Abstract
Highly accurate knockdown functional analyses based on RNA interference (RNAi) require the possible most complete hydrolysis of the targeted mRNA while avoiding the degradation of untargeted genes (off-target effects). This in turn requires significant improvements to target selection for two reasons. First, the average silencing activity of randomly selected siRNAs is as low as 62%. Second, applying more than five different siRNAs may lead to saturation of the RNA-induced silencing complex (RISC) and to the degradation of untargeted genes. Therefore, selecting a small number of highly active siRNAs is critical for maximizing knockdown and minimizing off-target effects. To satisfy these needs, a publicly available and transparent machine learning tool is presented that ranks all possible siRNAs for each targeted gene. Support vector machines (SVMs) with polynomial kernels and constrained optimization models select and utilize the most predictive effective combinations from 572 sequence, thermodynamic, accessibility and self-hairpin features over 2200 published siRNAs. This tool reaches an accuracy of 92.3% in cross-validation experiments. We fully present the underlying biophysical signature that involves free energy, accessibility and dinucleotide characteristics. We show that while complete silencing is possible at certain structured target sites, accessibility information improves the prediction of the 90% active siRNA target sites. Fast siRNA activity predictions can be performed on our web server at http://optirna.unl.edu/.
INTRODUCTION
It is a major challenge to select those target sites where a gene can be silenced most completely. Posttranscriptional regulation can silence tens of thousands of genes to different degrees (1). This indicates that whereas a wide spectrum of target sites responds to RNA interference, the knockdown remains incomplete for most of the sites. Opposing this diversity criterion, active siRNAs have to conform to requirements specific for the RNA-induced silencing complex (RISC) complex (2). As indicated by the 62% average activity of randomly selected siRNAs (3), these criteria are poorly satisfied by the majority of target sites. This paradox has inspired a number of researchers to capture these criteria in heuristic rules, statistical formulations or machine learning algorithms. Tuschl and his coworkers' rules (2,4) (http://www.rockefeller.edu/labheads/tuschl/sirna.html) specify a pattern of UU(N19)AA, limit the G + C content to a range of 30–70%, and suggest avoiding four or more consecutive A's or U's that act as terminator signals in vectors that utilize RNA polymerase III. Ui-Tei et al. (5) expressed preference for siRNAs with A/U at the 5′ end, G/C at the 3′ terminus at least 5 A/U nucleotides in the 5′ third of the antisense strand, and the absence of any G/C runs of 9 or more nucleotides. Amarzguioui and Prydz (6) propose an A/U differential between the 5′ and 3′ trinucleotides, C/G at position 1, A at 6 and A/U at 19, while associating the motifs U1 and G19 with lack of functionality. Translating these sequence patterns to changes in Gibbs free energy (ΔG) shows that most sequence rules correlate highly with thermodynamic profiles (7). In contrast to the wider acceptance of the above rules, the effects of secondary structures at the target site remain debated (2). While certain structures like stable hairpins have been shown to decrease or abolish silencing efficiency (8–10), many other structures do not seem to attenuate RNAi.
Machine learning methods select the best targets more accurately than the heuristic rules. Key to this success is rigorous optimization over high numbers of features. Support vector machines (SVMs) (11) perform accurate binary classifications (BCs) between low- and high-activity molecules and regression analyses (12) and helped to formulate the Stockholm rules (12). Long and degenerate sequence patterns are revealed by the GPboost genetic algorithm (13). Among the artificial neural networks, BIOPREDsi (1) was trained on the largest number of siRNAs, but the method was limited to undisclosed sequence features. Shabalina et al. (14) neural network model generated position-dependent consensus patterns from a smaller number of molecules by using both sequence and thermodynamic features. Unfortunately, these patterns remain to be disclosed.
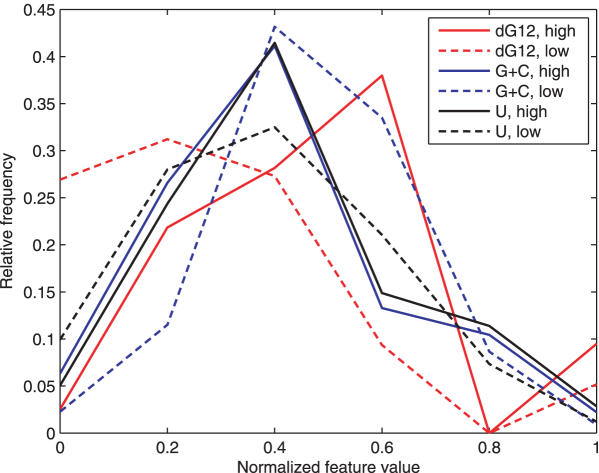
Here we present a practical, freely accessible and transparent tool for the identification of target sites with over 90% knockdown activity. Our work is based on two postulates. First, we expected that optimal selection from a significantly more comprehensive set of initial features may lead to the discovery of a complex and probabilistic signature. In turn, the signature(s) may lead to more sensitive and selective predictions. That Holen (15) needed to apply as many as 73 positional mononucleotide occurrence rules in order to achieve reliable predictions is evidence to support this postulate. We have compiled the possible most comprehensive set of 572 sequence, thermodynamic and accessibility features as further direct evidence. Global and positional mono- and dinucleotide frequencies, the number of longer runs of each nucleotide, C or G, or A or U were computed. Global and positional values of ΔG and change in enthalpy (ΔH) and entropy (ΔS) as well as the ΔH/ΔS ratio were calculated. Multiple predictors of the target site accessibility were computed (see Table 1; Supplementary Table S1 and Materials and Methods). Each of these individual features were correlated to the activities of the 2252 siRNAs in the Novartis dataset (1) (see Materials and Methods). No Pearson correlation coefficient exceeded r = 0.38 and only 15 features have r ≥ 0.2 or r ≤ −0.2 (Table 2). Several of these latter features represent the same phenomenon. For example, the decreased stability at the 5′ terminus of the antisense strand is represented in free energy, enthalpy, mono- or dinucleotide features, such as selection against extreme negative free energy, and GG, CC, GC and CG dinucleotides. The inferior performance of individual features is an even more serious issue. This performance is measured by the large overlaps in feature distributions between ≥90% and ≤80% active siRNAs (Figure 1). Because previous machine learning methods (1,13,14,16) used considerably less representative sets of features, significant improvements can be expected from their 86% prediction accuracy. This level is not satisfactory; even when applying multiple siRNA species, the risk of incomplete silencing remains substantial. However, to train a new method using 572 features over only 2252 siRNAs in the Novartis dataset would have led to overtraining; i.e. inferior performance on independent test sets. To avoid that, we applied constrained optimization models and SVMs for the optimal selection of a considerably smaller subset of features with the highest combined predictive value. We accomplished this objective by iteratively solving the models below with a stepwise elimination of the feature(s) using different methods. The comparability of diverse features was ensured by standardization to zero mean and unit SD.
Table 1.
Overview of the 572 sequence, thermodynamic and accessibility features of the siRNAs
| Both global and positional features: |
| • ΔG, ΔH and ΔS during the transition from double-stranded to single-stranded state of the RNA (18); |
| • The ratio ΔH/ΔS as above; |
| • Average probabilities of target site positions to form secondary structures (mono-, di- and tetranucleotides); |
| • G + C content. |
| Global features covering the complete antisense strand: |
| • ΔG during complex formation between the siRNA and the target mRNA; |
| • Relative frequencies of mono- or dinucleotides; |
| • Relative frequencies of homotri- and tetranucleotides; |
| • Maximal length of the G/C runs; |
| • Minimal free energy of the secondary structures at the mRNA target site; |
| • Melting temperature of the double-stranded siRNA; |
| • The probability and ΔG of forming a self-hairpin; |
| • Position of the target locus at the mRNA relative to the translation initiation site; |
| • Concentration of the siRNA. |
| Features specific to each position of the antisense strand: |
| • Presence or absence of mono- and dinucleotides; |
| • Presence of G or C mononucleotides; |
| • Probability of the target site positions to form secondary structures; |
| • Change in free energy during complex formation between the siRNA and the target mRNA. |
Both global and positional features were used. SVM and constrained optimization methods performed the iterative selection of the most predictive features shown in Table 2 and Supplementary Table S1.
Table 2.
The predictive performance of features
| Predictive performance | |||||
|---|---|---|---|---|---|
| Individuala | Combinedb | ||||
| Feature | Position | r | Feature | Position | Weight |
| ΔG | 1–2 | 0.38 | ΔG | All | 0.146 |
| U | 1 | 0.36 | CC | All | −0.134 |
| G | 1 | −0.31 | p3 | All | −0.128 |
| ΔH | 1–2 | 0.30 | U | 1 | 0.109 |
| ΔS | 1–2 | 0.27 | ΔH | 18–19 | −0.107 |
| U | All | 0.26 | A | 19 | −0.099 |
| ΔG | All | 0.25 | G | 1 | −0.094 |
| UU | 1 | 0.23 | UU | 18–19 | −0.086 |
| G | All | −0.22 | ΔH | 20–21 | 0.084 |
| ΔH | All | 0.22 | U | 2 | 0.068 |
| ΔΔG | 3–5 − 19–21 | 0.21 | A | 2 | 0.066 |
| ΔΔG | 1–3 − 19–21 | 0.21 | AU | 6–7 | −0.063 |
| ΔS | All | 0.21 | AA | 17–18 | −0.059 |
| GG | 1 | −0.20 | GG | 20–21 | 0.058 |
| GC | 1 | −0.20 | AA | 18–19 | −0.056 |
| UA | All | 0.18 | AU | 9–10 | −0.055 |
| U | 2 | 0.17 | ΔG | 3–4 | 0.055 |
| C | 1 | −0.17 | C | 1 | −0.054 |
| GG | All | −0.17 | GG | 16–17 | −0.053 |
| ΔΔG | 1–5 − 17–21 | 0.17 | CG | 1–2 | −0.052 |
| ΔG | 18 | −0.17 | AG | 20–21 | 0.052 |
| ΔG | 13 | 0.17 | G | 14 | −0.050 |
| ΔG | 2 | 0.17 | UG | 4–5 | −0.049 |
| GC | All | −0.16 | A | 20 | −0.047 |
| CC | All | −0.16 | UG | 20–21 | 0.046 |
| UU | All | 0.16 | CC | 13–14 | −0.044 |
| CG | 1 | −0.16 | GU | 5–6 | 0.040 |
| A | 19 | −0.16 | A | 1 | 0.039 |
| ΔH/ΔS | All | −0.15 | CC | 20–21 | −0.036 |
| CC | 1 | −0.15 | U | 7 | 0.035 |
Weights were optimized by an SVM with linear kernel. The absolute value of the weight indicates the contribution of that feature to the prediction in the linear kernel limited to 30 features. Note that the practical predictions use 142 features, shown in Supplementary Table S1 online. p3 is the probability of that each base of the tetranucleotide (i, i + 1, i + 2, i + 3) is paired as predicted by the sfold algorithm.
aThe 30 features with the strongest correlations to siRNA activity in the Novartis dataset.
bFeatures that in combination account for the most accurate predictions of the siRNA knockdown activity.
Figure 1.
Overlap between the distributions of ΔG at positions 1 and 2, global G + C content and U content between siRNAs with >90% (full-lines) and <80% activity (dotted lines) in the Novartis dataset (1).
MATERIALS AND METHODS
The comparability of the conditions of RNAi experiments underlying the prediction methods has to be ensured. Only experiments with a single siRNA species are useful to us since it is difficult to discern the effects of individual molecules from multi-siRNA experiments. Comparability may be violated by using 19mers (3) instead of 21mers (1). Knockdown activity has to be measured at the same time following transfection while maintaining similar cellular concentrations of siRNAs. The latter requirement can be approximated by using identical cell lines, transfection agents and extracellular siRNA concentration. These criteria are satisfied in two large datasets known to us. First, activities and sequences of 2252 siRNas targeted to 34 mRNA species were obtained from a Novartis study (1). These 21mers included two deoxynucleotide overhangs at the antisense strand complementary to the mRNA. NCI-H1299 and HeLa cells were transfected using combined Lipofectamine™ and Oligofectamine™ agents. Second, two hundred forty 19mer siRNA molecules designed to silence human or humanized targets were taken from Dharmacon (3). While this study targeted as few as eight genes, a major advantage is that all experiments were conducted in HEK293 cells using Lipofectamine™ maintained at 95% transfection efficacy or higher, and the siRNA concentration was held constant at 100 nM. Knockdown activity was measured after 24 h. Holen's (15) collection of 176 additional siRNAs and the database published by Sætrom (17) were also analyzed.
Features
SVMs and constrained optimization methods effectively selected the optimal subset of features from several hundred initial features in reasonable central processor unit (CPU) time. This allowed us to select from an unprecedented set of 572 sequence, thermodynamic and target accessibility features (Table 1). Sequence features included the global frequencies of mono- and dinucleotides and the presence or absence of mono- and dinucleotides at each of the 21 positions. Longer runs of identical bases were also considered since homotri- and tetranucleotides can act as termination signals for the RNA polymerase III enzyme used in certain vectors. Thermodynamic features, including the Gibbs free energy (ΔG), enthalpy (ΔH) and entropy (ΔS) differentials, and the ΔH/ΔS ratio, which is the major determinant of Tm (melting point), were calculated according to Xia et al. (18). Their derivative feature is the thermodynamic differential between the 5′ ends of the antisense and sense strands, which has been proposed as a distinctive feature of potent siRNAs (7). ΔG and the number of hydrogen-bonded nucleotide pairs characterize self-hairpins that can obstruct duplex formation. These features were predicted as described in (19). Target accessibility predictions require Bayesian sampling from a large number of alternative mRNA structures. The probability of the mRNA to form secondary structures and the free energy of these structures was calculated by the sfold tool (20–22) implemented at http://sfold.wadswort.org.
Feature selection required the compatibility of feature distributions. Therefore, feature values were standardized for the constrained optimization methods to a mean of zero and a SD of unity. For SVMs, feature values were normalized to the interval of [0,1].
Methods
We applied existing and created new machine learning methods for feature selection and predictions. Constrained optimization (mathematical programming or operations research) (23) is a powerful mathematical tool for maximizing or minimizing an objective function. Here we perform the optimal allocation of the regression plane to minimize the sum of deviations from this plane. Constrained optimization finds the globally optimal solution for a very large set of equations or inequalities in practically polynomial time (24).
SVMs are supervised learning methods used for classification and regression (25). SVMs transform the original data with nonlinear relationships into a higher dimension space to allow linear regression. SVMs have provided solutions to numerous biological problems as reviewed in Camps-Valls et al. (12). Support vectors were generated by the core vector machine (26) and the SVMlight (27) packages using linear, polynomial and Gaussian radial basis function kernels. To assess the robustness of the predictions and the underlying features, we implemented fundamentally different methods using constrained optimization. First, we created a BC model to separate above-average (>70% knockdown) siRNAs from those with <60% activity. A nontraditional multivariate regression was performed for the molecules predicted as above-average. Experimenting with other cutoffs for high- and low-activity siRNAs resulted in lower accuracy in the combined BC-MVR cross-validation analyses (data not shown).
Robust BC is performed by the iterative elimination of features and misclassified objects (28), a highly reliable method for feature selection, applying Misclassification Minimization models (29). The score zs for each sequence s is defined as the optimally weighted sum of values of the features f in the set of all features F:
| 1 |
where wf is the weight for feature f. Scores for the highly active molecules are expected to exceed the scores of less active molecules by a value not less than a positive threshold parameter δ, which is the width of the separating zone between the two classes. Increasing δ improves the robustness of the solution: when predicting untrained molecules, we can reduce the number of misclassified molecules. This comes at the cost of increasing the number of unpredicted molecules since scores within the separating zone are not significant enough to classify the underlying siRNA.
The sets of above-average and low-activity siRNAs are linearly inseparable. To make the solution of the model feasible, nonnegative error variables ɛh are introduced for each sequence h in the set H, sequences with experimentally determined high-activity:
| 2 |
where the geometric interpretation of γ is the intersection with the vertical axis. For each sequence l in the set L of low-activity sequences we require that
| 3 |
The sum of absolute values of weights wf must be limited to keep the model from growing unbound:
| 4 |
Here ‖w‖1 is the standard mathematical notation for the sum of the absolute values (first norm). We solve the system of the above inequalities and equations to minimize the sum of the error variables ɛh.
| 5 |
Here the user-defined parameter 0 < λ < 1 fine-tunes the balance between sensitivity and selectivity. When λ is set to a value higher than 0.5, errors related to above-average activity molecules are decreased by allowing more errors in the low-activity molecules. nH and nL are the number of the above-average and low-activity molecules in the training set, respectively. ψ is a small factor necessary for the calculation of the absolute values of the weights.
Solving the above system of linear inequalities by constrained optimization packages (e.g. CPLEX from ILOG, Incline Village, Nevada) leads to the minimization of errors by selecting the optimal values for the weights wf and the additive variable γ. Provided that the model has a unique, globally optimal solution, any of the simplex, dual or barrier algorithms (23,30) finds it in practically polynomial time (24).
Note that the solution for the above model is more sensitive to a few large errors than to several smaller ones. Incorrect experimental measurements of the knockdown activity may considerably exceed the magnitude of real prediction errors. Such incorrect input data may dislocate the separating zone, resulting in an unjustifiably large number of misclassified molecules. We reduce this effect by iteratively eliminating the siRNA with the largest error in the previous optimization. The saved basic solution allows solving the model about ten times faster than the first time. This is the key to the computational feasibility of several hundred iterations during feature selection (28).
For the numerical prediction of the knockdown activities, brute force traditional multivariate regression analysis has limited utility due to the high number of features. Robust Regression (31) was not as accurate as constrained optimization methods or SVMs (data not shown). In our regression model, for each sequence s, we minimize the absolute value distance from the regression plane:
| 6 |
where as is the experimentally determined knockdown activity of molecule s. Now we minimize the sum of the error variables ɛs and the sum of the absolute values of the wf weights:
| 7 |
Here ψ is a small factor for the contribution of absolute values.
Feature (property or variable) selection emerges as a highly successful new technique (32) for finding those biological or physical features that indicate or cause a certain effect; e.g. a disease. Selecting the most predictive features by traditional manual methods from among several hundred initial features over thousands of observations is prohibitively time-consuming. Fortunately, machine learning tools can perform such complex tasks in short processor time. Examples include differentially expressed genes as indicators and/or causative agents of cancer (33), semi-supervised learning for molecular profiling (34) and optimal selection of hydrophobicity-related, structural and other features determining protein secretion signals (28), physicochemical descriptors to discriminate protein–protein interactions (35), and automatic parsing of the biomedical literature (36). These studies revealed diagnostic combinations of features that frequently constituted some important biological signature. Feature selection also reduces overtraining. This is a fundamental issue when we do not have 5–10 times more observations than features (32).
For linear SVMs and constrained optimization models, we use a weight-based feature elimination algorithm (28). For comparability with related algorithms below, we abbreviate this algorithm as WFE. A feature's weight is proportional to its contribution to the prediction (Equations 2 and 3). Features with zero weights do not contribute to the model and therefore should be eliminated. In each of the subsequent iterations, the feature with the lowest absolute value is eliminated. This iteration is repeated until the number of features reaches a user-specified limit and the cross-validation accuracy decreases. Fortunately, the wf feature weights are transparent in constrained optimization models. In SVMs with linear kernels, wf = ∑v av rf,v, where av is the Lagrangian multiplier of support vector v and rf,v is the normalized value of feature f in support vector v (37). For the compatibility of features measured in different units, feature values are normalized in SVMs since SVMlight (38) and similar implementations limit feature values to the [0,1] interval. In constrained optimization, we standardize feature values to zero mean and unit SD. Standardization is less sensitive to a few outliers than the above normalization.
For nonlinear SVMs, the effect of leaving out a feature on the objective function is more informative than the weight itself (39). This justifies the computationally much more intensive recursive feature elimination (RFE) (33) method. Basically, in every iteration, a leave-one-out procedure is performed for each for the surviving features. The feature with the smallest effect on the objective function is removed.
Validation
Ten independent cross-validation experiments were used. In each experiment, the Novartis data were divided into a training set and a test set of equal size using a random number generator. siRNAs with 16 or more identities were eliminated. Blind tests were performed using a large enough dataset (either the Novartis or the Dharmacon data) for training and any other set for testing.
RESULTS
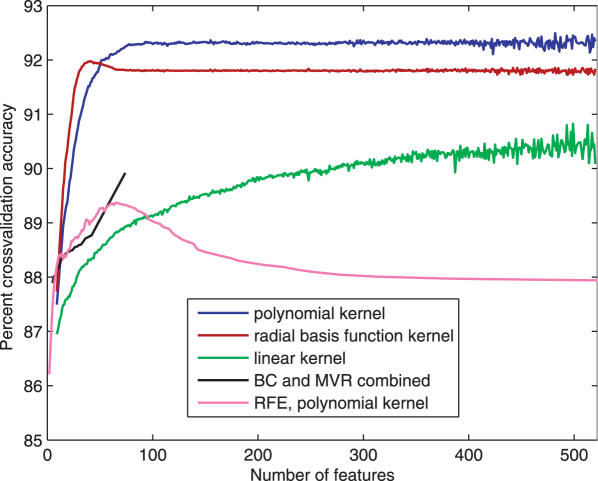
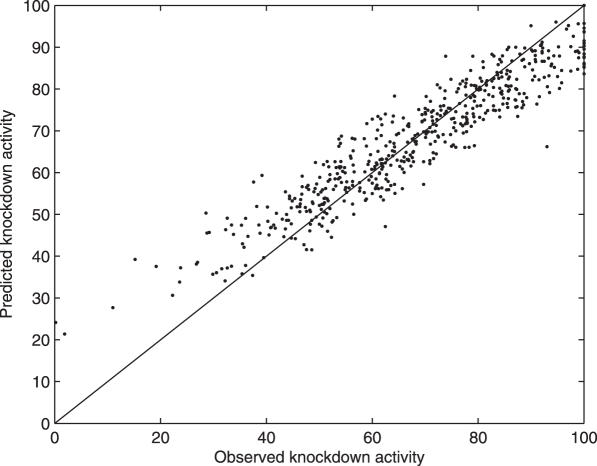
Predictions with 92.3% accuracy were achieved by SVMs with a polynomial kernel using WFE (28) in 10× cross-validation experiments (Figures 2 and 3). This accuracy is defined as 100 minus the average percentage difference between predicted and observed knockdown activities. SVMs with Gaussian radial basis function or linear kernel provided for less accurate predictions than the polynomial kernel. BC between <60% and >70% active siRNAs was 94% accurate. Here we set the parameter λ to 0.35 to reduce false positives. The subsequent MVR on the >70% active molecules is ∼95% accurate. Altogether, the BC-MVR combination predicted 89% of the ≥90% active siRNAs with a 12% false-positive rate. Regressing 19mers [from the Dharmacon (3), Holen's (15) and Sætrom's (17) sets] by any method trained on 21mers with deoxynucleotide overhangs in the Novartis set (1) or vice versa reduced the accuracy to 78% or lower (data not shown). Supplementing the missing two nucleotides did not lead to significant improvement.
Figure 2.
The accuracy of SVM using different kernels and constrained optimization methods as functions of the number of features. Results of 10× cross-validation experiments (see Materials and Methods) are shown. Note that constrained optimization eliminated all but 72 features in the first iteration.
Figure 3.
Observed versus predicted activities in the Novartis dataset (1). Predictions were performed by the polynomial kernel SVM using 142 features shown on Supplementary Table 1.
BC and MVR automatically reduced the number of features at the first iteration to 72 and 86, respectively. At identical feature numbers, WFE led to quite unexpected results: basically similar features were selected by constrained optimization methods and linear SVMs. This observation increases the confidence for finding the biological and thermodynamic signature for RNAi.
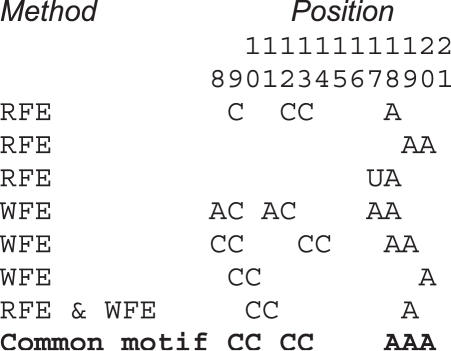
As a rule, either identical or analogous features are selected by WFE over linear methods and by RFE using a polynomial kernel (Supplementary Table S1). Although WFE requires as many as 142 features to reach maximal accuracy compared to 68 features with RFE/polynomial kernel, 30 features are shared between these two sets. More importantly, several remaining features form analogous combinations (Figure 4). As an example, the selection against AAA starting at 18 is expressed in WFE by selection against AA at positions 18 and 19. Analogously, RFE indicates selection against A at 18 and AA at 19. Another example is the negative preference for CC at 12, which is expressed in RFE by that single feature. However, WFE uses two features, AC at 11 and CC at 13, to the same effect. Yet another example is disfavoring C at 9 and CC at 10 in RFE, which is expressed by selection against AC at 8 and CC at 8, 9 and 10 in WFE.
Figure 4.
Three examples of aligned feature (dinucleotide) combinations selected by RFE and/or WFE with common sequence motifs. All of these features decrease siRNA activity. The selection against the dinucleotide CC at position 9 is expressed by disfavoring cytosines at position 9 in RFE and the dinucleotide CC at position 10 in both methods. In WFE, the selection against CC at 9 is expressed both directly (CC at 9) and indirectly by disfavoring AC and CC at 8, and CC at positions 8, 9, and 10 (see text).
As a more complex example, the global G + C content is selected by the polynomial kernels used in RFE, whereas WFE chooses a wide-array of local mono- and dinucleotide features that are clearly related to the global G + C content. We postulated that the features selected by WFE account for a more accurate prediction than the G + C content. To test this postulate, we complemented the feature set selected by WFE with G + C. As expected, adding G + C did not increase prediction accuracy, even with polynomial kernels.
However, the position of the target site was important for RFE but eliminated by WFE. We believe that the polynomial kernel uses this feature better since loci too close to or too far from the translation initiation site appear to decrease activity. To improve predictions, we overruled WFE and manually complemented it by the target site feature. The accessibility of the target site as measured by the sfold p3 feature is one of the heaviest weighted features of WFE both in MVF and SVM with a linear kernel. However, RFE with a polynomial kernel eliminated p3.
Although WFE outperformed RFE with a small margin in our study, this does not substantiate far-reaching conclusions. WFE with a linear kernel is more robust and better in handling a high number of features. However, RFE can identify features that have highly nonlinear effects on silencing activity. An example would be the distance of the target from the translation initiation site. Such features may be missed by WFE.
DISCUSSION
Highly active siRNA molecules, although diverse in sequences, appear to conform to a widespread dinucleotide, thermodynamic and accessibility signature. This signature is highly probabilistic, meaning that there are numerous exceptions to each ‘rule.’ Fortunately, appropriate methods allow accurate prediction, which in turn lets us identify the most active siRNAs for the gene to be silenced.
A total of 92.3% accuracy was achieved in weight-based feature elimination. The most accurate predictions in cross-validation experiments required as many as 142 features (Supplementary Table S1). For brevity, Table 2 shows the linear kernel that was limited to 30 features. Further indications include the need for ∼150 features and the lack of high weights (over 5% of the sum of the absolute values). RFE on polynomial kernels was somewhat less accurate (89.4%) than the weight-based feature elimination. However, this accuracy was achieved using as few as 68 features (Supplementary Table S1). Of these, 30 features are shared with the 142 obtained with weight-based feature elimination.
The lack of absolute criteria may be due to sequence diversity. Since a large number of genes are subject to posttranscriptional regulation, a wide spectrum of mRNA segments is sensitive to RNA interference. This diversity requirement can still accommodate probabilistic criteria specific for the RISC complex (see below). Silencing activity appears to be determined by a wide-range of flexible combinations of weighted sequence, thermodynamic and accessibility features.
A wide spectrum of sequences can fit this thermodynamic profile (40), which can provide a (partial) solution for the paradox of sequence diversity versus RISC-specific criteria. Accurate and rigorous analysis and prediction of RNAi in free energy terms may be a real possibility, akin to structural predictions of RNA (41) or proteins (42). Machine learning is also facilitated by the 16-fold reduction in dimensionality of ΔG profile as compared to dinucleotides.
Several key features are related to the change in free energy, enthalpy or entropy related to duplex formation. Global ΔG is assigned the highest weight by SVMs. For the 500 most active siRNAs, the average of ΔG is −164.43 kJ/mol, whereas for the 500 least active siRNAs it is −180.20 kJ/mol. In siRNAs with >90% activity, preference for lower stability is also indicated by the selection against CC and GG dinucleotides at the whole antisense strand. On the contrary to the expected antisense frequency of 0.0625, CC dinucleotides occur with a frequency of 0.0489 and GG with a frequency of 0.0540. CC was assigned a weight of −0.04503 and GG received a weight of −0.03433. The general preference for less negative global ΔG is fine-tuned by a preference at the 5′-terminus of the antisense for A and U and selection against G, C, CG and UG. The 3′ end shows a preference for C, G, GG, AG, UG, GU and a negative selection against A, UU, AA and CC. The putative cleavage site for the Argonaute-2 (43) or similar endonuclease at around position 7 is rich in U, but GU is preferred to AU. These results complement the thermodynamic profile reported earlier (7) and the proposition that the lower terminal stability is supposed to facilitate duplex unwinding by the topoisomerase enzyme (44).
Using WFE, the accessibility of the target site emerges as the most predictive of the 142 features (Supplementary Table S1) and the third most important feature among the 30 shown in Table 2. Extreme negative weight is assigned to p3, the probability that all bases of a tetranucleotide are involved in secondary structures. p3 is estimated by a Bayesian sampling from the Boltzmann probability distribution of conformations as implemented in the sfold algorithm (20). Therefore, it is not surprising that p3 consistently received more significant weights than the ΔG of the single most stable structure. However, for BC between <60% and >70% active siRNAs, all accessibility features receive zero weights (data not shown). This indicates that most structured target sites can be silenced by <70% efficacy. Whereas the correlation between activity and p3 is low (r = 0.0584), this is significant at the p = 0.0035 level. The considerable weight assigned to p3 indicates that the target sites of siRNAs with ≥90% activity are either highly accessible or other features must compensate for limited accessibility.
The formation of self-hairpins within a single strand may inhibit silencing action (45). SVMs with over 100 features (Supplementary Table S1), BC, and MLR assigned strong negative weights to this feature, which was estimated by the RNAup package (19). While self-hairpin probability received zero weights in the SVM models with <50 features, it was strongly penalized indirectly by p3 from the sfold predictions and sequence patterns that decrease the chances for Watson–Crick base pairing between the 5′ and the 3′ ends. Interestingly, while the 5′–3′ thermodynamic differential was eliminated during feature selection, high weights were assigned to sequence features that express the same thermodynamic differential. These include a preference for U and A at positions 1 and 2 but selection against these nucleotides at position 19. AG and UG are preferred at positions 20–21, whereas AA at 17–18, AA and UU at 18–19 and U at 20 are less frequent than expected on a random basis.
Contrary to some earlier rules (2), we found 12 siRNA molecules with ≥90% knockdown that contain GGGG tetranucleotide(s), which may form highly stable tetraplexes. Ten other highly active siRNAs contained overly stable runs of 7 or more G or C bases.
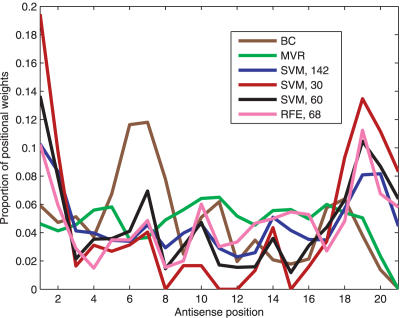
The distribution of weights along the sequence follows a consistent pattern across SVMs, BC and MVR with widely varying numbers of features (Figure 5). The first and second antisense positions dominate the predictions with the exception of BC and MVR. SVMs had another major peak at position 19, in line with the hypothesis that loose termini facilitate duplex unwinding by the topoisomerase enzyme (7). The importance of the possible Argonaute-2 (43) cleavage site at position 7 was pronounced only with BC and SVM with 60 features. The most accurate models specified preferences for all positions. However, when the number of features was limited to 30, all features at positions 8, 11, 12 and 15 were eliminated. The accuracy of predictions dropped at such a low number of features (Figure 2).
Figure 5.
Contributions of the individual antisense sequence positions to the predictions in MVR, BC, SVM/WFE with linear kernel, and SVM/RFE with a polynomial kernel. For the first four methods, we show the sum of weights (absolute values) for the features at that position. For RFE (magenta line), we display the total decrease in the prediction accuracy when features specific to a given position are eliminated.
Cross-validation experiments and blind tests on untrained data show the robustness (stable high-performance over new data) of the biophysical signature and the predictions. Dinucleotide preferences form a marked pattern that cannot be attributed purely to energetic or entropic factors. We postulate that these patterns are related to at least three sets of criteria. First, siRNAs need to be integrated into the RISC complex and have to facilitate helix unwinding by the topoisomerase and cleavage by Argonaute-2 enzymes. Second, accessible target sites are preferred or other features should compensate for reduced accessibility. Third, there is a selection against strands that can form self-hairpin structures.
Availability: fast siRNA activity predictions can be performed on our web server at http://optirna.unl.edu/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The author is grateful to Drs M. E. Fromm, W. W. Stroup and J. J. M. Riethoven and J. Gardner for comments and suggestions and Dr F. Ma for systems administration. The web page was implemented by M. Eirich, E. Moss and A. Guru. Special thanks to Drs T. Holen, A. Khvorova and P. Sætrom for their siRNA collections. Support from the National Science Foundation, Tobacco Settlement Fund, and a Cyberinfrastructure Development Grant from the University of Nebraska–Lincoln are gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation EPS-0346476.
Conflict of interest statement. None declared.
REFERENCES
- 1.Huesken D., Lange J., Mickanin C., Weiler J., Asselbergs F., Warner J., Meloon B., Engel S., Rosenberg A., Cohen D., et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 4.Yuan B., Latek R., Hossbach M., Tuschl T., Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Ueda R., Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 7.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 9.Bohula E.A., Salisbury A.J., Sohail M., Playford M.P., Riedemann J., Southern E.M., Macaulay V.M. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem. 2003;278:15991–15997. doi: 10.1074/jbc.M300714200. [DOI] [PubMed] [Google Scholar]
- 10.Kretschmer-Kazemi Far R., Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schölkopf B., Smola A.J., Williamson R.C., Bartlett P.L. New support vector algorithms. Neural Comput. 2000;12:1207–1245. doi: 10.1162/089976600300015565. [DOI] [PubMed] [Google Scholar]
- 12.Camps-Valls G., Chalk A.M., Serrano-Lopez A.J., Martin-Guerrero J.D., Sonnhammer E.L. Profiled support vector machines for antisense oligonucleotide efficacy prediction. BMC Bioinformatics. 2004;5:135. doi: 10.1186/1471-2105-5-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sætrom P. Predicting the efficacy of short oligonucleotides in antisense and RNAi experiments with boosted genetic programming. Bioinformatics. 2004;20:3055–3063. doi: 10.1093/bioinformatics/bth364. [DOI] [PubMed] [Google Scholar]
- 14.Shabalina S.A., Spiridonov A.N., Ogurtsov A.Y. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holen T. Efficient prediction of siRNAs with siRNArules 1.0: an open-source JAVA approach to siRNA algorithms. RNA. 2006;12:1620–1625. doi: 10.1261/rna.81006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalk A.M., Wahlestedt C., Sonnhammer E.L. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 17.Sætrom P., Snove J.O. A comparison of siRNA efficacy predictors. Biochem. Biophys. Res. Commun. 2004;321:247–253. doi: 10.1016/j.bbrc.2004.06.116. [DOI] [PubMed] [Google Scholar]
- 18.Xia T., SantaLucia J.,, Jr, Burkard M.E., Kierzek R., Schroeder S.J., Jiao X., Cox C., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 19.Muckstein U., Tafer H., Hackermuller J., Bernhart S.H., Stadler P.F., Hofacker I.L. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y., Lawrence C.E. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y., Lawrence C.E. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y., Lawrence C.E. A Bayesian statistical algorithm for RNA secondary structure prediction. Comput. Chem. 1999;23:387–400. doi: 10.1016/s0097-8485(99)00010-8. [DOI] [PubMed] [Google Scholar]
- 23.Dantzig G.B. Maximization of a linear function of variables subject to linear inequalities. In: Koopmans T.J.C., editor. Activity Analysis of Production and Allocation. NY: Wiley; 1951. pp. 339–347. [Google Scholar]
- 24.Goldfarb D., Todd M. Linear programming. In: Nemhauser G.L., Rinnooy Kan M.J., Todd M.J., editors. Optimization. Vol. 1. Amsterdam: Elsevier; 1994. pp. 73–170. [Google Scholar]
- 25.Schölkopf B., Smola A.J. Learning with Kernels. Support Vector Machines, Regularization, Optimization, and Beyond. Cambridge, MA: MIT Press; 2002. [Google Scholar]
- 26.Tsang I.W., Kwok J.T., Cheung P.-M. Core vector machines: fast SVM training on very large data sets. J. Mach. Learn Res. 2005;6:363–392. [Google Scholar]
- 27.Joachims T. Learning to Classify Text Using Support Vector Machines. Methods, Theory and Algorithms. Berlin: Springer; 2002. [Google Scholar]
- 28.Ladunga I. PHYSEAN: PHYsical SEquence ANalysis for the identification of protein domains on the basis of physical and chemical properties of amino acids. Bioinformatics. 1999;15:1028–1038. doi: 10.1093/bioinformatics/15.12.1028. [DOI] [PubMed] [Google Scholar]
- 29.Bennett K., Mangasarian O.L. Robust linear programming discrimination of two linearly inseparable sets. Optim. Meth. Software. 1992;1:23–34. [Google Scholar]
- 30.Chvàtal V. Linear Programming. NY: Freeman; 1983. [Google Scholar]
- 31.Huber P.J. Robust estimation of a location parameter. Ann. Math. Stat. 1964;35:73–101. [Google Scholar]
- 32.Kohavi R., John G.H. Wrappers for feature subset selection. Int. J. Digit. Libr. 1997;1:108–121. [Google Scholar]
- 33.Guyon I., Weston J., Barnhill S., Vapnik V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002;46:389–422. [Google Scholar]
- 34.Furlanello C., Serafini M., Merler S., Jurman G. Semisupervised learning for molecular profiling. IEEE/ACM Trans. Comput. Biol. Bioinform. 2005;2:110–118. doi: 10.1109/TCBB.2005.28. [DOI] [PubMed] [Google Scholar]
- 35.Block P., Paern J., Hullermeier E., Sanschagrin P., Sotriffer C.A., Klebe G. Physicochemical descriptors to discriminate protein–protein interactions in permanent and transient complexes selected by means of machine learning algorithms. Proteins. 2006;65:607–622. doi: 10.1002/prot.21104. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R., Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21:ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 37.Mladenic D., Brank J., Grobelnik M., Milic-Frayling N. Järvelin K., Allan J., Bruza P, Sanderson M. Feature selection using linear classifier weights: interaction with classifier models. 27th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; Sheffield, UK: ACM Press; 2004. pp. 234–241. [Google Scholar]
- 38.Joachims T. Making large-scale SVM learning practical. In: Schölkopf B., Burges C., Smola A.J., editors. Advances in Kernel Methods—Support Vector Learning. Cambridge, MA: MIT Press; 1998. pp. 41–56. [Google Scholar]
- 39.Le Cun Y., Denker J.S., Solla S.A. Optimum brain damage. In: Touretzky D., editor. Advances in Neural Information Processing Systems. Vol. 2. San Francisco, CA,: Morgan Kaufmann; 1990. pp. 598–605. [Google Scholar]
- 40.Boese Q., Leake D., Reynolds A., Read S., Scaringe S.A., Marshall W.S., Khvorova A. Mechanistic insights aid computational short interfering RNA design. Meth. Enzymol. 2005;392:73–96. doi: 10.1016/S0076-6879(04)92005-8. [DOI] [PubMed] [Google Scholar]
- 41.Mathews D.H., Turner D.H. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Eisenfeld J., Vajda S., Sugar I., DeLisi C. Constrained optimization and protein structure determination. Am. J. Physiol. 1991;261:C376–C386. doi: 10.1152/ajpcell.1991.261.2.C376. [DOI] [PubMed] [Google Scholar]
- 43.Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 44.Far R.K., Nedbal W., Sczakiel G. Concepts to automate the theoretical design of effective antisense oligonucleotides. Bioinformatics. 2001;17:1058–1061. doi: 10.1093/bioinformatics/17.11.1058. [DOI] [PubMed] [Google Scholar]
- 45.Patzel V., Rutz S., Dietrich I., Koberle C., Scheffold A., Kaufmann S.H.E. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat. Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]