. One set (Table 2) consisted of five sequences with a large number of nearby NHE cleavage positions (>12 within 500 bp), and the other set (Table 3) comprised five sequences in the promoter region of known genes (defined as 1000 bp upstream of the TSS), with three or more NHE cleavage positions within 50 bp (a tight definition given the limited range), and with some evidence of conservation in Mus musculus. The sequences were selected to include three bases at the 5′ and 3′ ends to eliminate artefacts resulting from having these ends proximal to the quadruplex itself. The sequences vary considerably in their base patterns, including the length of the putative loops, the number of GGG repeats and other parameters. The oligonucleotides were annealed under pseudo-physiological conditions [100 mM KCl and 10 mM Tris–HCl (pH 7.4)], heated to 90°C and allowed to slow cool.
Table 2.
Experimental results for PQS with frequent NHEs
| Name | No NHEa | Sequence | Tm (/°C) |
|---|---|---|---|
| 20h | 20 | tgtGGGGTCGGGGGAGGGGGGAGGGata | 71 |
| 14h1 | 14 | tgtGGGGTAGGGGGAGGGGGGAGGGata | 71 |
| 14h2 | 14 | gccGGGGCGGCTCGGGACGGGGCCCGGGGAGCGTGGGTGGGacc | 75 |
| 14h3 | 14 | cccGGGACGGGGGCCGGCGGGCCACGGGccc | n/o⊥ |
| 12h | 12 | gctGGGCGAGGGGTGGGAGCAGACGGGctg | 57 |
aNumber of NHE cleavage positions reported within 500 bp of the PQS midpoint ⊥ n/o signifies that no quadruplex melting transition was observed. A melting transition at 70°C was observed at 260 nm for this sequence.
Table 3.
Experimental results for Promoter PQS with frequent NHEs
| Name | No NHEa | Sequence | Tm (/°C) |
|---|---|---|---|
| GUK1 | 3 | agcGGGAGAAGACGGGCTGGGAGGGcgc | 52 |
| MCM2 | 3 | gaaGGGACACGGAGGGGCGGGCCAGAGGGtcc | 64 |
| NKFB2 | 3 | tcaGGGTGGGGGCCCCGAGGGCTGGGGccg | 69 |
| PSMA2 | 4 | gctGGGTTGGGGCGGGGGGAGCGGGacg | 69 |
| UTX | 3 | gccGGGCGGGGAGGGGGGGtca | >80°Cb |
Gene names: GUK1, Guanylate kinase 1; MCM2, DNA replication licensing factor; NKFB2, DNA-binding factor KBF2, Lyt10; PSMA2, Proteasome subunit alpha type 2; UTX, Ubiquitously transcribed X chromosome tetratricopeptide repeat protein.
aNumber of NHE cleavage positions reported within 50 bp of the PQS.
bThis sequence was further studied in 20 mM KCl and showed a complete melt with a Tm of 66°C.
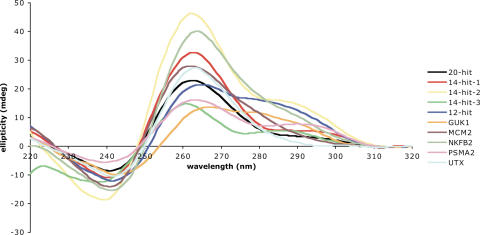
There are two classical CD traces described in literature for quadruplex structures, (35,36) corresponding to two distinct folding patterns-parallel (with all guanines anti), which has a peak at 260 nm and a trough at 240 nm, and anti-parallel (with guanines alternating syn and anti), which has a peak at 295 nm and a trough at 260 nm. As shown in Figure 4, all the sequences studied showed a peak at 260 nm (parallel), and many also showed some amount of peak at 295 nm, suggesting partial formation of anti-parallel structures. This polymorphism is frequent for intramolecular quadruplex structures, and has been observed before. UV-melting studies (26) can reveal the characteristic hypochromic transition at 295 nm that quadruplexes exhibit upon melting, and were also performed for these sequences (Tables 2 and 3).
Figure 4.
CD spectra of the sequences individually characterized in this study. Peaks at 260 nm indicate a parallel folded quadruplex, whereas peaks at 295 nm indicate an anti-parallel fold.
Nine of the ten PQS studies yielded both CD and UV data, which gave unambiguous support for quadruplex formation, supporting the algorithm used to predict these sequences.
CONCLUSIONS
We have addressed the quadruplex-promoter hypothesis on a human genome-wide scale revealing a number of striking observations. Quadruplexes are enriched in promoters with their probability density reaching a peak proximal to the TSS. Promoter quadruplexes are prevalent in the human genome, with >40% of the annotated genes containing one or more promoter quadruplexes. Quadruplexes are also enriched at nuclease hypersensitive sites, and quadruplexes at both nuclease hypersensitive regions and promoters are enriched by a factor of 230, compared to the rest of the genome. These multiple associations support the hypothesis that quadruplexes play an important role in gene regulation throughout the genome. This also opens up opportunities for novel chemical intervention strategies, especially given recent developments in small molecules that target the G-quadruplex structural motif.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Greg Crawford for helpful discussions and guidance, and Ashok Venkitaraman, Christine Bird and Caroline Wright for critical reading of the manuscript. J.L.H. is a Research Fellow at Trinity College, Cambridge. S.B. is a BBSRC Career Development Research Fellow. Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonsson T. G-Quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 3.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 4.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA:sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 6.Balagurumoorthy P., Bramachari S.K. Structure and stability of human telomeric sequence. J. Mol. Biol. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 7.Henderson E., Hardin C.C., Walk I., Tinoco I., Jr, Blackburn E.H. Telomeric DNA oligonucleotides form intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 8.Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nature Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 9.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd A.K., Johnstone M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying L.M., Green J.J., Li H.T., Klenerman D., Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.Y., Okumus B., Kim D.S., Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodford K.J., Howell R.M., Usdin K. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 1994;269:27029–27035. [PubMed] [Google Scholar]
- 14.Howell R.M., Woodford K.J., Weitzmann M.N., Usdin K. The chicken β-globin gene promoter forms a novel ‘cinched’ tetrahelical structure. J. Biol. Chem. 1996;271:5208–5214. doi: 10.1074/jbc.271.9.5208. [DOI] [PubMed] [Google Scholar]
- 15.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grand C.L., Han H., Muñoz R.M., Weitman S., Von Hoff D.D., Hurley L.H., Bearss D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 18.Howell R.M., Woodford K.J., Weitzmann M.N., Usdin K. The chicken β-Globin gene promoter forms a novel ‘cinched’ tetrahelical structure. J. Biol. Chem. 1995;271:5208–5214. doi: 10.1074/jbc.271.9.5208. [DOI] [PubMed] [Google Scholar]
- 19.Rankin S., Reszka A.P., Huppert J.L., Zloh M., Parkinson G.H., Todd A.K., Ladame S., Balasubramanian S., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernando H., Reszka A.P., Huppert J.L., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai J., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun D., Guo K., Rusche J.J., Hurley L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids. Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Armond R., Wood S., Sun D., Hurley L.H., Ebbinghaus S.W. Evidence for the presence of a Guanine quadruplex forming region within a polypurine tract of the Hypoxia Inducible Factor 1R promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 24.Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D., et al. 2005. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergny J.-L., Phan A., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 27.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross D.S., Garrard W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 29.Cross S.H., Bird A.P. CpG islands and genes. Curr. Opp. Gen. Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 30.Wolf S.F., Migeon B.R. Clusters of CpG dinucleotides implicated by nuclease hypersensitivity as control elements of housekeeping genes. Nature. 1985;314:467–469. doi: 10.1038/314467a0. [DOI] [PubMed] [Google Scholar]
- 31.Hazel P., Huppert J.L., Balasubramanian S., Neidle S. Loop-length dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 32.Risitano A., Fox K.R. Influence of loop size on the stability of intramolecular G-quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaffitzel C., Berer I., Postberg J., Hanes J., Lipps H.J., Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seenisamy J., Rezler E.M., Powell T.J., Tye D., Gokhale V., Joshi C.S., Siddiqui-Jain A., Hurley L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 35.Balagurumoorthy P., Brahmachari S.K., Mohanty D., Bansal M., Sasiskharan V. Hairpin and parallel quartet structure for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giraldo R., Suzuki M., Chapman L., Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7562. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.