Abstract
A series of antisense oligonucleotides (ASOs) containing either 2′-O-methoxyethylribose (MOE) or locked nucleic acid (LNA) modifications were designed to investigate whether LNA antisense oligonucleotides (ASOs) have the potential to improve upon MOE based ASO therapeutics. Some, but not all, LNA containing oligonucleotides increased potency for reducing target mRNA in mouse liver up to 5-fold relative to the corresponding MOE containing ASOs. However, they also showed profound hepatotoxicity as measured by serum transaminases, organ weights and body weights. This toxicity was evident for multiple sequences targeting three different biological targets, as well as in mismatch control sequences having no known mRNA targets. Histopathological evaluation of tissues from LNA treated animals confirmed the hepatocellular involvement. Toxicity was observed as early as 4 days after a single administration. In contrast, the corresponding MOE ASOs showed no evidence for toxicity while maintaining the ability to reduce target mRNA. These studies suggest that while LNA ASOs have the potential to improve potency, they impose a significant risk of hepatotoxicity.
INTRODUCTION
Synthetic oligonucleotides and their analogs are commonly used as research reagents to modulate gene expression in cell culture and in animal models. The most broadly utilized mechanism by which oligonucleotides are exploited for modulation of gene expression is through binding of the antisense oligonucleotide (ASO) to a specific mRNA or pre-mRNA by Watson–Crick base pairing. Upon binding, the oligonucleotide can modulate RNA processing, inhibit translation or promote degradation. Mechanisms of degradation include recruitment of RNase H, which cleaves the RNA strand of a DNA:RNA duplexes, and activation of the RNA interference (RNAi) pathway utilizing short RNA duplexes (siRNA) or hairpins (shRNA). In addition to the use of oligonucleotides as research reagents there is increasing interest in exploiting oligonucleotides as therapeutic agents. Currently there is one approved antisense product (Vitravene, fomivirsen) (1) and >30 products in active development.
The first generation of ASO therapeutics were 2′-DNA oligomers uniformly modified with the phosphorothioate (PS) backbone substitution and work predominantly through an RNase H-dependent mechanism. The substitution of sulfur for oxygen in the phosphate ester confers several properties onto ASOs which are crucial for their use as systemic drugs (2). Foremost, the PS linkage greatly increases stability to nucleolytic degradation (3), such that they possess sufficient stability in plasma, tissues and cells to avoid metabolism prior to reaching the target RNA after systemic administration to an animal. Additionally, the PS modification confers a substantial pharmacokinetic benefit by increasing the binding to plasma proteins, which prevents rapid renal excretion (4). While greatly increasing the stability of ASOs, PS modified drugs are still subject to metabolism, and have tissue half-lives of 1–3 days (4), which is sub-optimal for a parenterally administered drug. Furthermore, the PS modification reduces affinity for the target RNA (the ultimate biological receptor for ASO drugs) (3), which adversely effects potency.
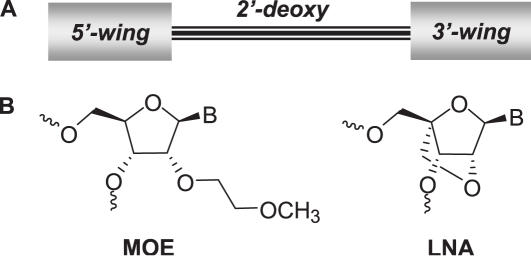
In order to improve upon the first generation ASO drugs, many different modifications to the core nucleoside monomer unit of the ASO have been evaluated for their effects on affinity for complementary RNA, nuclease resistance and ASO potency. Most of the modifications which enhance affinity and nuclease resistance, in particular the 2′-substituted nucleosides, also limit the ability of the ASO to support RNase H-mediated cleavage of the targeted RNA (5). Efforts to optimize the design of ASOs to retain the beneficial properties of 2′-modifications, yet maintain RNase H activity have led to the development of chimeric oligonucleotide designs which employ higher affinity 2′-substituted nucleosides combined with DNA regions which support RNase H activity. There are several designs for such chimeric oligonucleotides with ‘gapmer’ designs being most common in which a central DNA region of 7–14 nt is flanked on the 5′ and 3′ ends by 2–6 2′-modifications (Figure 1a) (6).
Figure 1.
(a) Gap design of current generation of ASO therapeutics having 2′-modified ‘wings’ at the 3′ and 5′ ends flanking a central 2′-deoxy gap region. (b) Structures of MOE and LNA nucleosides.
The most advanced second generation antisense designs are 2′-O-methoxyethylribose (7) (MOE) gapmer oligonucleotides (Figure 1). MOE modified ASOs show increased affinity toward a complementary RNA, and are highly resistant toward degradation by nucleases (8). These improvements result in a substantial (>20-fold) increase in oligonucleotide potency in cell culture, relative to first generation ASOs (9,10). In animals, MOE ASOs have been shown to possess both excellent pharmacokinetic properties (11–13) and robust pharmacological activity (14,15). Furthermore, the selective inhibition of gene expression with MOE ASOs elicits positive pharmacological activity in several animal models of human disease when given systemically with clinically relevant routes and schedules of administration (15–19). These beneficial properties have translated to human therapeutics. For example, a MOE ASO targeting ApoB has shown a dose-dependent reduction of target protein concurrent with lowering of LDL cholesterol. Doses as low as 100 mg per week produced statistically significant reductions in ApoB protein, and a dose of 200 mg per week reduced serum ApoB protein by 50% (20). Furthermore, to date MOE ASOs have an excellent safety record in human clinical trials (20–22).
The improvement in potency of MOE ASOs has, in part, been attributed to the increased affinity for target mRNA conferred by the MOE modification. Although MOE provides a substantial improvement in affinity, bicyclic nucleoside modifications such as 2′,4′-methylene bridged nucleic acids (23,24) commonly called ‘locked nucleic acid’ (LNA, Figure 1b) (25,26) have been shown to provide a further increase in affinity. LNA containing chimeric ASOs are just entering human clinical trials, and have recently been shown to inhibit growth in human tumor xenograft models (27). However, studies of the effects of LNA ASOs versus endogenous targets in liver, a tissue where antisense effects have been extensively characterized, have not been reported to date. In order to investigate whether LNA ASOs have the potential to improve upon MOE based ASO therapeutics, we directly compared the potency and therapeutic index of several designs of MOE and LNA containing ASOs in cell culture and in rodent liver after systemic administration. Our results indicate that although LNA modified ASOs have the potential to improve potency, they impose a significant risk of hepatotoxicity which must be considered when designing LNA containing antisense therapeutics.
MATERIALS AND METHODS
Oligonucleotide design and synthesis
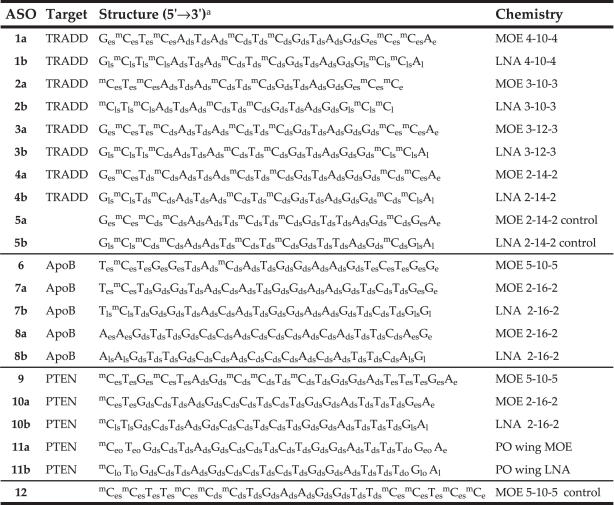
ASOs 1, 3 and 4 (sequence 5′-GCTCATACTCGTAGGCCA-3′, position 791–808) and 2 (sequence 5′-CTCATACTCGTAGGCC-3′, position 792–807) are complementary to Mus musculus TNFRSF1A-associated via death domain (TRADD) mRNA (Genbank accession no. NM_001033161). The ASO lead 1a is the murine homolog (a G to A base change at position 5) of the human TRADD lead reported previously (28). Control oligonucleotides 5 (5′-GCCCAATCTCGTTAGCGA-3′) were designed with six mismatches to 4, such that they contained ≥4 mismatches to all known mouse sequence. ASOs 6 and 7 (sequence TCTGGTACATGGAAGTCTGG, position 8232–8251) and 8 (sequence AAGTTGCCACCCACATTCAG, position 5586–5605) are complementary to Mus musculus apolipoprotein B (ApoB) mRNA (Genbank accession no. XM_137955.5). The sequences were identified by a screen of 5-10-5 MOE 20mer ASOs as described previously (29–31). ASOs 9, 10 and 11 (sequence 5′-CTGCTAGCCTCTGGATTTGA-3′, position 1931–1950) are complementary to M.musculus phosphatase and tensin homolog (PTEN), mRNA (Genbank accession no. NM_008960). ASO 9 (18) and control oligonucleotide 12 (19) have been described previously.
MOE phosphoramidites were prepared as described previously (7,32,33). LNA and 2′-deoxyribonucleoside phosphoramidites were purchased from commercial suppliers. Oligonucleotides were prepared similar to that described previously (34) on either an Amersham AKTA 10 or AKTA 100 oligonucleotide synthesizer. Modifications from the reported procedure include: a decrease in the detritylation time to ∼1 min, as this step was closely monitored by UV analysis for complete release of the trityl group; phosphoramidite concentration was 0.1 M; 4,5-dicyanoimidazole catalyst was used at 0.7 M in the coupling step; 3-picoline was used instead of pyridine for the sulfurization step, and the time decreased from 3 to 2 min. The oligonucleotides were then purified by ion-exchange chromatography on an AKTA Explorer and desalted by reverse phase HPLC to yield modified oligonucleotides in 30–40% isolated yield, based on the loading of the 3′-base onto the solid support. Oligonucleotides were characterized by ion-pair-HPLC-MS analysis (IP-HPLC-MS) with an Agilent 1100 MSD system. The purity of the oligonucleotides was ≥90% (Supplementary Table S1).
Cell culture assays
For determining potency in cell culture, mouse brain endothelial (bEND) cells (American Type Culture Collection) were transfected with the indicated concentration of oligonucleotide for 4 h using 3 μg/ml Lipofectin in OptiMEM. Transfection mixes were then replaced with normal growth media [DMEM-high glucose, 10% fetal bovine serum (FBS) containing penicillin-streptomycin]. Cells were harvested 24 h later and RNA was purified using Qiagen 96-well RNeasy plates. RNA was analyzed for TRADD and cyclophilin A RNA levels. TRADD RNA levels, normalized to those of cyclophilin A, are expressed as percent untreated control (% UTC). Each treatment was performed in triplicate. IC50 values were determined using GraphPad Prism software by fitting the data to a sigmoidal dose–response curve (variable slope) using a defined top of 100% and bottom of 0%. For caspase induction, A549 cells (American Type Culture Collection), a human lung carcinoma cell line, were seeded into 96-well plates and transfected the following day with 300 nm ASO for 4 h using Lipofectin (Invitrogen). Transfection mixes were subsequently replaced with normal growth media (Ham's F12K media containing 10% FBS). Cells were assayed 44 h later for caspase 3 activity and cell number (CyQuant) as described previously (35).
Animal treatment
All animal experiments were conducted according to the American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the Animal Welfare Committee. Male Balb/c mice, aged 6–8 weeks, were obtained from Charles River Laboratories. Compounds were suspended in phosphate-buffered saline (PBS), filter sterilized and administered by intraperitoneal (i.p.) injection according to the indicated dosing schedules in a volume corresponding to 10 μl/g animal weight. Animals were maintained at a constant temperature of 23°C and were allowed standard lab diet and water ad libitum and animal weights were monitored prior to dosing throughout the live phase of the study. Immediately prior to sacrifice, mice were anesthetized with isoflurane and terminal bleed was performed by cardiac puncture. Plasma or serum was isolated from whole blood and analyzed for clinical chemistries. Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were determined using an Olympus AU400e bioanalyzer. Immediately following terminal blood draw, mice were sacrificed by cervical dislocation while under anesthesia. In conjunction with necropsy, liver and spleen weights were determined. Effects of compounds on organ weights, normalized to body weight, are expressed relative to those of the saline treated group.
RNA analysis
Tissues were homogenized in 4 M guanidine isothiocyanate, 25 mM EDTA, 50 mM Tris–HCl, pH 6, containing 1 M β-mercaptoethanol immediately following sacrifice and homogenized. RNA was extracted using RNeasy columns (Qiagen) according to manufacturer's protocol. RNA was eluted from the columns with water. RNA samples were analyzed by fluorescence-based quantitative RT–PCR using an Applied Biosystems 7700 sequence detector. Levels of target RNAs as well as those of cyclophilin A, a housekeeping gene, were determined. Target RNA levels were normalized to cyclophilin levels for each RNA sample. Primers used for determination of TRADD RNA level are as follows: forward primer (FP) 5′-GGCCGCCTGCCAGAC-3′, reverse primer (RP) 5′-TGAAGAGTCAGTGGCCGGTT-3′ and probe (PR) 5′-6FAM-TTTCTGTTCCACGGGCAGCTCGTAGT-TAMRA-3′. Primers used for determination of PTEN RNA level are as follows: FP 5′-ATGACAATCATGTTGCAGCAATTC-3′, RP 5′ CGATGCAATAAATATGCACAAATCA 3′ and PR 5′-6FAM-CTGTAAAGCTGGAAAGGGACGGACTGGT-TAMRA-3′. Primers used for determination of ApoB RNA level are as follows: forward primer (FP) 5′-GAAAATAGACTTCCTGAATAACTATGCATT-3′, reverse primer (RP) 5′-ACTCGCTTGCCAGCTTGC-3′ and probe (PR) 5′-6FAM-TTTCTGAGTCCCCGTGCCCAACA-TAMRA-3′. Primers used for determination of cyclophilin A RNA level are as follows: FP 5′-TCGCCGCTTGCTGCA-3′, RP 5′-ATCGGCCGTGATGTCGA-3′ and PR 5′-6FAM-CCATGGTCAACCCCACCGTGTTC-TAMRA-3′.
Western blot analysis
Frozen tissue samples were homogenized in RIPA buffer (PBS containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing Complete™ protease inhibitors (Roche), and protein concentrations were determined by BioRad protein assay. Protein samples were separated on a 10% PAGE gel (Invitrogen) and subsequently transferred to a PVDF membrane (Invitrogen). Membranes were incubated at room temperature in blocking buffer consisting of 5% non-fat dry milk in TBS-T for 1 h. Rabbit polyclonal antibodies were obtained from commercial sources, and used at 1:1000 dilution. Phospho-eIF2alpha (Ser51) antibody was obtained from Cell Signaling Technology (Catalog no. 9721). HRP conjugated anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch and were used at 1:2500 dilution. Protein bands were visualized using ECL-plus reagent (Amersham).
Capillary gel electrophoresis
Immediately following removal, organs were frozen in liquid nitrogen and stored at −80°C until ASO extraction. Tissues were weighed and ASO was extracted as described previously (12,36). Briefly, tissue samples were homogenized in a BioSavant (Bio 101), and subjected to solid phase extraction using a phenyl bonded SPE column (Isolute). Concentrations of ASO in tissue as well as metabolite profiles were determined by capillary gel electrophoresis (CGE) using a Beckman P/ACE model 5010 capillary electrophoresis unit.
Hematoxylin and eosin (H&E) staining
Tissue samples were fixed in formalin for a minimum of 1 day followed by incubation in 70% ethanol for a minimum of 1 day. Tissue samples were further dehydrated and processed using a Leica ASP300 tissue processor. Tissues were embedded in paraffin and 4 μ sections were mounted on positive charged glass slides. Deparaffinized and rehydrated samples were stained for hematoxylin and eosin (H&E), using a Leica Autostainer XL.
Immunohistochemical staining
Immunohistochemical studies were performed to detect the cleaved form of caspase 3, as well as Bcl2-associated X protein (Bax) and growth arrest and DNA-damage-inducible beta (GADD45β) protein. Formalin-fixed, paraffin-embedded tissue sections were mounted on positive charged glass slides. Deparaffinized and rehydrated samples were heated for 20 min at 95°C in citrate buffer solution. The slides were cooled for 20 min and endogenous peroxidase was blocked with 3% hydrogen peroxide (H2O2) in methanol for 10 min at room temperature, followed by rinsing in distilled water. To detect cleaved caspase 3, Bax and GADD45β, the sections were incubated 1 h at RT with anti-cleaved caspase 3 polyclonal antibody, (1:50 dilution; Cell Signaling Technology, Danvers, MA), anti-Bax monoclonal rabbit antibody, E63 (1:200 dilution; Epitomics, Burlingame CA) and anti-GADD45β polyclonal antibody, C18 (0.5 μg/ml; Santa Cruz, CA), respectively. The HRP conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (1:200 dilution; West Grove, PA). The antigen was visualized with DAB (Dako Cytomation—Cat3K3466) for 5 min. For negative controls, primary antibodies were replaced with isotype matched normal IgG. The slides were then counterstained with hematoxylin.
RESULTS
Effect of LNA modification in cell culture
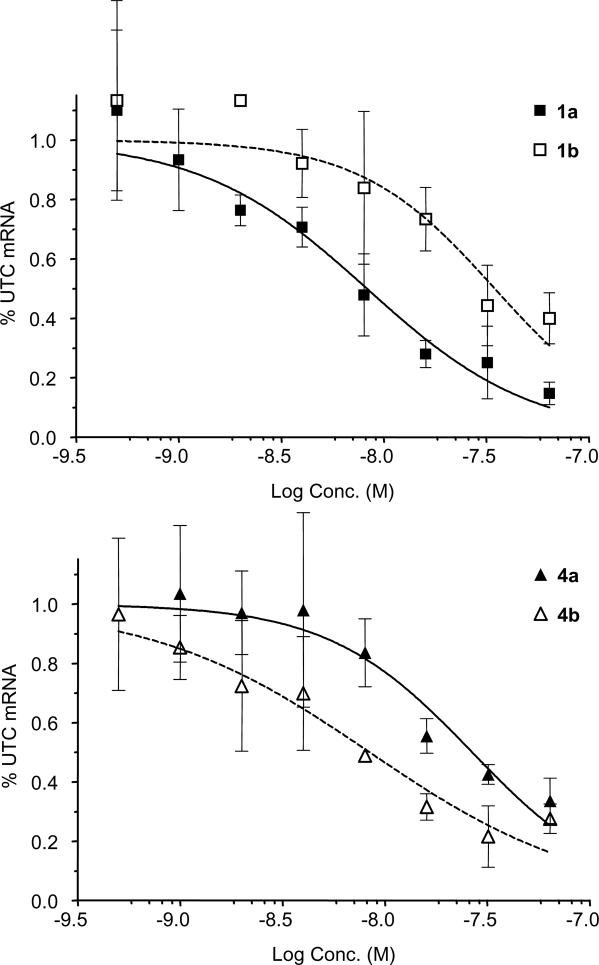
To directly compare the effects of LNA and MOE modifications, we designed a series of oligonucleotides targeting mouse TRADD mRNA. We have previously published the identification and characterization of MOE modified oligonucleotides targeting human TRADD, a death domain adapter protein that interacts with TNF receptor family members (28). The lead ASO from this study, a 4-10-4 MOE design, contains a single mismatch to murine TRADD mRNA, which was corrected to provide the mouse TRADD ASO 1a. Transfection of 1a (Table 1) into bEND cells reduces TRADD mRNA levels in a concentration-dependent manner as measured by quantitative RT–PCR (Figure 2 and Table 2), to give an IC50 of ∼8 nM. The 4-10-4 LNA version of this sequence (1b) reduced potency by ∼4-fold relative to the corresponding MOE ASO. Because LNA has an increased affinity relative to MOE, we hypothesized that the shorter ASOs with reduced LNA content may maintain or improve potency, and thus prepared the 3-10-3 MOE and LNA ASOs 2a and 2b. Interestingly, the MOE 16mer 2a had greatly reduced potency, whereas the potency of the 16mer LNA 2b was improved relative to the 18mer LNA ASO 1b. These results are consistent with a previous study of LNA ASOs (37), in which the high affinity of LNA facilitated good target reduction with 16mer LNA ASO designs.
Table 1.
Oligonucleotide targets, sequences and chemistry
aStructure code is nucleotide units 5′ to 3′. Capital letter is base code: G = guanine, A = adenine, C = cytosine, mC = 5-methylcytosine, T = thymine. Small subscript are sugar and linkage codes: e = MOE sugar, l = LNA sugar, d = deoxyribose sugar, s = PS linkage, o = PO linkage.
Figure 2.
Reduction of TRADD mRNA in bEND cells after transfection with MOE ASO 1a or LNA ASO 1b (top panel) or MOE ASO 4a or LNA ASO 4b (bottom panel).
Table 2.
Activity and liver levels of MOE (a series) and LNA (b series) ASOs
| ASO | IC50 (nM) in vitroa | Est. ED50 (mg/kg) in vivob | Liver conc. (μg/g tissue) ± SDc |
|---|---|---|---|
| 1a | 8.3 | 13 | 116 ± 20 |
| 2a | >60 | >25 | 54 ± 5.9 |
| 3a | 8.5 | 11 | 92 ± 11 |
| 4a | 27 | 9 | 96 ± 14 |
| 1b | 35 | 13 | |
| 2b | 15 | 6 | 64 ± 6.1 |
| 3b | 1.6 | 4 | 69 ± 4.5 |
| 4b | 8.4 | 2 | 48 ± 8.9 |
| 5a | ≫60 | ≫25 | |
| 5b | ≫60 | ≫25 |
aIC50values for reduction of TRADD mRNA in bEND cells after transfection with Lipofectin.
bED50 values for reduction of TRADD mRNA in mouse liver after dosing twice weekly for three weeks estimated by interpolation.
cDetermined by capillary gel electrophoresis for the 4.5 μmol/kg dose groups at end of study.
Since RNase H activity has been found to be an important factor in antisense potency both in vitro and in vivo (38), optimization of the gap region is of crucial importance. Because LNA has been shown to alter the conformation of adjacent DNA nucleotides for several residues (39), and duplex conformation is critical for RNase H activity (5), we included ASOs having increased gap sizes (3 and 4) in our SAR set. This replacement of high affinity modifications by additional 2′-deoxy units could increase RNase H activity by either adding potential cleavage sites, or by reducing the conformational transmission effects of LNA. We reasoned that the increased affinity of LNA relative to MOE would compensate for the reduced number of 2′-modifications, and may increase potency. Replacing two internal MOE modifications of 1a with deoxy nucleosides provided the 3-12-3 ASO 3a, which maintained potency relative to 1a. Further substitution of two additional MOE residues gave the 2-14-2 ASO 4a, which reduced potency of 3–4-fold. In contrast, replacing two LNA units of 1b with deoxy nucleosides resulted in a marked improvement, with 3b having an IC50 of 1–2 nM. As with the MOE series, further substitution to afford 4b reduced the potency, such that the LNA 2-14-2 4b was approximately equipotent relative to the parent 4-10-4 MOE 1a. As expected, both MOE (5a) and LNA (5b) four base mismatch control ASOs were inactive.
In vivo effects of 2′-MOE and LNA modified oligonucleotides
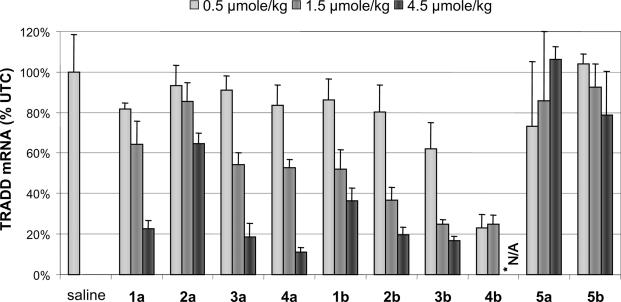
To determine if the behavior of LNA modified ASOs was similar in animals, mice were treated with ASOs at several dosage levels two times per week for 3 weeks. As expected, treatment of mice with the 4-10-4 MOE ASO 1a twice per week reduced TRADD mRNA in liver in a dose-dependent manner, producing a 77% reduction at the 4.5 μmol/kg dose (∼30 mg/kg, Figure 3). Doses of μmol per kg were employed as the compounds were of slightly different molecular weight, and we wished to directly compare potency on a molar basis. Consistent with cell culture results, the corresponding LNA analog 1b was less efficacious at this dose, resulting in only a 65% inhibition of mRNA. The potency of this ASO is difficult to assess from the limited dose–response data, but it appears to have a reduced efficacy, at least at the doses evaluated. Also consistent with cell culture results, the MOE 16mer ASO 2a was weakly active, while the corresponding LNA 2b showed ∼80 and 60% reductions in TRADD mRNA at the 4.5 and 1.5 μmol/kg doses, respectively.
Figure 3.
Reduction of TRADD mRNA for MOE (a series) and LNA (b series) ASOs. *Not Analyzed: due to severe toxicity at 8 days, the 4.5 μmol/kg dose group of 4b was terminated early.
The 3-12-3 and 2-14-2 MOE ASOs 3a and 4a produced a small increase in potency relative to the 4-10-4 1a, as well as an increase in efficacy at the 4.5 μmol/kg dose, with 2-14-2 design giving an 89% reduction in mRNA. The LNA ASO 3b (3-12-3 design) showed an increase in potency, achieving ∼80% reduction of mRNA at the 1.5 μmol/kg dose. In contrast to cell culture results, the 2-14-2 LNA ASO 4b further improved activity, giving a 75% inhibition of TRADD expression at the lowest dose of 0.5 μmol/kg (∼3 mg/kg). Interpolation of the TRADD mRNA reduction data allowed estimation of a rough ED50, which could be converted to mg/kg dosing for a comparison of in vivo potency (Table 2). For 3b, the data suggest an ∼3-fold increase in potency over the corresponding MOE 3a and its 4-10-4 parent 1a. The 2-14-2 LNA gapmer design 4b provided an even larger increase in potency, the magnitude of which could not be estimated from this experiment because the maximal effect was achieved at the lowest dose tested. The efficacy of inhibition of TRADD expression for the high-dose group could not be obtained, as the high-dose group of animals treated with 4b was sacrificed early due to severe toxicity observed as discussed below. Mismatch control MOE and LNA ASOs 5a and 5b showed no reduction in mRNA, indicating a specific in vivo antisense effect on the target mRNA.
LNA ASOs induce profound hepatotoxicity
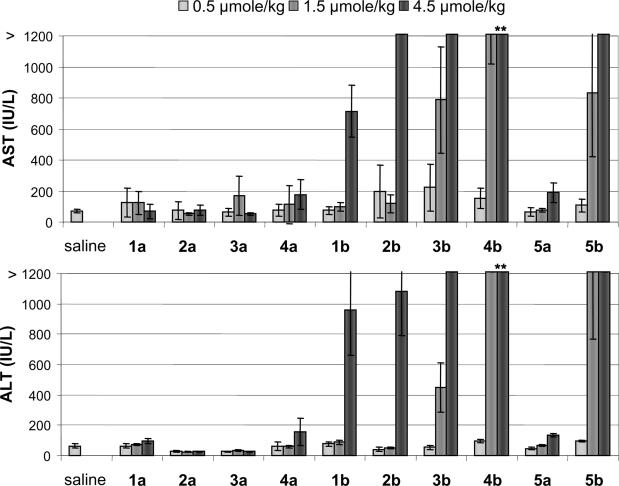
As part of the routine monitoring of animals during the study, plasma bilirubin and transaminase levels for all the high-dose groups were examined at Day 8 of the study in order to assess liver function. Bilirubin, ALT and AST levels were within the normal range for all animals, except for the 2-14-2 LNA ASOs 4b and 5b. LNA 4b showed elevations of 186-, 75- and 18-fold for ALT, AST and bilirubin, respectively, relative to saline treated animals. Notably, even the control LNA 5b, which showed no TRADD mRNA reduction, showed 46- and 25-fold increases in ALT and AST, respectively. Prior to receiving the fourth dose of ASO on Day 11 of the study, all the animals in the 4.5 μmol/kg group receiving LNA ASO 4b experienced significant weight loss, losing ∼25% of their body weight (Supplementary Figure S1). Because of this severe weight loss, coupled with the very large transaminase increases seen with 4b, the study was terminated early for this dose group. Upon necropsy, these animals showed severe hepatotoxicity by histopathological analysis, discussed further below.
The ALT and AST levels for all the remaining groups at the termination of study on Day 21 indicate a striking difference in the liver function profile of the MOE and LNA ASOs (Figure 4). All LNA ASOs studied showed at least a 10-fold increase in transaminases for the 4.5 μmol/kg dose groups, and some showed increases of >100-fold such as 3b, 4b (despite receiving only 3/6 doses) and 5b. In contrast, the transaminase levels in mice treated with MOE ASOs were within the normal range. The observed hepatotoxicity in the LNA ASO series was both compound and dose-dependent, with ASOs having a larger DNA gap increasing hepatotoxicity. Further evidence for the toxicity of the LNA ASOs was evident from the organ weights (Supplementary Figure S2). LNA ASOs produced large (45–62%, relative to saline) liver weight increases in a dose-dependent manner, except for 5b, which had a smaller (25%), non-dose-dependent increase. In contrast, the MOE ASOs showed much smaller (0–17%) liver weight increases. Increase in spleen weight was more variable, with 2b and 3b producing large (>100%, relative to saline) increases. The increase in spleen weights could be a direct proinflammatory effect of the oligonucleotide or secondary to the severe hepatotoxicity.
Figure 4.
Plasma transaminase levels for MOE (a series) and LNA (b series) ASOs. **Data from 8 days. AST 4020 ± 850, ALT 6470 ± 1450, and severe weight loss led to early termination of 4.5 μmol/kg dose group of 4b.
LNA ASOs induce apoptosis in vitro and in vivo
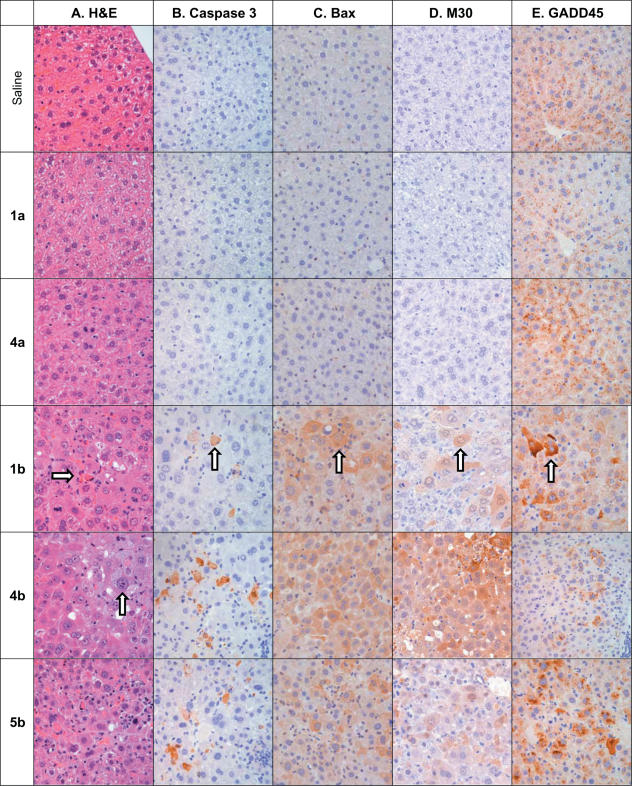
Examination of H&E stained liver sections from mice treated with LNA ASOs 1b and 4b, as well as control oligonucleotide 5b, confirmed hepatotoxic events. Histopathological observations included signs of apoptosis, profound eosinophilic cytoplasmic degeneration with glycogen depletion and hyperchromatic nuclei, as well as centrilobular coagulative necrosis surrounded with inflammatory infiltrate containing neutrophils, monocytes and lymphocytes (Figure 5). Lesions of intracytoplasmic microvesicular changes were also visualized in liver sections of 4b and 5b treated animals, suggesting early signs of steatosis. An increased number of mitotic hepatocytes were also observed in liver sections from LNA treated animals, likely indicating regeneration of damaged tissue.
Figure 5.
Histopathology of liver sections from mice treated MOE (a series) and LNA (b series) ASOs at 4.5 μmol/kg twice weekly for 3 weeks. Livers from animals treated with LNA ASOs 1b, 4b and 5b present with significant hepatotoxicities as demonstrated in (A), a routing H&E stain showing profound swollen eosinophilic degeneration, cell death (right arrow) and hyperchromatic nuclei (up arrow) of hepatocytes. The immunohistochemistry reveals that the injured hepatocytes appear with cytoplasmic staining (brown stain cells, up arrows) of cleavage caspase 3 (B), pro-apoptotic protein BAX (C) and M30 (D), a neo-epitope generated in epithelial cells as a result of caspase activation (cleavage). In addition a DNA damage and repair associated protein GADD45β (E) and peroxisome membrane protein PMP70 (Supporting Supplementary Figure S2) were both found to have increased cytoplasmic expression in LNA treated livers. IgG control slides of all four IHC markers were negative (data not shown).
To further characterize the hepatotoxicity associated with LNA ASO treatment, we conducted immunohistopathological evaluation to characterize the type of toxicity observed (Figure 5 and Supplementary Table S2). Liver sections from animals treated with LNA ASOs were stained for the expression of GADD45β, the activated form of caspase 3, peroxisome membrane protein 70 and Bax. Increased expression of activated caspase 3 and Bax suggested increased apoptosis in livers of LNA, but not MOE, oligonucleotide treated mice. GADD45β is a p53 and NF-κB regulated gene induced in response to cell stress. The LNA but not MOE modified oligonucleotides increased GADD45β expression dramatically. Evidence of peroxisome proliferation was also suggested by increased expression of peroxisome membrane protein 70 by IHC. Increased staining of each of these markers was dose-dependent and correlated well with ALT increases. To further confirm apoptosis involvement of liver injury mediated by LNA, liver sections of saline and LNA treated animals were stained with the monoclonal antibody M30, which is an apoptosis marker that monitors the neoantigen formed by caspase cleavage of cytokeratin 18. M30 immunoreactive cells appear at an early stage of apoptosis in epithelial cells, and are not detectable in vital or necrotic epithelial cells (40). Animals treated with LNA ASOs 1b, 4b or 5b all showed greatly increased M30 staining relative to saline treated animals (Figure 5), providing further evidence for apoptosis induced by treatment with the LNA ASOs.
To examine whether LNA ASOs could induce apoptosis in cell culture, we examined representative TRADD ASOs 2a, 4a at 300 nM in A549 cells for their ability to induce caspase 3 activity, a common marker of apoptosis, in cell culture after transfection (35). The LNA ASOs 2b and 4b resulted in a 3.1- and 6.2-fold induction of caspase 3 activity relative to control ASO 12, respectively, whereas the corresponding MOE ASOs gave no change relative to the control ASO (Supplementary Figure S3).
LNA ASO improves potency but also increases toxicity
To verify the activity and toxicity observed with 4b, it was tested in a repeat set of experiments along with 1a as a comparator (Figure 6). In addition to the previously utilized doses of 4.5, 1.5 and 0.5 μmol/kg, we also examined 0.9, 0.3 and 0.1 μmol/kg doses of 4b in order to assess to what extent potency was increased, as well as whether there was an increase in therapeutic index. In this experiment, mice treated with three doses of 4b at 4.5 μmol/kg (at 11 days) showed minimal weight loss, and the study was continued for the course of six administrations. At termination of the study, the animals treated with 4b had lost 10% of their body weight, as opposed to a 10% gain in the control group (Supplementary Figures S4 and S5). Furthermore, organ weights, transaminase increases and histopathological observations upon necropsy were consistent with the previous results, with >50-fold increases in AST and ALT at the 1.5 and 4.5 μmol/kg dose levels. The target reduction for 4b observed was similar to the first experiment, with 70 ± 8, 76 ± 6 and 66 ± 8% reduction of TRADD mRNA at the 4.5, 1.5 and 0.5 μmol/kg doses, respectively. The 0.9 μmol/kg dose (∼5 mg/kg) produced the maximal effect of 4b, providing an 81 ± 4% reduction of TRADD mRNA, but also appeared to be above the maximum non-toxic dose, as transaminases were increased ∼5-fold.
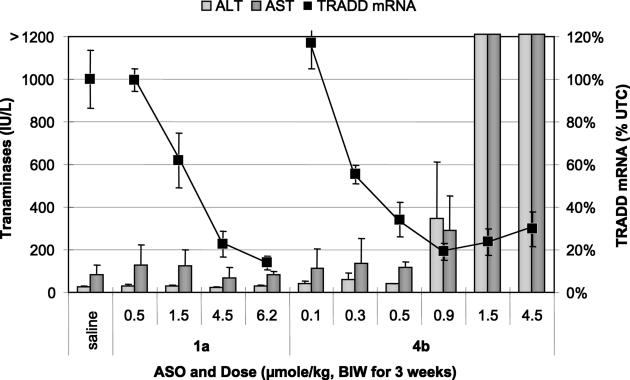
Figure 6.
Transaminases (bar graph, left scale) and reduction of TRADD mRNA (points on line graph, right scale) after treatment with the 4-10-4 MOE gapmer 1a or the 2-14-2 LNA gapmer 4b.
The 2-14-2 LNA 4b clearly demonstrated a dose-dependent reduction of TRADD mRNA, with an ED50 of ∼0.37 μmol/kg (corresponding to ∼2 mg/kg, Table 2). This suggests an improvement in potency of ∼4–5-fold relative to the corresponding 2-14-2 MOE 4a, and 6–7-fold relative to the comparator 4-10-4 MOE gapmer 1a. At dosage levels used for the previous study, the activity of 1a was essentially identical to that observed previously. A higher dose of 6.2 μmol/kg (∼40 mg/kg) of 1a produced a larger reduction (86 ± 3%) of TRADD mRNA. At all doses of 1a employed, there was no evidence for toxicity as measured by transaminase levels, organ weights, body weights or histopathological analysis of liver tissue samples. Thus, though LNA ASO 4b was more potent than the comparator MOE ASOs, it was not more efficacious, and furthermore did not produce maximal efficacy in the absence of observable toxicity. These results demonstrate that despite the increase in potency observed with the LNA ASOs, the therapeutic index is not increased, and is probably decreased relative to the corresponding MOE ASOs.
It is possible that both the improved potency and increased toxicity could be due to increased distribution of LNA modified oligonucleotides to liver. To determine if this was the case, the concentration of ASO in the liver was measured at the conclusion of the study for ASOs 1–4 (Table 2). The repeat group of 4b treated animals was used for this analysis, as the mice received the same number of total doses. The liver concentrations ranged from a high of 116 μg/g for 1a to a low of 48 μg/g for 4b. These results are consistent with expectations of higher metabolic stability for ASOs having greater numbers of MOE and LNA modifications, and also with increased distribution to liver for ASOs having more PS linkages. As the concentration trend of ASO in liver does not correlate with potency or toxicity, the increased potency and/or toxicity of LNA oligonucleotides is not due to more accumulation in the liver.
LNA ASO 4b shows hepatotoxicity with a single administration
To better characterize the nature of the hepatotoxicity, we administered a single dose of either MOE ASO 1a or LNA ASO 4b to mice, and examined effects at 2 and 4 days post-dosing. In addition to plasma transaminases and histopathology, levels of cleaved caspase 3 and phosphorylated eIF2α (p-eIF2α) were examined by western blot. Phosphorylation of eIF2α has been shown to mediate apoptosis, presumably via the inhibition of translation (41). Additionally, levels of Bax, GADD45β, PUMA, p53, TNFα and MDM2 mRNA were evaluated by RT–PCR. For all groups except for the 12 μmol/kg dosage level of 4b, there was no discernable difference from saline for any measured endpoint. In contrast, administration of 12 μmol/kg of the LNA ASO 4b gave a 7- and 2-fold elevation in ALT and AST, respectively, at the day 4, but not at the day 2 time-point. Concurrent with this increase in transaminases at day 4, an increase in p-eIF2α was observed by western blot (Supplementary Figure S6). No increases in other genes studied were evident by RT–PCR or western blot. However, weak staining of activated caspase 3, Bax and the M30 neoantigen were observed by immunohistochemistry (Supplementary Table S3). These results indicate that the hepatotoxicity induced by LNA 4b can occur in as little as 4 days, and are consistent with acute liver injury caused by induction of apoptosis.
LNA effects on potency and hepatotoxicity are independent of target
To help rule out a potential target related contribution to the hepatotoxicity and to verify the improved potency for other targets, we tested additional LNA ASOs targeting other mouse genes. We have previously published data demonstrating specific reduction of mouse ApoB (19) and PTEN (18) mRNA in mice treated with 20mer MOE ASOs. Three active ASO sequences (6, 8 and 9, Table 1) targeting either mouse ApoB or mouse PTEN were identified in cell culture assays using methods previously described for these targets. Because the LNA ASO design having two LNA nucleosides flanking a large deoxy gap region appeared to exhibit the greatest increase in potency and hepatotoxicity, we utilized a 2-16-2 design, and applied it to the previously identified 5-10-5 MOE sequences.
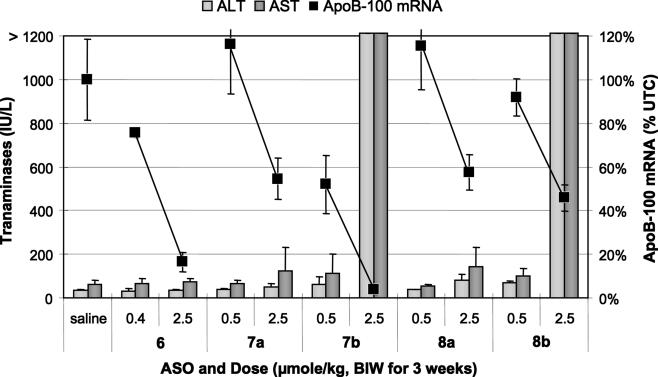
For the ApoB target, ASOs were dosed at 2.5 and either 0.5 or 0.4 μmol/kg twice weekly for 3 weeks (Figure 7). The 5-10-5 MOE ASO 6 produced a modest 24% inhibition of ApoB mRNA at the 0.4 μmol/kg dose and an 84% inhibition at the 2.5 μmol/kg dose. The 2-16-2 MOE ASOs 7a and 8a gave no inhibition at the low dose, and produced ∼50% reduction in ApoB-100 mRNA at the 2.5 μmol/kg (∼18 mg/kg) dose. The potency increase of the LNA ASOs was variable, with LNA 7b providing a 5-fold estimated increase in potency over the corresponding 2-16-2 MOE 7a, but not the parent 5-10-5 MOE ASO 6. LNA 8b was only marginally more active than MOE 8a, and substantially less active than the MOE 6. None of the MOE ASOs showed evidence for toxicity as measured by organ weights and serum transaminase increases. In contrast the LNA ASOs 7b and 8b both resulted in >20-fold increases in AST and ALT, along with increased organ weights at the 2.5 μmol/kg dose.
Figure 7.
Transaminases (bar graph, left scale), and reduction of ApoB mRNA (points on line graph, right scale) after treatment with MOE (a series) and LNA (b series) ASOs.
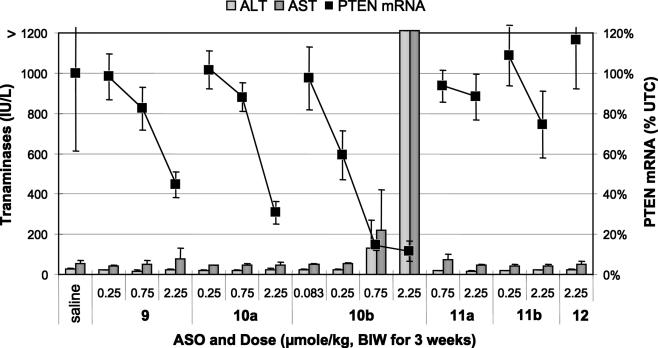
ASOs targeting murine PTEN were dosed at 0.083–2.25 μmol/kg twice weekly for 3 weeks (Figure 8). The parent MOE 5-10-5 ASO 9 as well as its gap widened counterpart 10a reduce target mRNA in a dose-dependent manner without evidence for toxicity as measured by transaminase levels, organ weights and body weights. The LNA ASO 10b was significantly more potent (estimated 5–10-fold) than either MOE ASO. However, once again, this increase in potency correlated with an increase in hepatotoxicity. The 0.75 μmol/kg dose group started to show mild transaminase elevations, while the higher dose group resulted in large (>50-fold) increases in both AST and ALT, increases in liver and spleen weights and caused significant weight loss in treated animals. A non-targeted control 5-10-5 MOE ASO 12 had no effect on either target or toxicity measures.
Figure 8.
Transaminases (bar graph, left scale), and reduction of PTEN mRNA (points on line graph, right scale) after treatment with MOE (a series) and LNA (b series) ASOs.
LNA ASO hepatotoxicity is not likely due to LNA degradation products
To help determine if the increased toxicity of the LNA ASOs was due to the oligonucleotide or to degradation products, we prepared mixed backbone versions of the PTEN 2-16-2 ASOs containing phosphodiester (PO) linkages between LNA nucleosides as well as at the LNA/DNA junction (Table 1). These ASOs are much more rapidly metabolized in vivo, and presumably will release either free MOE or LNA nucleosides or nucleotides. If these nucleosides or nucleotides were the source of the observed hepatotoxicity, these ASOs should be more toxic, while if the intact ASO was causing the hepatotoxicity, the mixed backbone versions should be less toxic. Neither the MOE (11a) nor LNA (11b) ASO caused significant target reduction at the doses tested. Importantly, the LNA containing ASO showed no evidence of hepatotoxicity, and analysis of drug levels in liver confirmed near complete metabolism of the intact drug 11b. These results suggest that the intact LNA oligonucleotides are responsible for the observed hepatotoxicity.
LNA ASO shows hepatotoxicity in rats
As the sequence of the PTEN ASOs was homologous to rat, we were able to examine if LNA 10b was more potent in rat, and importantly if the toxicity observed in mouse translated to another species. Accordingly, rats were treated with ASOs 9, 10b, or control 12 at dose of 0.83, 2.5 or 7.5 μmol/kg twice weekly for 3 weeks (Supplementary Figures S7–S9). The MOE ASO 9 showed a dose-dependent reduction of PTEN mRNA in rat liver, with an ED50 of ∼2.5 μmol/kg. In contrast to the mouse data, the LNA ASO 10b was only slightly more potent in rat liver, with an estimated ED50 of ∼1.5 μmol/kg. However, the LNA ASO was also hepatotoxic in rats. At the 7.5 μmol/kg dose, body weights were decreased 25% relative to control ASO 12, AST (but not ALT) was increased 5- and 10-fold, respectively in two of the four animals, and bilirubin was increased dramatically in the same two animals. Histopathological evaluation of H&E stained liver sections confirmed hepatotoxicity, showing moderate eosinophilic cytoplasmic degeneration with focal single cell apoptosis and mild mononuclear cell infiltration (Supplementary Table S4). No hepatotoxicity was observed by histopathological evaluation, ALT, AST, bilirubin, organ weights or body weights for either of the MOE ASOs. This data suggest that the potency increase of LNA ASOs relative to MOE ASOs is more pronounced in mouse than in rat, though the observed hepatotoxicity is still present in rat.
DISCUSSION
The main goal of our study was to determine if LNA containing ASOs would improve potency and therapeutic index relative to the current generation of MOE ASOs. Our assumption entering the work was that an improvement in potency would yield an improved therapeutic index, since it has been generally believed that many of the toxicities of ASOs are due to class effects as a result of the PS backbone. However, this proved not to be the case with the LNA ASOs studied.
Our results clearly demonstrate the ability to improve potency with some, but not all, LNA containing ASO designs, particularly for the TRADD and PTEN targets. This improvement was occasionally fairly large, as much as 5–10-fold, and was most pronounced for LNA ASOs of length 18–20 nt which contained 2–3 LNA residues at each end. As little as 0.75–1 μmol/kg (∼5–6 mg/kg) of these ASOs given twice weekly for 3 weeks reduced target mRNA by 80%. The optimal LNA ASO design in vivo appeared to be different than that observed in cell culture, where we found that two LNA nucleotides on each end of the ASO provided the largest potency increase. This is evident from a comparison of 3b and 4b, where 3b was 5-fold more potent in cell culture, but less potent in vivo. It is unlikely that the improved potency is due solely to increased affinity of the ASO for target RNA, as adding more LNA to the ASO actually decreased potency both in cell culture and in vivo (compare 2b and 3b with 1b). Because of these trends, combined with the lack of increased distribution of LNA ASOs to liver, it is likely that other factors are contributing to the increased potency of LNA modified ASOs observed in our studies. Additional investigations will be required to further characterize the nature of this potency improvement.
Unfortunately, the increased activity of LNA containing ASOs was also accompanied by the observation of severe hepatotoxicity, such that there was little or no separation between toxic doses and those that produced significant levels of mRNA reduction. Hepatotoxicity was chemistry-, sequence- and design-dependent, as it was only observed with LNA containing ASOs, and the onset occurred at slightly different dose levels for different compounds. The fact that the MOE ASOs in some cases (compare 2a with 2b, and 6 with 7b and 8b) produced similar reductions in target RNA without producing observable toxicity suggest that the toxicity is not secondary to reduction in target gene expression. This is further supported by the observation of severe hepatotoxicity with control 5b, which has >3 mismatches to all known mouse sequences. Hepatotoxicity also seemed to be the most severe for the more potent LNA ASO designs regardless of target (see 4b, mismatch 5b, 7b and 10b). Thus, therapeutic index was not improved, and was likely decreased relative to the MOE ASOs.
The hepatotoxicity was evident from the observation of multiple parameters, including histopathological evaluation of liver tissue upon necropsy as well as large increases in plasma levels of aminotransferases (ALT and AST). Furthermore, the toxicity was commonly accompanied by large increases in liver and/or spleen weights, likely as a consequence arising from a response to hepatic injury induced by the LNA ASOs. In several cases, the toxicity was severe enough to cause extensive weight loss in the animals. Histology data clearly demonstrated both LNA oligonucleotide-indiced liver necrosis and activation of apoptosis pathways, as evidenced by H&E staining, as well as cleaved caspase 3, increased Bax expression and increased expression of the M30 neo-epitope. The upregulation of the pro-apoptotic protein Bax suggests involvement of the p53-mediated apoptosis pathway, as Bax is a key response gene to p53 activation (42,43). Furthermore, GADD45β, a key downstream target gene of p53 during DNA damage and repair process (44), was highly up-regulated in the injured hepatocytes. GADD45β appears to help protect cells against programmed cell death through blocking the c-jun N-terminal kinase cascade, and is probably induced in response to cellular damage.
It is unclear why LNA oligonucleotides cause this level of hepatotoxicity though the corresponding MOE oligonucleotides do not. One possibility is that antisense effects on genes partially complementary to the hepatotoxic ASOs are playing a role in the toxicity, as LNA ASOs have been shown to decrease the selectivity for a perfectly complementary target relative to the corresponding MOE ASOs (45). However, this seems unlikely because multiple unrelated LNA sequences cause similar toxicities. All oligonucleotides were prepared and purified in the same laboratory using the identical methods. The impurity profiles of LNA and MOE ASOs were nearly identical, and contained only the expected impurities resulting from PS oligonucleotide synthesis (PO, N-1, etc.). This makes it extremely unlikely that the toxicity is due to impurities resulting from LNA, but not MOE oligonucleotide synthesis. Additionally, since the metabolically unstable PO containing ASO 11b (which should be metabolized in vivo to LNA nucleosides and nucleotides) was non-toxic, it is not likely that the toxicity is due to the LNA monomers. This suggests that the intact LNA containing PS oligonucleotides are responsible for the observed toxicity. There are distinct structural differences between MOE and LNA which may allow LNA containing oligonucleotides to selectively effect hepatotoxicity. Perhaps importantly, the rigid acyclic 2′-methoxyethyl side chain of MOE protects the corresponding 3′-phosphorothioate linkage from interactions via increased steric bulk and hydration (8), relative to the compact and more hydrophobic cyclic structure of LNA. This could cause selective binding affinity differences between MOE and LNA oligonucleotides for as yet unknown macromolecular binding partners, and/or result in differential compartmentalization of the two classes of oligonucleotides within liver tissue.
The mild hepatotoxicity induced 4 days after a single administration of LNA ASO 4b occurred concurrently with apoptosis and activation of Bax and caspase 3 in hepatocytes as evidenced by histopathological evaluation. Furthermore, an increase in p-eIF2α was observed to coincide with the onset of toxicity. Phosphorylated eIF2α inhibits translation initiation, and has been shown to mediate apoptosis, possibly by preventing the synthesis of short lived anti-apoptotic factors (41,46). There are four known kinases which phosphorylate eIF2α: PKR, which is activated by binding of double stranded RNA (dsRNA); GCN2, which is activated by amino acid deprivation; HRI, which is activated by low heme levels; and PERK, which responds to stress in the endoplasmic reticulum. It is unclear from our data how treatment with hepatotoxic LNA oligonucleotides results in increased phosphorylation of eIF2α; however, it is tempting to speculate that PKR could be involved. PKR is activated by binding of dsRNA to distinct dsRNA binding domains which serve as allosteric inhibitors of the kinase domain (47,48). It remains to be determined if LNA oligonucleotides interact with PKR, and more extensive mechanistic work is required to confirm both p-eIF2α mediation of an acute apoptotic response in hepatocytes, and elucidate the precise biochemical mechanism responsible for the observed hepatotoxicity.
In conclusion, we have shown that optimization of size and gap configuration employing LNA containing ASOs can improve potency in mouse liver. However, this potency increase is strongly correlated with the onset of a severe hepatotoxicity not seen with the corresponding MOE ASOs, and therapeutic index is not improved. These results suggest that while LNA ASOs have the potential to improve potency of antisense therapeutics, they impose a significant risk of hepatotoxicity. The toxicities observed herein also pose challenges for interpreting target validation and pharmacology experiments using LNA ASOs in rodent models. It is possible more extensive screening efforts may identify less toxic LNA sequences, or that other ASO designs utilizing LNA may not elicit hepatotoxicity. Additionally, bicyclic nucleosides similar to LNA such as amino-LNA, thio-LNA (27) and ENA (which contains one extra methylene unit in the bicyclic bridge) (49) have been reported to show antisense effects similar to LNA and MOE ASOs. Although optimized designs of one or more of these modifications may ultimately improve potency while maintaining the safety profile of MOE ASOs, no detailed characterization of their toxicological properties has yet been reported. It is clear that additional structure–activity and structure–toxicity relationship studies will be required to more fully assess the therapeutic potential of LNA and other bicyclic nucleoside modified oligonucleotides. Ultimately, the value of LNA oligonucleotide drugs will be determined in human clinical trials, where they are currently being developed for the treatment of cancer. Our results are based only on studies in rodents; however, caution should be exercised in the clinical development of LNA modified oligonucleotides, especially for chronic, non-life threatening indications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Charles Allerson, Amy Dan and Rick Carty for synthesis of oligonucleotides, Megan Pearce for determination of ASO levels in liver tissue, Denis Drygin for the caspase 3 activation data, Richard H. Griffey for numerous thought provoking discussions and Stanley T. Crooke and Arthur A. Levin for a critical read of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Isis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vitravene Study Group. Randomized dose-comparison studies of intravenous fomiversen for treatment of cytomegalovirus retinitis that has reactivated or is persistently active despite other therapies in patients with AIDS. Am. J. Ophthalmol. 2002;133:475–483. doi: 10.1016/s0002-9394(02)01326-0. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 3.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geary R.S., Yu R.Z., Levin A.A. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr. Opin. Investig. Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- 5.Lima W.F., Nichols J.G., Wu H., Prakash T.P., Migawa M.T., Wyrzykiewicz T.K., Bhat B., Crooke S.T. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 6.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 7.Martin P. Ein neuer Zugang zu 2-O-Alkylribonucleosiden und Eigenschaften deren Oligonucleotide. Helv. Chim. Acta. 1995;78:486–504. [Google Scholar]
- 8.Teplova M., Minasov G., Tereshko V., Inamati G.B., Cook P.D., Manoharan M., Egli M. Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nature Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 9.McKay R.A., Miraglia L.J., Cummins L.L., Owens S.R., Sasmor H., Dean N.M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 10.Altmann K.H., Martin P., Dean N.M., Monia B.P. Second generation antisense oligonucleotides—inhibition of pkc-alpha and c-raf kinase expression by chimeric oligonucleotides incorporating 6′-substituted carbocyclic nucleosides and 2′-O-ethylene glycol substituted ribonucleosides. Nucleosides Nucleotides. 1997;16:917–926. [Google Scholar]
- 11.Geary R.S., Watanabe T.A., Truong L., Freier S., Lesnik E.A., Sioufi N.B., Sasmor H., Manoharan M., Levin A.A. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001;296:890–897. [PubMed] [Google Scholar]
- 12.Yu R.Z., Geary R.S., Monteith D.K., Matson J., Truong L., Fitchett J., Levin A.A. Tissue disposition of 2′-O-(2-methoxy) ethyl modified antisense oligonucleotides in monkeys. J. Pharm. Sci. 2004;93:48–59. doi: 10.1002/jps.10473. [DOI] [PubMed] [Google Scholar]
- 13.Geary R.S., Yu R.Z., Watanabe T., Henry S.P., Hardee G.E., Chappell A., Matson J., Sasmor H., Cummins L., Levin A.A. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab. Dispos. 2003;31:1419–1428. doi: 10.1124/dmd.31.11.1419. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., Cook J., Nickel J., Yu R., Stecker K., Myers K., Dean N.M. Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat. Biotechnol. 2000;18:862–867. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- 15.Yu R.Z., Zhang H., Geary R.S., Graham M., Masarjian L., Lemonidis K., Crooke R., Dean N.M., Levin A.A. Pharmacokinetics and pharmacodynamics of an antisense phosphorothioate oligonucleotide targeting Fas mRNA in mice. J. Pharmacol. Exp. Ther. 2001;296:388–395. [PubMed] [Google Scholar]
- 16.Liang Y., Osborne M.C., Monia B.P., Bhanot S., Watts L.M., She P., Decarlo S.O., Chen X., Demarest K. Antisense oligonucleotides targeted against glucocorticoid receptor reduce hepatic glucose production and ameliorate hyperglycemia in diabetic mice. Metabolism. 2005;54:848–855. doi: 10.1016/j.metabol.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 17.Zinker B.A., Rondinone C.M., Trevillyan J.M., Gum R.J., Clampit J.E., Waring J.F., Xie N., Wilcox D., Jacobson P., Frost L., et al. PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose, and improves insulin sensitivity in diabetic mice. Proc. Natl Acad. Sci. USA. 2002;99:11357–11362. doi: 10.1073/pnas.142298199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler M., McKay R.A., Popoff I.J., Gaarde W.A., Witchell D., Murray S.F., Dean N.M., Bhanot S., Monia B.P. Specific inhibition of PTEN expression reverses hyperglycemia in diabetic mice. Diabetes. 2002;51:1028–1034. doi: 10.2337/diabetes.51.4.1028. [DOI] [PubMed] [Google Scholar]
- 19.Crooke R.M., Graham M.J., Lemonidis K.M., Whipple C.P., Koo S., Perera R.J. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Kastelein J.J.P., Wedel M.K., Baker B.F., Su J., Bradley J.D., Yu R.Z., Chuang E., Graham M.J., Crooke R.M. Potent reduction of apolipoprotein B and LDL cholesterol by short-term administration of an antisense inhibitor of apolipoprotein B. Circulation. 2006;114:1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- 21.Chi K.N., Eisenhauer E., Fazli L., Jones E.C., Goldenberg S.L., Powers J., Tu D., Gleave M.E. A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2′-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer. J. Natl Cancer Inst. 2005;97:1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- 22.Sewell K.L., Geary R.S., Baker B.F., Glover J.M., Mant T.G., Yu R.Z., Tami J.A., Dorr F.A. Phase I trial of ISIS 104838, a 2′-methoxyethyl modified antisense oligonucleotide targeting tumor necrosis factor-alpha. J. Pharmacol. Exp. Ther. 2002;303:1334–1343. doi: 10.1124/jpet.102.036749. [DOI] [PubMed] [Google Scholar]
- 23.Obika S., Nanbu D., Hari Y., Morio K.-I., In Y., Ishida T., Imanishi T. Synthesis of 2′-O-4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 24.Obika S., Nanbu D., Hari Y., Andoh J.-I., Morio K.-I., Doi T., Imanishi T. Stability and structural features of the duplexes containing nucleoside analogs with a fixed N-type conformation, 2′-O-4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 25.Wengel J. Synthesis of 3′-C- and 4′-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA) Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 26.Koshkin A.A., Nielsen P., Meldgaard M., Rajwanshi V.K., Singh S.K., Wengel J. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 27.Fluiter K., Frieden M., Vreijling J., Rosenbohm C., De Wissel M.B., Christensen S.M., Koch T., Orum H., Baas F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide. Chem. Biochem. 2005;6:1104–1109. doi: 10.1002/cbic.200400419. [DOI] [PubMed] [Google Scholar]
- 28.Siwkowski A.M., Madge L.A., Koo S., McMillan E.L., Monia B.P., Pober J.S., Baker B.F. Effects of antisense oligonucleotide-mediated depletion of tumor necrosis factor (TNF) receptor 1-associated death domain protein on TNF-induced gene expression. Mol. Pharmacol. 2004;66:572–579. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 29.Chiang M.Y., Chan H., Zounes M.A., Freier S.M., Lima W.F., Bennett C.F. Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J. Biol. Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- 30.Dean N.M., McKay R. Inhibition of protein kinase C-alpha expression in mice after systemic administration of phosphorothioate antisense oligodeoxynucleotides. Proc. Natl Acad. Sci. USA. 1994;91:11762–11766. doi: 10.1073/pnas.91.24.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 32.Ross B., Song Q. 2004. US U.S. Pat. Appl. Publ. 20040082775. [Google Scholar]
- 33.Ross B.S., Song Q., Han M. Kilo-scale synthesis process for 2′-O-(2-methoxyethyl)-pyrimidine derivatives. Nucleosides Nucleotides Nucleic Acids. 2005;24:815–818. doi: 10.1081/ncn-200060295. [DOI] [PubMed] [Google Scholar]
- 34.Ravikumar V.T., Andrade M., Carty R.L., Dan A., Barone S. Development of siRNA for therapeutics: efficient synthesis of phosphorothioate RNA utilizing phenylacetyl disulfide (PADS) Bioorg. Med. Chem. Lett. 2006;16:2513–2517. doi: 10.1016/j.bmcl.2006.01.081. [DOI] [PubMed] [Google Scholar]
- 35.Drygin D., Barone S., Bennett C.F. Sequence-dependent cytotoxicity of second-generation oligonucleotides. Nucluic Acids Res. 2004;32:6585–6594. doi: 10.1093/nar/gkh997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leeds J.M., Graham M.J., Truong L., Cummins L.L. Quantitation of phosphorothioate oligonucleotides in human plasma. Anal. Biochem. 1996;235:36–43. doi: 10.1006/abio.1996.0088. [DOI] [PubMed] [Google Scholar]
- 37.Frieden M., Christensen S.M., Mikkelsen N.D., Rosenbohm C., Thrue C.A., Westergaard M., Hansen H.F., Orum H., Koch T. Expanding the design horizon of antisense oligonucleotides with alpha-l-LNA. Nucleic Acids Res. 2003;31:6365–6372. doi: 10.1093/nar/gkg820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 39.Bondensgaard K., Petersen M., Singh S.K., Rajwanshi V.K., Kumar R., Wengel J., Jacobsen J.P. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chem.-Eur. J. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 40.Grassi A., Susca M., Ferri S., Gabusi E., D'Errico A., Farina G., Maccariello S., Zauli D., Bianchi F.B., Ballardini G. Detection of the M30 neoepitope as a new tool to quantify liver apoptosis: timing and patterns of positivity on frozen and paraffin-embedded sections. Am. J. Clin. Pathol. 2004;121:211–219. doi: 10.1309/UK62-1LFJ-4FX0-7KDE. [DOI] [PubMed] [Google Scholar]
- 41.Scheuner D., Patel R., Wang F., Lee K., Kumar K., Wu J., Nilsson A., Karin M., Kaufman R.J. Double-stranded RNA-dependent protein kinase phosphorylation of the alpha-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J. Biol. Chem. 2006;281:21458–21468. doi: 10.1074/jbc.M603784200. [DOI] [PubMed] [Google Scholar]
- 42.Chipuk J.E., Kuwana T., Bouchier-Hayes L., Droin N.M., Newmeyer D.D., Schuler M., Green D.R. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 43.Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U.M. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 44.Xiao G., Chicas A., Olivier M., Taya Y., Tyagi S., Kramer F.R., Bargonetti J. A DNA damage signal is required for p53 to activate gadd45. Cancer Res. 2000;60:1711–1719. [PubMed] [Google Scholar]
- 45.Simoes-Wust A.P., Hopkins-Donaldson S., Sigrist B., Belyanskaya L., Stahel R.A., Zangemeister-Wittke U. A functionally improved locked nucleic acid antisense oligonucleotide inhibits Bcl-2 and Bcl-xL expression and facilitates tumor cell apoptosis. Oligonucleotides. 2004;14:199–209. doi: 10.1089/oli.2004.14.199. [DOI] [PubMed] [Google Scholar]
- 46.Saelens X., Kalai M., Vandenabeele P. Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J. Biol. Chem. 2001;276:41620–41628. doi: 10.1074/jbc.M103674200. [DOI] [PubMed] [Google Scholar]
- 47.Nanduri S., Rahman F., Williams B.R., Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S., Kaufman R.J. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 49.Koizumi M., Takagi-Sato M., Okuyama R., Araki K., Sun W., Nakai D., Tsutsumi S., Kawai K. Direct comparison of in vivo antisense activity of ENA oligonucleotides targeting PTP1B mRNA with that of 2′-O-(2-methoxy)ethyl-modified oligonucleotides. Oligonucleotides. 2006;16:253–262. doi: 10.1089/oli.2006.16.253. [DOI] [PubMed] [Google Scholar]