Abstract
Expression of selenocysteine (Sec)-containing proteins requires the presence of a cis-acting mRNA structure, called selenocysteine insertion sequence (SECIS) element. In bacteria, this structure is located in the coding region immediately downstream of the Sec-encoding UGA codon, whereas in eukaryotes a completely different SECIS element has evolved in the 3′-untranslated region. Here, we report that SECIS elements in the coding regions of selenoprotein mRNAs support Sec insertion in higher eukaryotes. Comprehensive computational analysis of all available viral genomes revealed a SECIS element within the ORF of a naturally occurring selenoprotein homolog of glutathione peroxidase 4 in fowlpox virus. The fowlpox SECIS element supported Sec insertion when expressed in mammalian cells as part of the coding region of viral or mammalian selenoproteins. In addition, readthrough at UGA was observed when the viral SECIS element was located upstream of the Sec codon. We also demonstrate successful de novo design of a functional SECIS element in the coding region of a mammalian selenoprotein. Our data provide evidence that the location of the SECIS element in the untranslated region is not a functional necessity but rather is an evolutionary adaptation to enable a more efficient synthesis of selenoproteins.
INTRODUCTION
Selenocysteine (Sec)-containing proteins serve important oxidoreductase functions in a variety of organisms in all three domains of life (1). Although the mechanism of Sec synthesis and incorporation into nascent peptide chains has been well characterized in bacteria, a much different strategy for selenoprotein biosynthesis has evolved in archaea and eukaryotes (2,3). To support Sec insertion into nascent peptide chains, bacterial selenoprotein mRNA has to contain cis-acting elements in the form of a UGA codon and a stem–loop structure immediately downstream, which allows recoding of the codon from stop to Sec insertion (4). This secondary structure is known as Sec insertion sequence (SECIS) element (5). In bacteria, the essential region for Sec insertion is located in the apical loop of the SECIS element, which includes the binding site for a specialized elongation factor SelB. SelB in turn recruits Sec tRNA and mediates elongation at the in-frame UGA by competing with a release factor (3). As a result, the process of Sec incorporation is relatively slow and inefficient.
In archaea and eukaryotes, a different mechanism has evolved, which has puzzled researchers for the past 15 years. Most strikingly, the essential cis-acting SECIS element is not located immediately downstream of the corresponding UGA codon but has evolved in the 3′-untranslated regions (3′-UTRs) of the selenoprotein-encoding mRNAs (6). The location of the SECIS element immediately downstream of UGA stalls the ribosome complex during elongation thereby increasing efficiency of Sec insertion (3,7). The 3′-UTR location requires a different mechanism for the access of Sec tRNA to the ribosomal A-site (8,9). A kink-turn structure located in the stem-region of the SECIS element is the binding site of SECIS-binding protein 2 (SBP2) (8). However, in eukaryotic SECIS elements additional nucleotides have been identified in the stem region as being essential for the binding of SBP2, a protein belonging to the ribosomal protein L7Ae/L30e/S12e/Gadd45 family (10). Mutations in this region have been shown to abrogate selenoprotein synthesis, indicating that SBP2 is an essential factor for Sec insertion. In eukaryotes, a Sec tRNA-specific elongation factor (EFSec) has been identified which is a functional homolog of the bacterial SelB gene product and has been found to be recruited by and to interact with SBP2 (2,11). In addition, archaeal and eukaryotic genomes code for several other factors implicated in selenoprotein synthesis which are described elsewhere (12).
Selenoproteins have been conserved throughout evolution, most likely because of their unique physico-chemical properties as compared with their sulfur-containing homologs. However, identification of these proteins in sequence databases is challenging due to recognition of Sec-encoding UGA codons as stop signals by sequence annotators. Recently, bioinformatics tools have been developed that detect selenoprotein genes by searching for SECIS elements, cysteine-containing homologs and coding nature of UGA codons (13–15). Application of these tools identified full sets of selenoproteins in many prokaryotic and several eukaryotic organisms (15–18).
In 1998, a widely recognized study reported that even a virus has taken advantage of a selenoprotein by acquiring the human gene for glutathione peroxidase 1 (GPx1) in order to protect itself and the host cell from environmental stress (19). To date, no additional scientifically proven Sec-containing proteins have been identified in viral genomes. Here, we applied sensitive computational approaches to characterize viral selenoproteomes. Three SECIS elements have been detected in the searches. Interestingly, a fowlpox virus structure was found to be residing in the coding region of a GPx4 homolog. We demonstrated that mammalian cell lines support the expression of in-frame SECIS element-containing mRNAs of both viral and mammalian origin. This study reports for the first time that the location of the SECIS element in the 3′-UTR is not a functional necessity but rather an adaptation to the more complex translation mechanism of higher organisms. The results should be of interest for future studies investigating the mechanism of selenocysteine insertion.
MATERIALS AND METHODS
Databases and programs
Nucleotide sequences of viral genomes were downloaded from NCBI (http://ncbi.nlm.nih.gov/) using Batch Entrez. Human genome sequences and non-redundant protein sequences (ftp://ftp.ncbi.nih.gov/genbank/) were also obtained from NCBI. SECISearch was used for the identification of candidate SECIS elements (20). BLAST and FASTA packages were used for similarity searches (21).
Identification of homologs of known selenoprotein genes
A full set of known eukaryotic selenoproteins, including human selenoproteins and several selenoproteins with restricted occurrence (i.e. Chlamydomonas MsrA, Gallus gallus SelU and protein disulfide isomerase from Emiliania huxleyi) were used as query sequence (20,22–24). A stand-alone version of TBLASTN program was utilized for the detection of nucleotide sequences corresponding to known selenoprotein families. In addition, all known prokaryotic selenoproteins, including recently discovered selenoprotein families from Sargasso Sea organisms were used in the searches (20,25).
Searches for SECIS elements
A loose pattern of SECISearch with relaxed settings was utilized. A step-by-step description of the search procedure is as follows:
Analysis of primary nucleotide sequence. PatScan was used to search for the NTGA__AA__GA pattern, which was optimized for SECIS elements as described previously (20,26). Additional requirements included (a) the distance between the Quartet (NTGA) and the unpaired AA/CC in the apical loop of 10–13 nt; (b) length of the apical loop without the unpaired AA/CC of 6–23 nt; (c) no more than one insertion, one deletion and two mismatches in the stem preceding the unpaired AA/CC; and (d) the presence of an additional stem upstream of the Quartet.
Prediction and analysis of secondary structure. Each SECIS candidate was examined for consistency with the eukaryotic SECIS consensus model. Additional filters excluded Y-shaped SECIS elements and SECIS elements with >2 consecutive unpaired nucleotides.
Estimation of the free energy for each SECIS candidate structure. Using RNAfold from Vienna RNA package, the free energies for the whole structure and the upper stem–loop were calculated, with the threshold value of −12.6 kcal/mol for the former and −3.7 kcal/mol for the latter (27). Only thermodynamically stable structures were further considered.
Protein identification. This step includes analyses of SECIS location and identification of open reading frames (ORFs). The purpose was to filter out SECIS candidates located on the complementary strand.
The final step included sequence analyses of predicted ORFs to identify candidate Sec-encoding TGA codons.
Design of in-frame SECIS elements
Design of in-frame SECIS elements was carried out using the following procedure:
Nucleotide sequences of mouse GPx1 and GPx4 were analyzed with RNAfold for occurrence of stem–loop structures resembling SECIS elements in the overall shape and conservation of functional regions. The best candidates located downstream of the Sec-encoding TGA codons were selected in each case.
Reverse-translation of the corresponding coding regions using ambiguous nucleotide codes was carried out.
Minimal differences between the existing nucleotide sequences and the sequences that satisfied eukaryotic SECIS consensus were identified using SECISearch.
Necessary mutations were subsequently introduced by site-directed mutagenesis to generate structures with high SECISearch scores.
Cloning strategies
All constructs used in this study were generated using pEGFP-N1 (Clontech, Mountain View, CA). Initially, a V5 epitope encoding sequence, including an ATG initiation codon, was cloned into the XhoI and HindIII sites. Next, the vector was cut with HindIII and AflII, which removed the EGFP ORF and the vector-encoded polyadenylation signal. PCR products for mouse genomic GPx1, mouse GPx4 cDNA and fowlpox GPx containing endogenous 3′-UTRs including poly(A) signals were cloned into the HindIII and AflII sites. Control constructs lacking functional SECIS elements were generated by site-directed mutagenesis by changing the conserved Quartet region ATGA to AAAA and/or the apical loop region AA to TT. These changes are indicated in the manuscript as ΔATGA and ΔAA, respectively. The construct GPx1 ΔTAA was generated by deleting the TAA stop signal. The construct GPx1 FPV SECIS was generated by introducing a BamHI restriction site immediately after the stop codon. This construct was subsequently cut with BamHI and AflII to remove the GPx1 3′-UTR. The fowlpox virus SECIS element-encoding region and 3′-UTR were amplified with PCR primers containing BamHI (and one additional nucleotide in frame with the GPx1 ORF) and AflII restriction sites and cloned into the corresponding sites of the vector. In the final step, the stop codon and the BamHI restriction site were deleted by site-directed mutagenesis. Constructs GPx1 1.IFS, GPx1 2.IFS and GPx4 IFS (for In-Frame SECIS) were generated by disrupting the SECIS element in the 3′-UTR by site-directed mutagenesis; in addition, changes in the sequences were introduced to stabilize the structures as shown in Figure 6A and B. All Cys mutants used in this study were also generated by site-directed mutagenesis. Constructs containing a functional SECIS element coding for sequences corresponding to the N-terminal portion of an ORF were generated by cloning the ORF containing the fowlpox virus SECIS region into the NheI and XhoI sites of pEGFP-N1. The sequence containing V5 tag and GPx1 ORF with disrupted 3′-UTR SECIS element was cut from the control construct GPx1 ΔATGAΔAA with HindIII and AflII and pasted downstream of the fowlpox virus SECIS sequence (Figure 8B). In order to be in frame with the downstream region, the forward primer for the fowlpox virus SECIS contained an ATG initiation codon and one additional nucleotide. Modified constructs of this type were generated by deleting the HindIII restriction site and the ATG codon of the V5 tag by site-directed mutagenesis. These modifications resulted in higher expression levels, likely by preventing alternative translation initiation. Primer sequences are available on request.
Cell culture, transfections and metabolic labeling
Mouse hepatoma cell line Hepa 1–6 and human embryonic kidney HEK293 cells (ATCC, Manassas, VA) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin and 100 U/ml streptomycin. Transfections were carried out in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. An aliquot of 4 μg cDNA per well was used in a ratio of 1:2 in OptiMEM (Invitrogen, Carlsbad, CA) for 5 h. The transfection medium was replaced with regular culture medium and cells were harvested after an additional 48 h. Metabolic labeling was done by supplementing the cell culture medium with 75Se (specific activity 1000 Ci/mmol; Research Reactor Facility, University of Missouri, Columbia, MO) with 2.5 μCi/ml for 48 h, without the addition of unlabeled selenium.
SDS–PAGE and Western blot analyses
Forty-eight hours post-transfection, the cells were washed three times with phosphate-buffered saline (PBS) and lysed in 300 μl lysis buffer (Sigma–Aldrich, St Louis, MO), and 1/10 volume was electrophoresed on NuPage 10% polyacrylamide gels and transferred on to polyvinylidene difluoride membranes (Invitrogen, Carlsbad, CA). Membranes were then incubated with mouse monoclonal anti-V5 antibody (1:5000) (Invitrogen, Carlsbad, CA) and horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (1:10 000). After washing, the membranes were incubated with chemiluminescent peroxidase substrate-1 (Sigma–Aldrich, St Louis, MO) and exposed to an X-ray film. For the detection of metabolically labeled selenoproteins, the PVDF membrane was exposed to a PhosphorImager screen which was then scanned using a Storm PhosphorImager system (GE Healthcare, formerly Amersham Biosciences, Piscataway, NJ).
RESULTS
Computational analysis of viral genomes for selenoprotein genes
To identify viral selenoprotein genes, we initially scanned all available viral sequences in the NCBI database against representatives of all known prokaryotic and eukaryotic selenoprotein families using TBLASTN. The search was carried out to identify ORFs having in-frame UGA codons corresponding to Sec in known selenoprotein sequences. Only two selenoproteins, both members of the glutathione peroxidase family, were found. One of the detected selenoproteins, from Molluscum contagiosum virus, has been described previously (19). It is essentially a typical eukaryotic selenoprotein with the SECIS element located in the 3′-UTR. Homology and phylogenetic analyses revealed close similarity to human GPx1, consistent with acquisition of this gene from the host [(19) and data not shown].
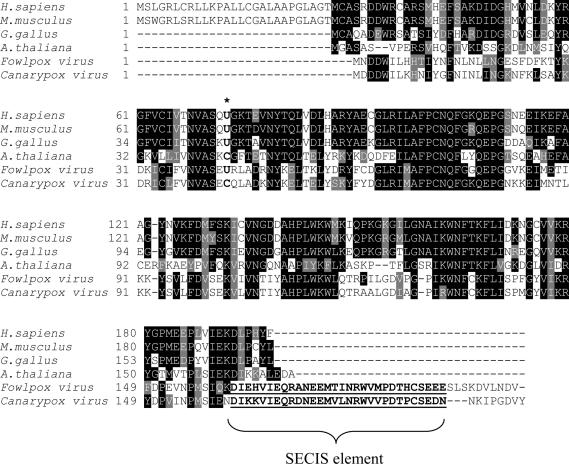
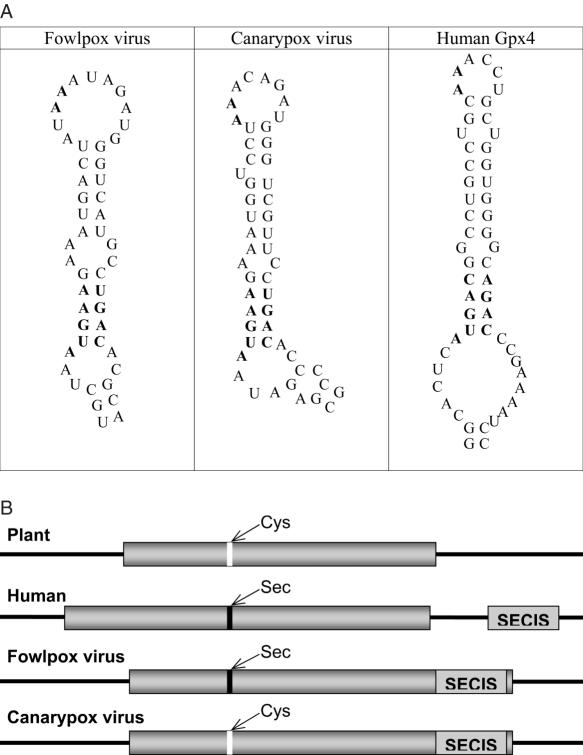
The second selenoprotein gene was detected in fowlpox virus and was most homologous to vertebrate GPx4, which belongs to a different glutathione peroxidase family (Figure 1 and Supplementary Figure S1). Thus, the fowlpox virus selenoprotein gene was acquired from the host independent of the M.contagiosum virus GPx1. Although the fowlpox virus selenoprotein gene had an in-frame UGA in the appropriate position that coded for Sec, SECISearch analysis revealed no candidate SECIS element in the 3′-UTR. Instead, a strong candidate SECIS structure was detected in the coding region (Figure 2 and Supplementary Figure S2). This structure satisfied the eukaryotic SECIS consensus, including the conserved SECIS core (Quartet of non-Watson–Crick base pairs) and the conserved AA motif in the apical loop.
Figure 1.
Amino acid sequence alignment of glutathione peroxidases of the GPx4 family. Accession numbers of the sequences used in the alignment are as follows: NP_566128.1 (Arabidopsis thaliana), AAH22071.1 (Homo sapiens), NP_032188.3 (Mus musculus), AAM18080.2 (Gallus gallus), AAR83433.1 (canarypox virus), CAE52609.1 (fowlpox virus). In-frame SECIS elements in the C-terminal regions of viral proteins are underlined. Location of catalytic Sec and the corresponding Cys is indicated by an asterisk.
Figure 2.
SECIS elements in viral glutathione peroxidases. (A) SECIS structures. Left structure, fowlpox virus GPx4 SECIS element. Middle structure, canarypox virus GPx4 SECIS element. Right structure, H.sapiens GPx4 SECIS element. SECIS core and unpaired AA nucleotides in apical loop are shown in boldface. (B) Location of viral GPx4 SECIS elements within mRNAs and comparison with GPx4 genes from other sources. Cys-containing GPx4 homologs from higher plants lack a SECIS element. In the human Sec-containing GPx4, SECIS element is in the 3′-UTR. In the fowlpox virus Sec-containing GPx4 homolog, SECIS element is the in the coding region. In the canarypox virus Cys-containing GPx4, a fossil SECIS element is in the coding region. Location of Cys is shown in white and Sec in black. SECIS elements are indicated by light-gray boxes and ORFs by dark-gray boxes. Left and right black lines represent 5′- and 3′-UTRs, respectively.
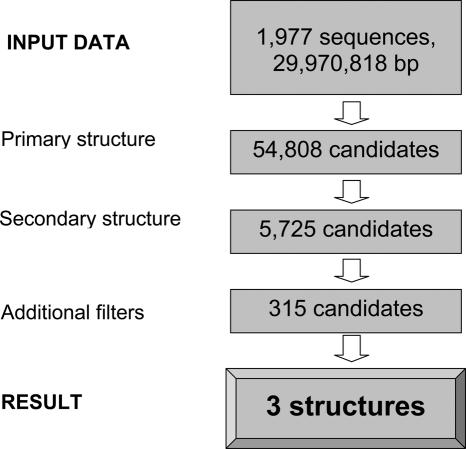
In addition to BLASTN analyses of viral genomes, we searched these genomes for SECIS elements. A stand-alone version of SECISearch was utilized as described in Materials and Methods. A relatively small size of the query sequence (i.e. combined sequences of all viral genomes; the full list of viral genomes is provided in the form of Supplementary Table 1) allowed us to use relaxed parameters in the searches. Results of the search are shown in Figure 3. After the final step, all candidate SECIS elements could be filtered out except for three structures. Two of them corresponded to fowlpox virus GPx4 and M.contagiosum virus GPx1 described above. The third one, while satisfying all criteria of the SECIS model, was detected in the coding region of canarypox virus GPx4 (Figure 2), a protein highly homologous to fowlpox GPx4. Thus, provided that sequences deposited into the sequence database are correct, the canarypox virus SECIS element is likely a fossil structure that remained in the GPx4 gene following the conversion of Sec to Cys in the protein. Importantly, two independent methods that searched for (i) homologs of known selenoproteins and (ii) SECIS elements, arrived with the same set of two selenoprotein genes in the scanned viral genomes.
Figure 3.
Analysis of viral genomes for selenoprotein genes. Details of the search are provided in Materials and Methods. The search of 1977 viral sequences revealed three SECIS element structures.
Expression of mammalian selenoproteins using in-frame SECIS elements
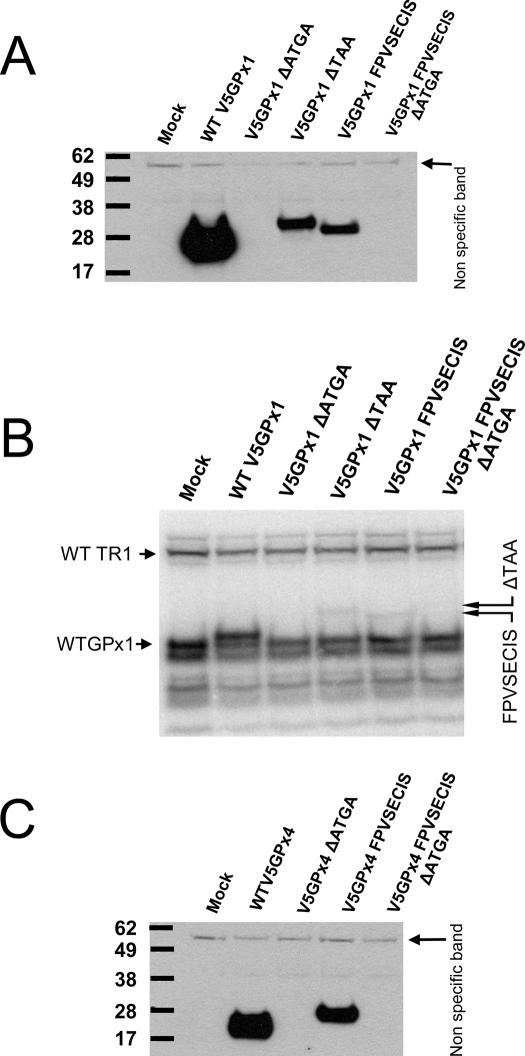
To test the surprising finding of the SECIS element within the ORF in our computational screen, we first examined whether mammalian selenoprotein transcripts can be expressed utilizing a coding region SECIS element. Several constructs were generated based on the mouse GPx1 gene, containing the endogenous intron (thereby subjecting it to the spliceosomal machinery), as well as mouse GPx4 cDNA. In the case of GPx1, deletion of the TAA stop codon allowed a theoretical readthrough until after the SECIS element since no additional in-frame stop codons were present in the message upstream of the SECIS element. In addition to deleting the endogenous stop codon, a construct was generated wherein the 3′-UTR of GPx1 was replaced with the sequences containing the fowlpox virus GPx4 SECIS element (including the viral 3′-UTR). In the latter construct, the stop codon was deleted by site-directed mutagenesis, thereby fusing the viral-encoded sequence within the mammalian ORF. Control constructs containing mutations within the SECIS region were also generated. In the case of mouse GPx4, the viral SECIS was fused within the mouse GPx4 ORF such that the coding SECIS region replaced non-homologous sequences of the mouse GPx4 gene. Again, control constructs with mutations in the conserved SECIS regions were also generated. As shown in Figure 4A and C, all selenoproteins encoded by transcripts containing in-frame SECIS elements could be expressed in mammalian cells, although at lower levels compared with the constructs expressing wild-type proteins. Mutations in the SECIS core region, however, abrogated Sec insertion completely. In a separate experiment, the transfected cells were metabolically labeled with 75Se to directly observe Sec insertion into proteins. As shown in Figure 4B, 75Se insertion was clearly evident in proteins derived from GPx1 and having either mammalian or fowlpox SECIS in the coding region (see lanes GPx1 ΔTAA and GPx1 FPV SECIS). Consistent with the Western blot data, wild-type GPx1 could be expressed more efficiently than the corresponding proteins containing SECIS elements in the coding region and the mutation in the SECIS core disrupted Sec insertion.
Figure 4.
Eukaryotic SECIS elements located in coding regions are functional. (A) Western blot analysis of HEK293 cells transfected with GPx1 constructs containing functional in-frame SECIS elements. The indicated constructs were generated by either deleting the stop codon TAA or replacing the GPx1 3′-UTR with the fowlpox virus SECIS element thereby generating a GPx1-FPV fusion protein. (B) The same samples as shown in A were metabolically labeled with 75Se and exposed to a PhosphorImager cassette. Location of selenoproteins expressed with the help of coding region SECIS elements is shown by arrows on the right. (C) Western blot analysis of HEK293 cells transfected with GPx4 constructs containing in-frame SECIS elements. GPx4-based constructs were generated by replacing non-homologous sequences with the fowlpox SECIS element thereby generating a GPx4-FPV fusion protein. Lane designations are as follows: Mock, control transfection; WT GPx1, wild-type GPx1 gene; GPx1 ΔATGA, mouse GPx1 gene with disrupted SBP2-binding region; GPx1 ΔTAA, mouse GPx1 gene with its natural stop codon deleted (allowing the ORF to extend beyond the SECIS element which becomes part of the ORF); GPx1 FPV SECIS, the fowlpox SECIS element replacing the 3′-UTR region of the mouse GPx1 gene; GPx1 FPV SECIS ΔATGA, the fowlpox virus SECIS was fused in frame with GPx1 and its SBP2-binding site was disrupted; WT GPx4, cells were transfected with wild-type mouse GPx4 cDNA; GPx4 ΔATGA, the SBP2-binding site in the wild-type GPx4 cDNA was disrupted; GPx4 FPV SECIS, the fowlpox virus SECIS element was fused in frame with GPx4 lacking its own 3′-UTR; GPx4 FPV SECIS ΔATGA, the fowlpox virus SECIS fused to GPx4 was mutated to disrupt the SBP2-binding site.
Mammalian cells support expression of the fowlpox GPx4 gene
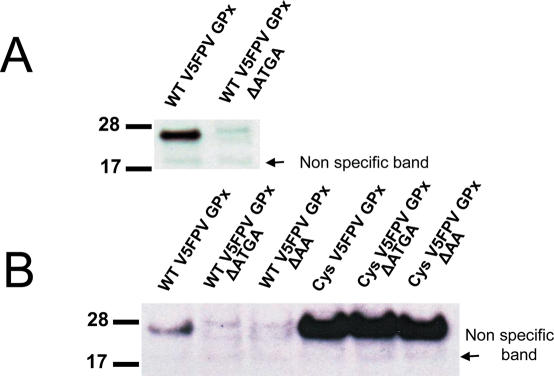
We further tested if the wild-type fowlpox GPx4 gene containing the in-frame SECIS element can be expressed in mammalian cells. In addition to the wild-type construct, a corresponding construct coding for a Cys-containing mutant and several control constructs containing mutations within the SECIS element were generated. Mammalian Hepa1-6 (mouse hepatoma cell line) (Figure 5A) and HEK293 (human embryonic kidney cell line) cells (Figure 5B) supported Sec insertion into fowlpox virus GPx4. Mutations within conserved regions of the SECIS element, either within the conserved Quartet core region that binds SBP2 or within the apical loop previously shown to be important for Sec insertion, abrogated expression of the selenoprotein. However, compared with endogenous or recombinant mammalian selenoproteins, the level of expression of fowlpox virus GPx4 was very low. Surprisingly, even the Cys-containing mutant of this protein did not reach the expression level of Cys mutants of mammalian selenoproteins expressed in transfected cells (Figure 5B). The low expression level of the viral GPx4 in transfected mammalian cells precluded detection of this selenoprotein by metabolic labeling of cells with 75Se.
Figure 5.
Expression of wild-type fowlpox virus GPx4 in mammalian cell lines. (A) Western blot analysis of mouse hepatoma cell line Hepa1-6 transfected with wild-type fowlpox virus GPx4 construct (WT FPV GPx) or the corresponding construct in which the SBP2-binding site was disrupted (WT FPV GPx ΔATGA). (B) Transfection of HEK293 with wild-type fowlpox virus GPx4 (FPV GPx), its mutants lacking SBP2-binding site (WT FPV GPx ΔATGA) or the conserved AA in the apical loop (WT FPV GPx ΔAA), and the corresponding Sec-to-Cys mutants.
Expression of mammalian selenoproteins using designed in-frame SECIS elements
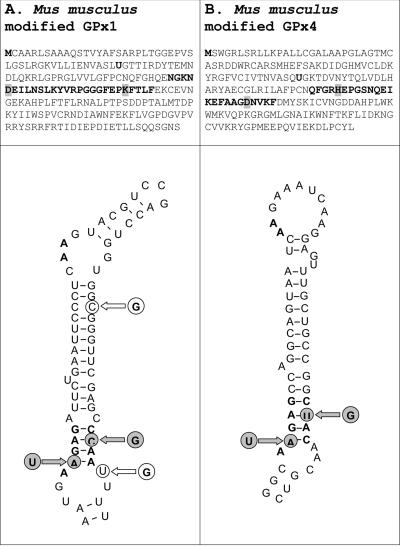
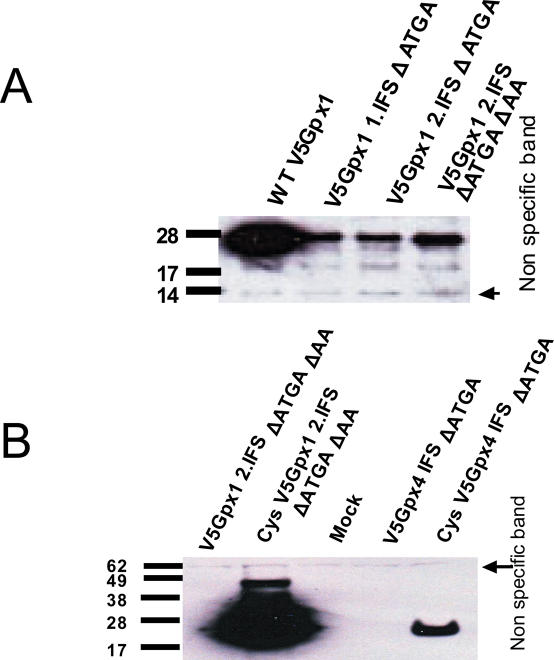
Occurrence of SECIS elements within coding regions may result in significant sequence restrictions as the same mRNA segments would code for polypeptide and be engaged in stem–loop structure (i.e. efficient Sec insertion by the coding region SECIS element may need to be balanced by the use of appropriate amino acid residues encoded by the SECIS sequence that support selenoprotein function). To examine this situation, we attempted to design SECIS elements within coding regions of endogenous mammalian selenoprotein genes, again choosing GPx1 and GPx4 genes as model selenoproteins. Following the de novo computational design procedure described in Materials and Methods and selection of candidate structures, single nucleotide exchanges were introduced into GPx1 and GPx4 expression constructs, resulting in structures within the ORFs resembling SECIS elements as shown in Figure 6A and B, respectively. The mutations resulted in two amino acid changes in GPx1 (GPx1 1.IFS), as well as two amino acid changes in GPx4 (GPx4 IFS). In the case of GPx1, a construct was further modified to improve the stem of the structure (GPx1 2.IFS). To inactivate the native SECIS element in the 3′-UTR of the GPx1 gene, the constructs were further modified to disrupt the SBP2-binding site, the conserved sequences in the apical loop, or both of these regions. As shown in Figure 7A, all three GPx1 constructs containing the coding region SECIS element expressed protein products in Hepa 1–6 cells. Although the expression levels were low compared with the construct expressing wild-type GPx1, the data clearly showed that the designed coding region SECIS element supported readthrough at the in-frame UGA codon. In the case of GPx4, however, no protein was detectable when the construct containing the designed SECIS element in the coding region was expressed, suggesting that the designed structure, while closely mimicking the SECIS element consensus, was not sufficient for efficient Sec insertion.
Figure 6.
Design of SECIS elements in the coding regions of mammalian selenoprotein genes. (A) Designed GPx1 SECIS element. (B) Designed GPx4 SECIS element. The first amino acids (methionine), Sec, and the SECIS elements are shown in boldface. The amino acids that were mutated to accommodate the designed SECIS elements are highlighted. The newly introduced residues are shown by gray circles, and further changes in the designed GPx1 SECIS are shown by white circles.
Figure 7.
Expression of GPx1 containing a designed in-frame SECIS element. (A) Hepa1-6 cells were transfected with WT GPx1 and different in-frame SECIS element containing constructs. The construct GPx1 2.IFS ΔATGA contains an additional modification in the stem-region as shown in Figure 6A. To disrupt the function of the natural SECIS element, mutations were made in either the SBP2-binding region or the apical loop. (B) Transfection of HEK293 cells. No expression was detectable for GPx4 containing a designed in-frame SECIS element. Note the low expression level of the Cys mutant with the designed in-frame SECIS element compared with GPx1. Expressed proteins were detected by western blot analysis using antibodies specific for V5 tag.
We further generated Cys mutants of GPx1 and GPx4 that contained designed SECIS elements in the coding region. Interestingly, while in the case of GPx1 the expression level was increased as expected for Cys-containing mutants of selenoproteins, expression of the GPx4-based Cys mutant was relatively low (Figure 7B).
Expression of mammalian selenoproteins using upstream SECIS elements
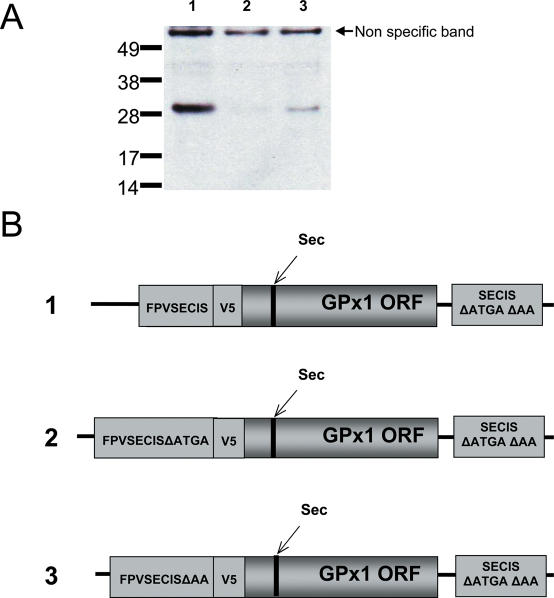
The data described above suggest that the location of SECIS element is not as much a functional necessity but rather a matter of efficient Sec insertion. To further examine the function of SECIS elements located in coding regions, we tested whether SECIS elements could function if present upstream of the Sec-encoding UGA. We generated the constructs wherein the fowlpox virus SECIS element was cloned upstream of the wild-type GPx1 gene containing an initiation codon plus one additional nucleotide to preserve the reading frame of GPx1. In addition, the constructs contained a V5 tag to allow detection of the expressed proteins, which was cloned between the fowlpox virus SECIS element and GPx1 ORF. The sequence coding for the tag contained another translation initiation ATG codon, which was deleted. This modification allowed higher levels of expression suggesting that this ATG could be used as an alternative initiation codon. As shown in Figure 8A, readthrough at the in-frame TGA codon was detectable when a functional SECIS element was located upstream of the Sec codon. Since the use of the control construct that had mutations in the SECIS did not result in detectable protein, it is possible that the upstream SECIS could also support Sec insertion. However, since the expression level was low, the use of 75Se was not feasible. Therefore, definitive conclusions on whether this SECIS inserted Sec or only supported readthrough could not be made. Interestingly, GPx1 could be detected at low levels when the apical loop of the upstream coding SECIS element was mutated from AA to TT. Expression of the protein from the constructs, in which the SECIS element was in the 5′-UTR (i.e. absence of the ATG in the fowlpox virus SECIS region and translation initiation at V5 tag) could not be detected (data not shown).
Figure 8.
An upstream SECIS element allows readthrough at in-frame UGA codons. (A) Constructs were generated in which the fowlpox SECIS element was cloned upstream and in-frame with the ORF of GPx1 separated by a V5 tag. Control constructs contained mutations in either the SBP2-binding region or in the conserved loop region. (B) Schematic illustration of the constructs used in (A). Numbers on the left correspond to lane designations in A.
Computational analysis of the human genome for in-frame SECIS elements
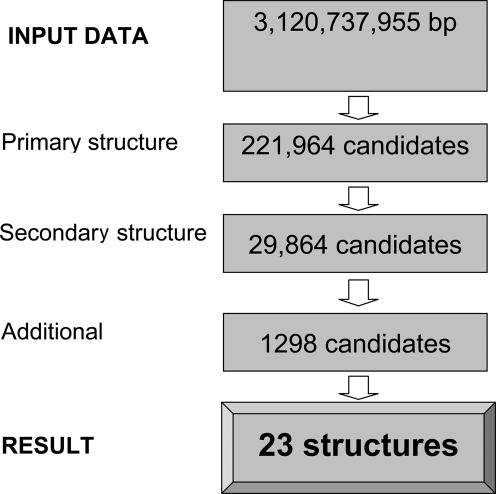
We have previously analyzed human and mouse selenoproteomes for SECIS elements and selenoprotein genes (20). However, this search focused on SECIS elements in the 3′-UTR. The data that SECIS elements could also function if present in the coding regions raised the possibility that additional selenoprotein genes might exist in mammals. Therefore, we carried out an additional search of the human genome for SECIS elements, extending the search to the coding regions (Figure 9). While known selenoprotein genes could be efficiently identified by this procedure, no selenoprotein genes with coding SECIS elements were identified. Thus, the use of the coding SECIS element appears to be unique to viral selenoproteins.
Figure 9.
Computational search of the human genome for selenoprotein genes containing SECIS elements in the coding region. A total of 23 selenoprotein genes could be detected using this procedure; however, no additional selenoprotein genes containing SECIS elements in the coding region were found.
DISCUSSION
Viruses are known for their ability to acquire new genomic information by integrating host-derived genes. In 1998, this was shown for a homolog of human GPx1 of M.contagiosum, a virus known to cause skin neoplasms in immunocompromised humans. M.contagiosum belongs to the family of poxviridae (19). This protein still remains the only known true viral selenoprotein. In this study, we carried out the most comprehensive computational analysis of currently available viral genomes. Following the analysis of 1977 viral sequences (29 970 818 bp), we identified three SECIS elements, including the formerly identified GPx1 homolog. Surprisingly, detailed analysis of the other candidates revealed an unusual location of the SECIS element within the ORF of a fowlpox selenoprotein gene. In contrast, the M.contagiosum gene had the SECIS element in the 3′-UTR, similar to its mammalian counterpart. Interestingly, the two new SECIS element-containing genes also belonged to the glutathione peroxidase family; however, both were derived from the GPx4 subfamily. Both candidates have also evolved in avian pox viridae, i.e. fowlpox virus and canarypox virus (28,29). Recombinant fowlpox virus is currently being investigated as a promising candidate vector in the search for a suitable delivery system of HIV antigens and other vaccines, since it is not known to cause disease in humans (30,31). However, in chickens it results in skin lesions, or in a more devastating diphteric form causing inflammation of the upper respiratory tract. Infections with fowlpox virus result in considerable economic loss for the poultry industry (28).
Although both new SECIS candidates were present within the coding region, currently available sequencing data suggest that only the fowlpox transcript encodes a Sec-containing protein, while the canarypox virus-encoded transcript codes for a Cys-containing homolog (29). Thus, it appears that there was a recent mutation of Sec to Cys codon in canarypox virus. Occurrence of a fossil SECIS element indicates that the GPx4 selenoprotein gene was first acquired from the host and recently converted to the Cys form. The fact that at least two selenoproteins are encoded by viral genomes suggests that these proteins provide substantial advantage for viruses. In mammals, GPx4 is an essential protein (32). Similar to GPx1, the fowlpox virus GPx4 may provide survival benefits for the virus, either within or outside of its host.
The mechanism of eukaryotic selenoprotein synthesis is not fully understood. While in recent years much information has been added to our understanding of how Sec is incorporated into nascent peptide chains, several key observations are still not explained. For example, it is not clear why the mechanism of Sec insertion is different in prokaryotes, archaea and eukaryotes. Our study shows for the first time that mammalian cell lines support the expression of selenoproteins using an in-frame SECIS element as is the case in prokaryotes. However, the level of expression decreases dramatically compared with the SECIS present in the 3′-UTR. Perhaps, binding of SBP2 or other SECIS-binding proteins to the overall structure interferes with the rate of translation elongation. Since secondary RNA structures are generally melted by helicases, the melting would inhibit association of SECIS element with its binding partners thereby impeding Sec insertion. Evolution of SECIS elements in the 3′-UTR allows linearization of the ORF while preserving the functional structure in the 3′-UTR. Although bacterial SECIS elements are located in the coding regions, they have to be close to the Sec codon because of the coupling between transcription and translation.
Thus, it seems that the 3′-UTR location of SECIS elements in eukaryotes is a novelty that was possible due to separation of these processes in eukaryotes (i.e. transcription in the nucleus and translation in the cytosol). In this regard, the use of the coding region SECIS in fowlpox may be due to cytosolic replication of this virus. Whereas coding region SECIS elements could support Sec insertion in other selenoprotein genes, the SECIS elements are maintained in the 3′-UTRs due to evolutionary pressure that maximizes efficiency of Sec insertion.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
Acknowledgments
We thank Drs W. M. Schnitzlein and D. N. Tripathy, University of Illinois at Urbana-Champaign, IL for providing fowlpox DNA, Dr D. L. Rock, Plum Island Animal Disease Center, Greenport, NY for his advice, and Dr D. L. Hatfield for comments on the manuscript. This work is supported by NIH GM061603 (to V.N.G.). Funding to pay the Open Access publication charges for this article was provided by GM061603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hatfield D.L., Gladyshev V.N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry M.J., Tujebajeva R.M., Copeland P.R., Xu X.M., Carlson B.A., Martin G.W.,, III, Low S.C., Mansell J.B., Grundner-Culemann E., Harney J.W., et al. Selenocysteine incorporation directed from the 3′UTR: characterization of eukaryotic EFsec and mechanistic implications. Biofactors. 2001;14:17–24. doi: 10.1002/biof.5520140104. [DOI] [PubMed] [Google Scholar]
- 3.Driscoll D.M., Copeland P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 4.Bock A., Rother M., Leibundgut M., Ban N. Selenium metabolism in prokaryotes. In: Hatfield D.L., Berry M.J., Gladyshev V.N., editors. Selenium: Its molecular biology and role in human health, 2nd edn. Springer; 2006. pp. 9–28. [Google Scholar]
- 5.Low S.C., Berry M.J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 6.Berry M.J., Banu L., Chen Y.Y., Mandel S.J., Kieffer J.D., Harney J.W., Larsen P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′-untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher J.E., Copeland P.R., Driscoll D.M. Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA. 2000;6:1573–1584. doi: 10.1017/s1355838200000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher J.E., Copeland P.R., Driscoll D.M., Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 9.Kinzy S.A., Caban K., Copeland P.R. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 2005;33:5172–5180. doi: 10.1093/nar/gki826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatfield D.L., Carlson B.A., Xu X.M., Mix H., Gladyshev V.N. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 13.Guigo R. Computational gene identification: an open problem. Comput. Chem. 1997;21:215–222. doi: 10.1016/s0097-8485(97)00008-9. [DOI] [PubMed] [Google Scholar]
- 14.Lescure A., Gautheret D., Krol A. Novel selenoproteins identified from genomic sequence data. Methods Enzymol. 2002;347:57–70. doi: 10.1016/s0076-6879(02)47008-5. [DOI] [PubMed] [Google Scholar]
- 15.Kryukov G.V., Gladyshev V.N. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lescure A., Gautheret D., Carbon P., Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 17.Castellano S., Morozova N., Morey M., Berry M.J., Serras F., Corominas M., Guigo R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra G., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigo R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shisler J.L., Senkevich T.G., Berry M.J., Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279:102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 20.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novoselov S.V., Rao M., Onoshko N.V., Zhi H., Kryukov G.V., Xiang Y., Weeks D.P., Hatfield D.L., Gladyshev V.N. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellano S., Novoselov S.V., Kryukov G.V., Lescure A., Blanco E., Krol A., Gladyshev V.N., Guigo R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obata T., Shiraiwa Y. A novel eukaryotic selenoprotein in the haptophyte alga Emiliania huxleyi. J. Biol. Chem. 2005;280:18462–18468. doi: 10.1074/jbc.M501517200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Fomenko D.E., Gladyshev V.N. The microbial selenoproteome of the Sargasso Sea. Genome Biol. 2005;6:R37. doi: 10.1186/gb-2005-6-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dsouza M., Larsen N., Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 27.Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonso C.L., Tulman E.R., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of fowlpox virus. J. Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tulman E.R., Afonso C.L., Lu Z., Zsak L., Kutish G.F., Rock D.L. The genome of canarypox virus. J. Virol. 2004;78:353–366. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coupar B.E., Purcell D.F., Thomson S.A., Ramshaw I.A., Kent S.J., Boyle D.B. Fowlpox virus vaccines for HIV and SHIV clinical and pre-clinical trials. Vaccine. 2006;24:1378–1388. doi: 10.1016/j.vaccine.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher A.D., Puls R.L., Bebbington M., Boyle D., Ffrench R., Kent S.J., Kippax S., Purcell D.F., Thomson S., Wand H., Cooper D.A., Emery S. A randomized, placebo-controlled phase I trial of DNA prime, recombinant fowlpox virus boost prophylactic vaccine for HIV-1. Aids. 2006;20:294–297. doi: 10.1097/01.aids.0000199819.40079.e9. [DOI] [PubMed] [Google Scholar]
- 32.Yant L.J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J.G., Motta L., Richardson A., Prolla T.A. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.