Abstract
The homotrimeric fusion protein nucleocapsid (NC)-dUTPase combines domains that participate in RNA/DNA folding, reverse transcription, and DNA repair in Mason-Pfizer monkey betaretrovirus infected cells. The structural organization of the fusion protein remained obscured by the N- and C-terminal flexible segments of dUTPase and the linker region connecting the two domains that are invisible in electron density maps. Small-angle X-ray scattering reveals that upon oligonucleotide binding the NC domains adopt the trimeric symmetry of dUTPase. High-resolution X-ray structures together with molecular modeling indicate that fusion with NC domains dramatically alters the conformation of the flexible C-terminus by perturbing the orientation of a critical β-strand. Consequently, the C-terminal segment is capable of double backing upon the active site of its own monomer and stabilized by non-covalent interactions formed with the N-terminal segment. This co-folding of the dUTPase terminal segments, not observable in other homologous enzymes, is due to the presence of the fused NC domain. Structural and genomic advantages of fusing the NC domain to a shortened dUTPase in betaretroviruses and the possible physiological consequences are envisaged.
INTRODUCTION
dUTPases have an essential role in regulating cellular dUTP/dTTP ratios by catalyzing the hydrolysis of dUTP into the dTTP-precursor dUMP and pyrophosphate. Lack of enzymatic activity leads to high levels of deoxyuridine incorporation into DNA. Uracil-DNA transforms base-excision repair into a hyperactive futile cycle resulting in thymine-less cell death (1). All free-living organisms, as well as several DNA and retroviruses encode dUTPase. The enzyme is essential for viability in pro- and eukaryotes (2,3) and contributes to host range preferences and infectivity of viruses (4).
Betaretroviral genomes contain the dUTPase gene adjacent to the gene of the nucleocapsid (NC) polypeptide. In betaretroviruses Mouse mammary tumour virus (MMTV) and Mason-Pfizer monkey virus (M-PMV), ribosomal frameshifts between gag and pro as well as between pro and pol frames yield Gag-Pro and Gag-Pro-Pol polyproteins. The first frameshift gives rise to a transframe fusion protein joining the NC and dUTPase polypeptides (NC-dUTPase). Recently, NC-dUTPase was shown to be present in stable form resisting proteolysis by retroviral and cellular proteases in M-PMV virion as well as in M-PMV virus-infected cells (5). The fusion protein is the only physiological form of M-PMV dUTPase. M-PMV NC-dUTPase retains both nucleic acid binding and dUTP hydrolyzing catalytic activity. The catalytic rate constant of recombinant M-PMV NC-dUTPase is, however, 10-fold decreased as compared to lentiviral or other dUTPases. Enzyme kinetic analyses of recombinant NC-dUTPase and a truncated protein segment comprising only the dUTPase domain suggested that (i) the NC domain has no inverse effect on enzymic activity and (ii) oligonucleotide binding to the NC domain may modulate enzymatic activity. These results failed to provide an explanation for the 10-fold lowered catalytic rate constant. A significant shortening of linker region between conserved Motif IV and V (Figure 1) was observed in all betaretroviral NC-dUTPases. A hypothesis was put forward arguing that lack of these connecting residues may give rise to a steric constraint thereby lowering catalytic efficiency (5).
Figure 1.
Alignment of betaretroviral NC-dUTPase sequences. Grey background indicates Zn-knuckle motifs, white lettering on black background indicates the five conserved dUTPase motifs. Asterisks show preferential tryptic sites of M-PMV NC-dUTPase, as identified by mass spectrometry. Dots indicate point mutation sites: Gly160Pro, Ser163Gly, Gln220STOP, Arg223Lys, Ser227Gly, and Asp232Gly. M-PMV, (GenBank™ accession M12349); SRV-1, simian retrovirus 1 (GenBank™ accession U85506); SRV-2, simian retrovirus 2 (GenBank™ accession M16605); JSRV, Jaagsiekte sheep retrovirus (GenBank™ accession M80216); MMTV, (GenBank™ accession 15122).
Retroviral proteins are usually cleaved out of viral polyproteins by the retroviral protease to result in separate polypeptides. The transframe NC-dUTPase fusion protein of M-PMV, however, does not get cleaved into separate NC and dUTPase proteins, although both NC and dUTPase proteins are present in separate forms in other retroviruses. The resistance of NC-dUTPase against retroviral protease is due to a highly conserved sequence segment at the gag-pro frameshift site that connects the two protein domains and that is not susceptible to proteolysis by the retroviral protease (5).
Why do betaretroviruses contain this intriguing fusion protein while other retroviruses manage themselves with separate NC and dUTPase polypeptides? What is the reason for the low catalytic activity of NC-dUTPase? Analysis of structure and function relationships may provide some clues to these problems. Here, high-resolution 3D structural investigations by X-ray crystallography in combination with a gallery of structural investigations in the solution phase as well as computational methods were employed to characterize the full-length fusion protein with a special focus on the roles of the flexible N- and C-terminal segments. Mutant constructs were designed to explain the 10-fold decreased catalytic efficiency of M-PMV NC-dUTPase. Modeling studies revealed that the C-terminal arm in M-PMV dUTPase folds back upon its own monomer and reaches the active site in a novel fashion, possibly causing steric constraints. Results show that the protein N- and C-terminal segments of the same subunits shape co-folding and function of NC-dUTPase in a concerted fashion. The dUTPase homotrimeric core efficiently organizes the NC domains that also adopt some symmetrical orientation induced by oligonucleotide binding. By unique co-folding, the fusion protein offers an advantageous novel way for dUTPase active site organization within a shortened polypeptide and increased symmetry in NC domains.
MATERIALS AND METHODS
Electrophoresis materials were from BioRad, resins and columns from Amersham Biosciences, UK. Phenol Red indicator was from Merck, Germany. Nucleotides, buffer substances, and other materials of analytical grade purity were from Sigma, US, α,β-imino dUTP was from Jena Bioscience. Restriction enzymes and other molecular biology materials were from New England BioLabs or Stratagene, USA, unless stated otherwise. Wild type and mutant M-PMV NC-dUTPases were constructed using standard molecular biology protocols, and were purified as in (5).
Protein concentration was measured by Bradford's assay (6) or spectrophotometrically using A0.1% 1 cm, 280 nm = 0.74 for NC-dUTPase.
Discontinuous activity assay by thin layer chromatography was used to check the catalytic competence of protein crystals and for following the slow enzymatic hydrolysis of α,β-imino dUTP (5).
Continuous activity assay was performed at enzyme concentrations 0.5–1 μM in 1 mM TES/HCl, pH 7.5 containing 40 μM dUTP, 5 mM MgCl2, 150 mM KCl, and 40 μM Phenol Red at 25°C, using a JASCO-V550 spectrophotometer. Initial velocity, kcat and KM were determined as in (7,8).
SDS–PAGE was performed according to Laemmli (9) using 12–14% polyacrylamide gels stained by colloidal Coomassie Brilliant Blue (Bio-Rad).
Small-angle X-ray scattering studies were performed at EMBL on the storage ring DORIS III of the Deutsches Elektronen Synchrotron (DESY) using beamline X33. The samples contained 1.3–7.4 mg/ml NC-dUTPase or 3.4–5.5 mg/ml NC protein, freshly prepared before the measurements, with and without equimolar (TG)4 oligonucleotide in 500 mM Tris–HCl buffer pH 8.0, also containing 150 mM NaCl. Low resolution shape analysis of all solutes was done using the ab initio program DAMMIN (10) which represents the macromolecule by an assembly of densely packed beads. For rigid body modeling of NC-dUTPase, the atomic models of the individual domains were employed and the program BUNCH was used (11) on SAXS data. The high-resolution models were retrieved from the Brookhaven Protein Data Bank (12) (PDB): dUTPase 2AKV, NC domain with oligonucleotide 1A1T, model 1 (13) and free NC 1AAF, model 1 (14). Sofwares used are detailed and PDB files of the models are included in the supplementary data.
Dynamic modeling of the C-terminal 219QQPYRGQGSFG229 segment—Based on the structure of the human dUTPase with α,β-imino dUTP substrate (PDB 2HQU), three distance constraints were applied: 4.9 Å between the ring centers of the α,β-imino dUTP pyrimidine and the Phe228 benzene rings, 2.5 and 3.5 Å between the γ-phosphate O2G and O3G and the Arg223 NH2, respectively. The initial model has been generated by the Modeller program (15) using the structure PDB 2D4N (cf Table 1). The arm was computed for one monomer, the other two segments were generated by symmetry operations. For details, see supplementary data.
Table 1.
Crystallographic data collection and refinement statistics
| WT-dUTPase (PDB: 2D4M) | Q220Stop mutant dUTPase (PDB: 2D4L) | WT-dUTPase: α,β-imino dUTP:Mg2+ (PDB: 2D4N) | |
|---|---|---|---|
| Space group | P63 | P63 | P63 |
| Cell parameters a, c (Å) | 61.2, 63.3 | 61.2, 64.2 | 61.1, 64.3 |
| Wavelength (Å) | 0.8031 | 0.8430 | 0.8128 |
| Resolution (Å) | 20.0–1.83 (1.94–1.83) | 20.0–1.70 (1.80–1.70) | 20.0–1.52 (1.61–1.52) |
| Measured reflections | 31 928 (4585) | 45 739 (7137) | 55 279 (7005) |
| Unique reflections | 11 637 (1804) | 14 813 (2308) | 20 787 (3282) |
| Completeness (%) | 97.4 (95.0) | 98.3 (98.0) | 98.7 (98.2) |
| <I/σ(I)> | 8.9 (2.7) | 21.2 (3.3) | 12.3 (2.9) |
| Rmeas (%)a | 9.1 (50.3) | 3.7 (53.3) | 6.7 (44.2) |
| Refinement statistics | |||
| Resolution (Å) | 20.00–1.85 | 20.00–1.7 | 20.00–1.53 |
| Nonhydrogen atoms | 982 | 975 | 1029 |
| Water molecules | 147 | 124 | 145 |
| Data:parameter:restraint | |||
| Rmsd. bonds (Å)b | 0.02 | 0.02 | 0.019 |
| Rmsd. angles (°)b | 1.862 | 1.948 | 1.911 |
| Rwork (%)c | 16.02 | 16.23 | 16.44 |
| Rfree (%)c,d | 19.19 | 17.88 | 18.71 |
WT indicates wild type enzyme.
Values in parentheses correspond to the highest resolution shell.
a, where Ij is the recorded intensity of the jth reflection and 〈I〉 is the average intensity over multiple recordings and n is the multiplicity (34).
bRoot mean square deviation from ideal/target geometries.
cRwork, free = ∑‖Fobs|−|Fcalc‖/|Fobs|.
dRfree values are calculated for a randomly selected 5% of the data that was excluded from the refinement.
Crystallization and structure determination—Conditions used to crystallize wild type and C-terminally truncated dUTPases were as described previously (16). Structure determination was carried out by rigid body refinement (CCP4i) (17) using the structure of the heavy-atom derivative complex (16). The asymmetric unit contained one monomer. In each case, the resultant initial maps were of exceptional quality, allowing residues 107–219 and entire ligands to be built with ease. Generation of monomer libraries for the ligand (α,β-imino dUTP) and refinement were carried out using CCP4i and Refmac5 (17). Positional and B-factor refinement rounds were altered with manual rebuilding steps using the graphics program Coot (18) and ARP/wARP (19) was used for water building. Residues belonging to the C-terminal Motif V (residues 220–234) are hardly visible even in the final maps and are therefore mostly omitted from the model. A summary of the crystallographic data collection and refinement statistics is given in Table 1. Coordinates and structure factor data have been deposited in the Protein Data Bank with accession codes 2D4M, 2D4N and 2D4L for the wild type apoenzyme, wild type enzyme:α,β,-imino dUTP:Mg2+ complex, and the Gln220STOP mutant enzyme, respectively.
RESULTS
Several constructs of the NC-dUTPase fusion protein were designed to allow a detailed characterization of its structure and function (Table 2). Flexible segments were identified by DISPROT and IUPred prediction methods (20,21) as well as by limited proteolysis. Results showed in agreement that most of the NC domain and as well as the segments containing residues 8–130 and 220–234 are characterized by highly flexibility (cf Supplementary Figures S1 and S2).
Table 2.
Description of NC-dUTPase constructs
| Construct | Methods of characterization | Results |
|---|---|---|
| Wild type full-length NC-dUTPase (Ala1–Tyr234) | Kinetics X-ray crystallography (visible segment Val85–Val219) Dymanic modeling SAXS Fluorescence spectroscopy Isothermal calorimetry titration Limited proteolysis | Table 3, 4 Figure 2, Table 1 Figures 3 and 5 Figures 4 and Supplementary Figure S5 Supplementary Figure S4 Text Figure 1 and Supplementary Figure S2 |
| Truncated mutant Gln220STOP (Ala1–Gln219) | Kinetics X-ray crystallography (visible segment Val85–Val219) | Table 3 Table 2 |
| Trypsinolysed construct (Val85–Arg223) | Kinetics | Table 3 |
| Point mutants (Ala1–Tyr234) Arg223Lys Ser227Gly/Asp232Gly Gly160Pro/Ser163Gly/Ser227Gly/Asp232Gly | Kinetics | Table 3 |
| C-terminal arm (Gln219–Gly229) | Dynamic modeling | Figures 3 and 5 |
| NC domain (Ala1–Asn75) | SAXS | Figure 4 and Supplementary Figure S5 |
Enzymatic role of the flexible C-terminus
Among the five conserved motifs, characteristic for all dUTPases, betaretroviral dUTPase sequences show some variations within Motifs II and V (22). The usual consensi for Motif II and Motif V are ProArgSerGly and ArgGlyXxxGlyGlyPheGlySerThrGly; while for M-PMV dUTPase these segments are 160GlyArgSerSer163 and 223ArgGlyGlnGlySerPheGlySerSerAsp232, respectively (altered residues in bold face) (Figure 1). Both of these Motifs are essential for enzyme function and Motif V is characterized with an increased flexibility (8,23,24). To investigate in detail the role of the flexible Motif V and to decide if the residue alterations may explain the low catalytic efficiency of M-PMV dUTPase, several mutations were designed (Table 2). Kinetic analysis with mutant constructs (Ser227Gly/Asp232Gly and Gly160Pro/Ser163Gly/Ser227Gly/Asp232Gly) indicates that the differences in the amino acid sequence do not contribute to low catalytic efficiency (Table 3, cf also Table 4). Further constructs focused on Motif V. Both truncated constructs (Gln220STOP and the Val85–Arg223 tryptic segment) are characterized with a ≥105-fold decrease in catalytic rate constant (Table 3), arguing for the critical importance of the intact Motif V for protein function. It is also evident that the presence of Arg223, one of the most important Motif V residues does not reconstitute catalytic activity in lack of other Motif V residues. The role of the Arg223 residue was further explored in the mutant Arg223Lys where the only difference is that the guanidine group of the arginine side chain is replaced by the amino group of lysine. This mutant showed again a 105-fold decrease in catalytic rate constant (Table 3), suggesting that the guanidine group is crucial for catalytic function. The mutational analysis proved the essential role of Motif V and also showed that the 10-fold decrease in kcat of M-PMV dUTPase is not due to alterations of conserved Motif II and V residues.
Table 3.
Kinetic constants of M-PMV NC-dUTPase constructs
| kcat (s−1) | |
|---|---|
| Gln220STOP mutant (Ala1–Gln119) | <10−5 |
| Arg223Lys mutant | <10−5 |
| Trypsinolysed construct (Val85–Arg223) | <10−5 |
| Ser227Gly/Asp232Gly (double mutant) | 0.8 ± 0.2 |
| Gly160Pro/Ser163Gly/ Ser227Gly/Asp232Gly (quadruple mutant) | 0.9 ± 0.3 |
Kinetic data were determined in three to five parallel independent experiments.
Table 4.
Kinetic constants of M-PMV dUTPase in the absence and presence of Mg2+
| kcat (s−1) | kcat/KM (s−1M−1) × 106 | |
|---|---|---|
| M-PMV dUTPase in the absence of added Mg2+ | 0.45 | 0.38 |
| M-PMV dUTPase in the presence of 1 mM Mg2+ | 0.78 | 1.60 |
| M-PMV NC-dUTPase in the absence of added Mg2+ | 0.50 | 0.33 |
| M-PMV NC-dUTPase in the presence of added Mg2+ | 0.80 | 1.65 |
Kinetic data were determined in 3–5 parallel independent experiments. Data represent the mean of the measured value (with SD ± 18%).
Crystal structure of the folded protein core
Wild type and C-terminally truncated NC-dUTPase were crystallized to determine the high-resolution 3D structure of the protein, to estimate potential effects of the C-terminus on the folding of the protein and to provide a detailed description of the active site that may account for the 10-fold decreased enzymic activity (Table 1). The crystals contained only the dUTPase domain, as determined by N-terminal microsequencing and mass spectrometry of redissolved crystals, the N-terminus corresponded to the Val85 (cf Figure 1), although the full-length NC-dUTPase protein was crystallized. This fact is most probably due to some proteolytic degradation during crystallization that specifically targeted the flexible linker region between the NC and the dUTPase domains. Importantly, the C-terminus of the dUTPase domain, containing the conserved dUTPase Motif V (cf Figure 1), remained intact in the crystals, as determined by mass spectrometric analyses (data not shown).
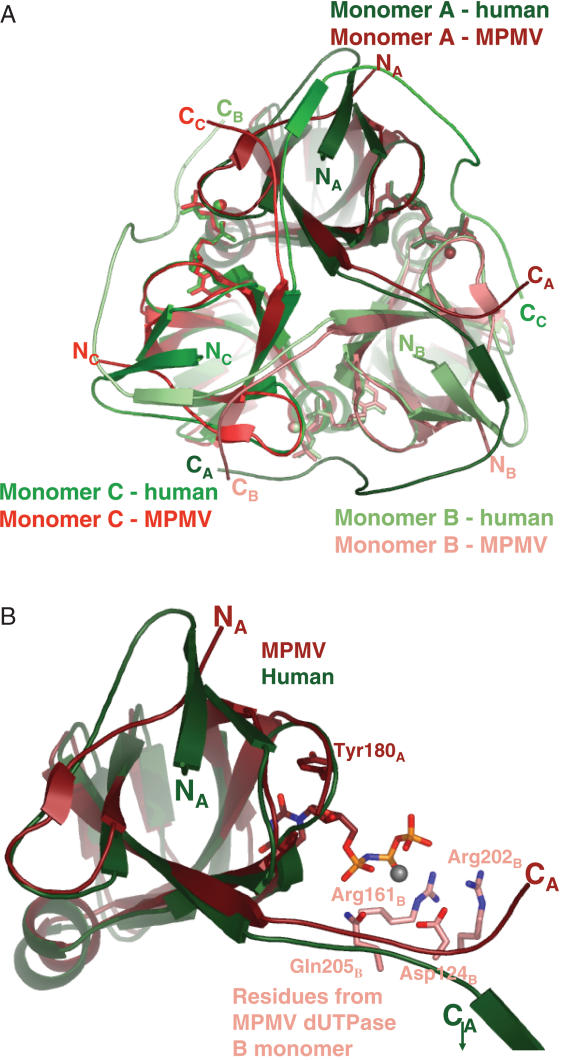
Apoenzyme and substrate-enzyme complexes were crystallized to gain information on substrate binding induced conformational changes that may explain the low catalytic efficiency. The dUTPase fold (jelly-roll subunits entwined in a homotrimer) was found in all these crystals providing yet another strengthening example for the conservation of this quaternary structure in homotrimeric dUTPases (Figure 2A and B). For formation of the enzyme–substrate complex, the physiological dUTP substrate was replaced by the slowly hydrolysable isosteric analogue α,ß-imino dUTP by Escherichia coli dUTPase (25). M-PMV dUTPase also catalyzes the hydrolysis of α,ß-imino dUTP (although kcat is decreased by four orders of magnitude, data not shown). This substrate analogue is accommodated in a manner highly reminiscent to other dUTPases. The uracil and deoxyribose rings are contacted within a ß-hairpin around Motif III of subunit A, e.g. a conserved tyrosine (Tyr180 in M-PMV NC-dUTPase) stacks to the deoxyribose ring. Residues from the conserved Motif II (Arg161 in M-PMV NC-dUTPase) and Motif IV (Arg202 and Gln205 in M-PMV NC-dUTPase) of subunit B reach towards the phosphate chain of the substrate (cf also Figure 1 for sequences and Supplementary Figure S3). Conserved Motif I of subunit B donates an aspartate residue (Asp124 in M-PMV NC-dUTPase) that contributes to coordination of the catalytic co-factor Mg2+ (Supplementary Figure S3). The intimate pair-wise interactions between subunits A and B donating conserved residues for jointly building the active site are found in an equivalent fashion in other dUTPases, as well as in the M-PMV dUTPase. Motif V at the C-terminus could not be localized in the density maps of the M-PMV dUTPase structure, similarly to several other cases due to the increased flexibility in this region. The electron density map was only traceable till Gln219 (16). In the authentic homotrimeric dUTPase fold, this Motif is donated by subunit C (for human dUTPase cf Figure 2A).
Figure 2.
3D crystal structures of M-PMV dUTPase and its complexes. Panel A. Superimposed structures of dUTPase- α,β-imino dUTP from M-PMV (determined in the present study, PDB 2D4N) and human dUTPase (manuscript in preparation, PDB 2HQU). Subunits are colored in shades of red or green for M-PMV or human dUTPase, respectively. The substrates and magnesium atoms are shown in subunit-color code, with ball-and-stick model. Panel B. Altered backbone conformation at the N- and C-terminal segments of M-PMV dUTPase. Monomer A is superimposed from human and M-PMV dUTPases. Conserved residues from monomer B, involved in substrate accommodation are shown in atomic coloring, with carbons colored to the monomer code shades. Substrate is in atomic coloring, magnesium is gray.
Despite the overall similarities, inspection of the active site and subunit interactions revealed some intriguing differences as compared to previously determined dUTPase and dUTPase-ligand complex structures. First, substrate binding induced ordering of two Arg side chains (Arg161, Arg202) of M-PMV dUTPase, both contributing to phosphate chain coordination (Supplementary Figure S3). No ordering was observed in the N- and C-terminal segments.
Second, in the quaternary fold of M-PMV dUTPase, a significant alteration is evident at both N- and the C-terminal regions (Figure 2A and B). In the usual homotrimeric dUTPase fold, the N-terminal β-strand contributes to the apolar core of its own monomer and participates in intersubunit interactions by donating main chain H-bonds to the C-terminal β-strand of the neighbouring subunit to realize the β-strand swapping. The continuing segment containing the conserved Motif V closes over the active site of the neighbouring monomer, as suggested by the crystal structures that could partially localize this segment in the electron density maps [PDB 1Q5H (24) and 1F7R (26)]. In the M-PMV dUTPase structure, the N-terminal β-strand is absent since the N-terminus needs to have an outgoing orientation to leave the monomer core and connect to the NC domain (Figure 2B). In lack of this N-terminal β-strand, the most important interaction for arm-swapping is lost. Moreover, although the full length of the C-terminal segment cannot be localized due to its increased flexibility, the initial main chain conformation clearly dictates an orientation highly different from the usual dUTPase fold (Figures 2A and B and 3). Given this initial orientation, it seems that the C-terminus in M-PMV dUTPases, even in fully extended conformation, cannot cover the ∼36 Å to contact the ligand in the active site of the neighbouring subunit, as it is the case in other dUTPases.
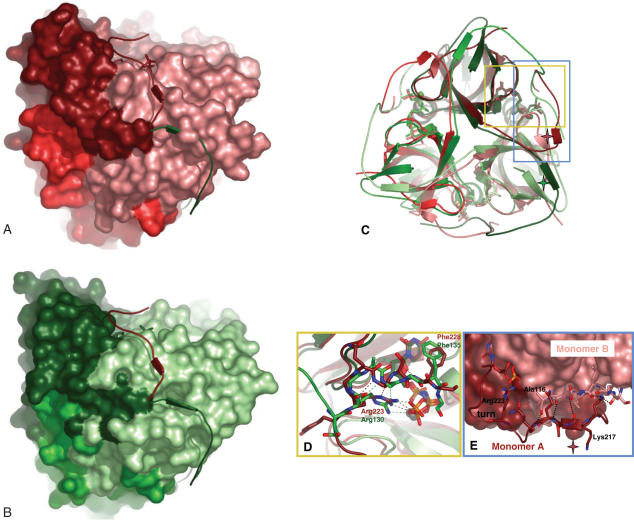
Figure 3.
Altered orientation of the C-terminal segment in human and M-PMV dUTPases. Panels A and B M-PMV (shades of red) and human (shades of green) dUTPases structures are shown in space-filling models. The C-terminal segments of both enzymes are shown superimposed (human dUTPase C-terminal segment (green) onto M-PMV dUTPase structure (red) (Panel A) and vice versa (Panel B). Panel C. Superimposed human and M-PMV dUTPase structures, the latter also containing the modeled C-terminus. Note the β-stranded interactions in both cases between the N- and C-terminal regions (red and green stars, respectively). Yellow and blue rectangles indicate regions shown in detail in Panels D and E. Panel D. Stability of contacts between α,β-imino dUTP and protein atoms from the C-terminus. Human (green carbons) and M-PMV (red carbons) dUTPase C-terminal segments (residues Glu129–Gly136 and Tyr222–Gly229, respectively) are superimposed. Important side chains are colored according to atoms, with carbons retaining the monomer color code. H-bonds present in the M-PMV dUTPase complex structure are red; those in the human structure are green. The coordinated Mg-ion is colored gray. Note close similarity in H-bonding in the different structures (Arg130/223 side chain terminal amino-groups H-bond to the phosphate chain of the ligand, and this same Arg residue and main chain nitrogens also participates in H-bonding to Gly133/226 main chain oxygen, numbers reflect human and M-PMV dUTPase sequences, respectively). Panel E. β-stranded interactions between N- and C-terminal (modeled) β-strands of M-PMV dUTPase. Stick model segments for Asn216–Gly229 and Ser111–Ala116 are shown, for monomers A and B, respectively. Main chain H-bonds within the segments Lys217–Gln220 and Arg115–Ala116, for monomers A and B, respectively, are indicated.
The mutational analysis (cf above) proved the essential functional role of Motif V. This role may either be to induce an alteration of protein fold in the core of the dUTPase domain or to contact the nucleotide ligand in the active site of the same monomer. To investigate if the C-terminal segment may induce a conformational alteration in the dUTPase core domain, the crystal structure of Gln220STOP mutant (Table 2) was determined. The superimposed structures of wild type and mutant enzymes (rms value of 1.5 Å) indicates that there is no significant conformational alteration in the truncated mutant. This result excludes the possibility that the C-terminus play a role in modulating the fold of the core dUTPase domain. The second possibility to account for the functional role of the C-terminus is to assume that it contributes to catalysis by providing interactions for the substrate in the active site. It was also ascertained by the mutations that the interactions require the Arg223 side chain in a way that is impossible to reconstruct by a lysine at this position.
Dynamic modeling of the C-terminal arm
Based on the mutational analysis and relying on literature data for human dUTPase (24), we constructed a homology model covering most of the C-terminal arm of M-PMV dUTPase, invisible in the crystal structure (residues 219GlnGlnProTyrArgGlyGlnGlySerPheGly229, conserved Motif V residues in bold face). As a first step, we investigated the possible contacts of the C-terminus with both its own subunit (monomer A) as well as with the neighbouring subunit (monomer B), assuming that the interactions of the well-conserved Arg223 and Phe228 with the α,β-imino dUTP substrate are persistent. Accordingly, we probed whether the guanidine group of Arg223 can form a hydrogen bond with the γ phosphate group of α,β-imino dUTP or Phe228 can establish van der Waals interaction with the pyrimidine ring of the substrate. Based on the calculated distances we concluded that the arm in M-PMV dUTPase continues in the direction towards the active site of its own monomer (cf also Supplementary data).
An initial model was built by Modeller when distances between Arg223 and Phe228 to the respective groups of α,β-imino dUTP were reinforced. Then we probed the stability of these contacts and the flexibility of the arm by molecular dynamics simulations. After equilibration the protein was very stable: with 1.5 and 1.9 Å RMS deviations in average from the backbone and all heavy atoms of the initial structure, respectively with very small fluctuations (0.06 Å). Figure 3 shows a representative model of the structure containing the C-terminal arm. The flexibility of the arm as compared to the rest of the protein structure that could be resolved by X-ray crystallography, was demonstrated by significantly higher deviations from the initial structure and by larger fluctuations: RMS deviations for the backbone is 2.8 ± 0.20 Å and 1.3 ± 0.07 Å, for the arm and for the rest of the protein, respectively. The hydrogen bond between Arg223 and the γ phosphate of α,β-imino dUTP was maintained in all the three subunits, with even a shorter contact distance than in the initial structure (the distance between CZ of Arg223 and P of the γ phosphate decreased from 4.7 to 4.3 Å). Phe228 turned out to be more floppy, the distance between the center of the benzene ring and the pyrimidine ring of α,β-imino dUTP varies between the three subunits from 4.6 to 5.6 Å (compared to the 4.9 Å in the original human dUTPase model). This contact also exhibits large variations around the average structure ranging from 0.5 to 0.7 Å, indicating larger flexibility associated with it. Thus dynamic analysis of the structure suggests that the contact between Arg223 and α,β-imino dUTP is primarily conserved. Figure 3D shows details of the interactions between the protein atoms of the C-terminus and the ligand α,β-imino dUTP.
In addition to contacts with the substrate, the modeled C-terminus showed several characteristic interactions with other protein atoms from the neighbouring monomer (cf Supplementary Figure S3E).
The N-terminal NC segment
This segment was shown to be highly flexible (cf prediction and proteolysis methods, Supplementary Figures S1 and S2), so its functional role was characterized by solution studies. Interaction of the NC domain with its cognate Zn2+ ligand was assessed by fluorescence spectroscopy (Supplementary Figure S4). Binding of the Zn2+ ligand was clearly detected to be the functional property of the NC-dUTPase fusion protein, not perturbed by the presence of the dUTPase domain. The interaction of the dUTPase active site ligand α,β-imino dUTP with the fusion protein was quantified by isothermal titration calorimetry (Supplementary data). The dissociation constant of the complex formed between NC-dUTPase and α,β-imino dUTP was estimated to be (1.2 ± 0.02) μM or (1.0 ± 0.07) μM in the absence or presence of the hexanucleotide ACTGCC, a cognate ligand for the NC domain (5). For all dUTPases investigated so far, very similar values of respective dissociation constants have been determined (27,28). Lack of functional interaction between the NC domain in the absence of the oligonucleotide and the active site was also reinforced by kinetic data (Table 4) where kcat and KM values for the dUTPase catalyzed hydrolysis of dUTP were determined both for the fusion protein NC-dUTPase and for the dUTPase protein by itself. No change was seen in the kinetic parameters when the NC domain got fused to the enzyme domain. It is also evident that Mg2+ enhances catalytic activity but is not an obligate co-factor [just as in the case of E.coli dUTPase cf (29)].
SAXS analysis of the bifunctional NC-dUTPase—The high flexibility and solvent-exposed character of the NC segment precluded crystallographic investigations on this protein domain. Solution-phase SAXS studies were therefore initiated. The experimental SAXS patterns from free NC-dUTPase and bound to the oligonucleotide are presented in Supplementary Figure S5 (curves 1 and 2), and the structural parameters computed from the SAXS data are given in Table 5. Comparison of the estimated molecular masses (MMs) of the solutes with the values predicted for the monomers indicates trimeric NC-dUTPase assembly in both cases. Significant decrease of the hydrated volume of bound NC-dUTPase as well as somewhat smaller values of Dmax and Rg compared to free NC-dUTPase point to compactisation of the protein upon binding of the oligonucleotide. Additionally, SAXS data were also collected from the NC domain alone and this domain complexed with ligand (Supplementary Figure S5, curves 3 and 4). The structural parameters summarized in Table 5 demonstrate that both NC samples are monomeric in solution and that NC domain also undergoes compactisation after ligand binding. This suggests that the structural changes observed in the full-length NC-dUTPase upon binding of the oligonucleotide are likely caused by the compactisation of its NC domain.
Table 5.
Parameters determined from SAXS data
| Sample | Rg (nm) | Dmax (nm) | MM (kDa) | MMmon (kDa) | Vp (nm)3 | χs | χ |
|---|---|---|---|---|---|---|---|
| NC-dUTPase | 5.10 ± 0.15 | 19.0 | 88 ± 10a | 25.0 | 180 ± 10 | 0.79/0.97 | 1.71/1.86 |
| NC-dUTPase with bound oligonucleotide | 4.73 ± 0.12 | 18.0 | 99 ± 10 | 31.5 | 130 ± 10 | 0.54/0.63 | 1.28/1.31 |
| NC | 2.45 ± 0.05 | 8.5 | 9.0 ± 1 | 8.9 | 14 ± 3 | 0.39 | 0.55 |
| NC with bound oligonucleotide | 2.15 ± 0.07 | 7.5 | 12.8 ± 2 | 15.4 | 17 ± 2 | 0.46 | 1.62 |
Rg, MM, Dmax and Vp are, respectively, the radius of gyration, molecular mass, maximum size and excluded volume, calculated from the scattering data. MMmon is the theoretical MM of monomeric constructs computed from the primary structure. χs and χ are discrepancies between the experimental data and computed scattering curves from the ab initio and rigid body models, respectively. For NC domain (free and bound) χ values are computed from the appropriate portions of the full-length rigid body models.
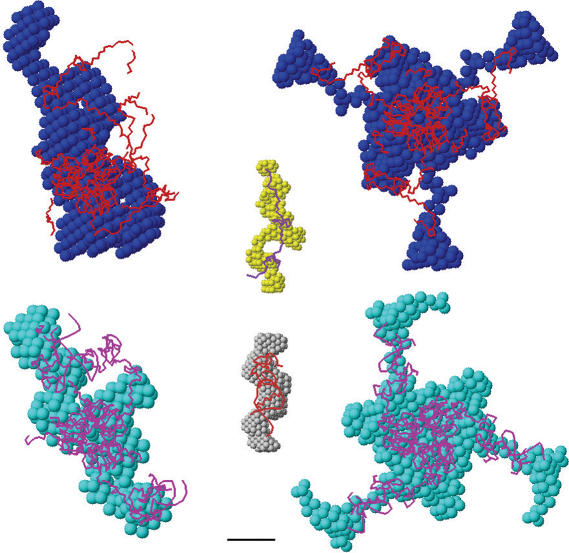
Shape determination—Typical ab initio low resolution models of free NC-dUTPase and with bound oligonucleotide generated by DAMMIN are displayed in Figure 4. As primary data analysis suggests trimeric assembly for both samples, reconstructions were done either with no symmetry restrictions or assuming P3 symmetry and the curves computed from these models neatly fit the experimental data (Supplementary Figure S5; the corresponding χS values are given in Table 5). All models, especially those obtained in P3 symmetry, demonstrate globular core compatible with the crystal structure of trimeric dUTPase surrounded by protuberances representing the NCs. The ab initio models of the complex are similar to those of the free NC-dUTPase but display a somewhat more compact shape in agreement with the above analysis of the overall parameters of the samples.
Figure 4.
Results of the modeling of NC-dUTPase against SAXS data. Structural models of free (top) and bound (bottom) NC-dUTPase. Ab initio shapes are presented as beads (blue or cyan for bound or free states, respectively), rigid body models as backbones (red and magenta for bound and for states, respectively). The models in left column were built with no symmetry restrictions (P1); those in the right column were constructed using 3-fold symmetry (P3). The central panel represents superposition of ab initio models of the free NC domain in solution and of this domain in complex with the oligonucleotide (yellow and grey beads, respectively) and NMR-derived models of these constructs (purple and red backbones for first models in the PDB files 1AAF and 1A1T, respectively). Bar length, 3 nm.
The ab initio shapes of the free NC and this domain complexed with oligonucleotide are superimposed with the corresponding NMR models in Figure 4, central part. The shape of the free NC displays an extended configuration compatible with an unfolded protein. In contrast, NC in complex with oligonucleotide has a much bulkier appearance, larger excluded volume and is well superimposed with the relatively compact atomic model of this complex obtained by NMR (1A1T, 1AAF) (30).
Rigid body modeling—To further characterize the structure of NC-dUTPase in solution, rigid body modeling against the experimental data were performed using program BUNCH. Crystallographic model of dUTPase domain and the NMR model of Zn-knuckle domain of both free NC and NC with bound oligonucleotide were employed as rigid bodies for the modeling. As the results of ab initio shape determination are compatible with the crystallographic trimer of dUTPase, the dUTPase domains were fixed to keep the trimer structure. The search was done in terms of the position and orientation of the NC domain (or NC with oligonucleotide) and conformation of the linkers represented by chains of dummy residues that fits the appropriate data set. Typical BUNCH reconstructions of free and bound NC-dUTPase in P1 and P3 are presented in Figure 4. They all yield the overall shapes similar to the corresponding ab initio models with NC tails protruding from the dUTPase core (Figure 4) and provide good fits (Supplementary Figure S5 and Table 5) to the experimental data. These models also agree with the compactisation of the NC domains upon oligonucleotide binding suggested by the ab initio shape determination and primary data analysis. Interestingly, the computed scattering of the NC portion extracted from of the NC-dUTPase model in the absence of the oligonucleotide yields a very good fit to the experimental scattering from free NC in solution (Supplementary Figure S5, curve 3). For the oligonucleotide-bound NC, the same procedure yields a somewhat worse fit (Supplementary Figure S5A, curve 4), indicating that the bound NC in the full-length NC-dUTPase is more compact than the isolated NC in solution complexed with the ligand.
DISCUSSION
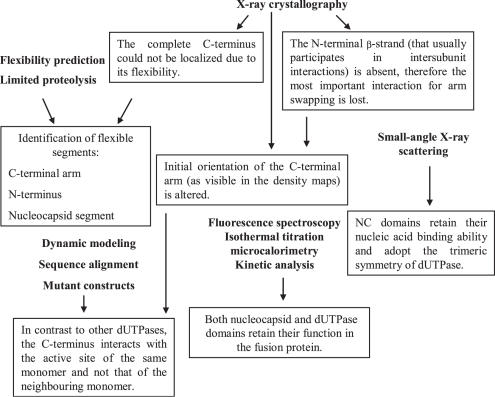
The integrated application of varied techniques during the present study and several of the key results are summarized in Figure 5. Importantly, crystal structure determination of M-PMV dUTPase has revealed a significant alteration in the orientation of C-terminus as compared to the human dUTPase. Since the complete C-terminal arm could not be localized in the density maps due to its high flexibility, dynamic modeling was applied to determine its possible position in the structure. Modeling results suggested that the C-terminal segment interacts with the active site of the same monomer and not the neighbouring monomer. This result was reinforced by mutational studies within Motif V of the C-terminus indicating that residues Arg223 and Phe228 have to contact the ligand in the active site while distance calculations prove that even the fully extended conformation of C-terminus is not capable to interact with the ligand in the active site of the neighbouring monomer. Structure of the fusion protein has indicated a unique conformation for the N-terminal segment of the dUTPase domain, as well. The N-terminal β-strand (that usually participates in intersubunit interactions) is absent, therefore the most important interaction for arm-swapping is lost. Despite these structural alterations, dUTPase retains its adequate function as reinforced with kinetic analyis and isothermal titration microcalorimetry. Fluorescence spectroscopy and small-angle X-ray scattering measurements confirm that NC domain within covalently joined fusion protein can bind nucleic acid and also Zn2+ ion. According to SAXS results, oligonucleotide binding to the fusion protein induces a significant ordering in the protein conformation and formation of trimeric symmetry of NC domains (Figure 5).
Figure 5.
Flowchart of multidisciplinary experiments and summary of several results. Techniques are listed in bold face, results are defined within text boxes.
C-terminus orientation
Trimeric dUTPases usually form their active sites in a unique manner where arm-swapping between monomers have an essential role in active site architecture; it provides active site closure by interactions among residues in conserved dUTPase Motif V and the uracil ring as well as the phosphate chain of the substrate. However, in recent structural investigations, some members of the dUTPase trimeric superfamily were shown to lack this arm-swapping interaction (Figure 6). Namely, the bifunctional dCTP-deaminase-dUTPase from Archea, the Plasmodium falciparum dUTPase, as well as the more distantly related E.coli dCTP-deaminase proteins all display homotrimeric organization with similar active site architecture as most dUTPases, but arm-swapping is not present in any of these structures (Figure 6) (31–33). The present investigation on M-PMV dUTPase clearly demonstrates that despite lack of arm-swapping, the C-terminal segment retains a major role in active site architecture: the truncated mutant is characterized by a 105-fold decrease in catalytic efficiency (kcat/KM). These data provide explanation for the conserved presence of the Motif V residues in this segment. The dynamic model constructed for this segment argues for the possibility of conserved interactions between (i) the phenylalanine (Phe228) and the uracil ring and (ii) the phosphate chain oxygens and the conserved Arg223 residue. In the specific case of M-PMV dUTPase, the altered C-terminal arm orientation is probably due to the decreased number of residues between conserved Motifs IV and V. The shortening of this spacer makes it impossible for the C-terminus to bridge out to the active site ligand in the neighbouring subunit. Therefore, the arm folds back upon its own monomer and efficiently reaches towards the substrate accommodated within the same monomer. The altered orientation of the arm is also dictated by a profound folding difference at the N-terminus where M-PMV dUTPase lacks the first β-strand usually present in other representatives of the dUTPase fold. The folding difference is very probably directly related to the fact that the N-terminal segment of M-PMV dUTPase reaches out from the folded monomer core to connect to the NC domain in the full-length fusion protein.
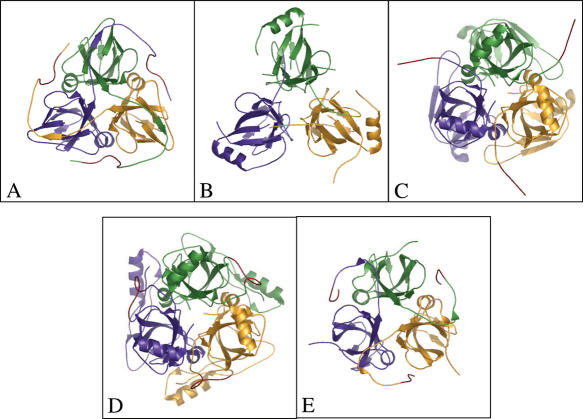
Figure 6.
C-terminal arm comparisons. (A) Classical arm-swapping in feline immunodeficiency virus dUTPase [PDB 1F7Q, (26)]. (B) Intersubunit interactions ensure the trimer organization in absence of C-terminal arm in case of Plasmodium falciparum dUTPase (PDB 1VYQ). (C) Methanococcus jannaschii dCTP-deaminase-dUTPase (PDB 1PKH). (D) E.coli dCTP-deaminase (PDB 1×S1). (E) Partial swapping and same active site contact in M-PMV dUTPase (PDB 2D4N, present study). Main chain ribbon models are colored according to subunits (yellow, green and purple), red segments indicate the position of C-terminal dUTPase conserved Motif V, or related sequences.
The dynamic model comprises nine C-terminal residues, among which the last six belong to the conserved Motif V. The conserved residues of Motif V were found to contact the substrate-analogous ligand in the active site pocket of the same subunit in a manner practically identical to other dUTPases where the C-terminus closes upon the active site of the neighbouring subunit (cf Figure 3). In this latter case, the orientation of the C-terminus is facilitated by interactions between main chain atoms belonging to residues preceding the conserved Motif V segment and main chain atoms of the neighbouring subunit N-terminal β-strand (Figure 2). Interestingly, the dynamic model suggests that a highly similar main chain H-bonding pattern is created by the M-PMV dUTPase residues 217LysValGlnGln220 (just preceding the C-terminal conserved Motif V) and the residues 115ArgAla116 (N-terminal segment of the neighbouring monomer) (Figure 3E). It seems therefore that the main chain interaction manner of guiding the conserved functional residues into the respective active site pocket is conserved also, even if the specific path that the C-terminus needs to trace along the neighbouring subunit surface is largely different. It is, of course, highly advantageous that the protein surface of the neighbouring subunit N-terminal segment presents the required H-bonding partners for the C-terminus. The concerted alterations in the N- and C-termini suggest that these segments were shaped to find respective complementary interacting partners with the final goal of correctly creating the active site.
Full-shape trimeric symmetry induced by oligonucleotide binding and capability in the bifunctional protein for oligonucleotide binding
It is not possible to deduce from the SAXS data alone whether the full-length NC-dUTPase maintains the 3-fold symmetry of the dUTPase core. The modeling was therefore done with and without symmetry restrictions, and in the latter case, give n more degrees of freedom available; the models did not display the 3-fold symmetry. On the other hand, symmetric models were also compatible with the data, and, the conclusion about compactisation of NC and NC-dUTPase follows from the models independently on the symmetry assumption. The presence of the fused dUTPase domain involves significant ordering upon binding of oligonucleotide to the NC domain (cf Figure 4).
Oligonucleotide binding to NC protein was already shown to induce significant ordering in the protein conformation (13). The present SAXS results reinforce these effects and, in addition, show that this ordering is significantly enhanced if NC becomes fused to trimeric dUTPase. This result is the first experimental indication arguing for the contribution of the trimeric dUTPase core to nucleic acid binding in the NC domains. Such an architectural effect, if realized within the cellular milieu, may positively influence the chaperoning role NC has to perform to encapsulate retroviral RNA within the viral capsid. This enhanced chaperoning machinery may be especially required for betaretroviruses that assemble into capsids before budding.
Comparison of betaretroviral dUTPases with numerous other dUTPases investigated to date reveal that the betaretroviral enzymes have evolved to support the proper function of the NC protein fused to their N-terminus. This evolution affected the whole sequence except for the conserved motifs, as shown by the low sequence similarity (<30%) between betaretroviral and other dUTPases. The modifications, however, do not provoke changes in the protein's overall fold, but do alter the electrostatic character of the surface. The usual rather acidic isoelectric point observed in most dUTPases (Homo sapiens 6.1, E.coli 4.2, equine infectious anemia virus 6.1, feline immunodeficiency virus 5.7, puma lentivirus 5.4) would obviously perturb nucleic acid binding in NC-dUTPase, therefore M-PMV and other betaretroviruses have evolved a much higher, basic pI for their dUTPase proteins (M-PMV 8.6, MMTV 8.2, JSRV 9.0, SRV-1 8.8, SRV-2 8.7, cf Figure 1). This electrostatic modification allows the fusion protein to bind RNA/DNA and thereby supports NC function but also localizes dUTPase at the polynucleotides. Since dUTPase activity is supposedly predominantly required during reverse transcription, the enzyme proximity at this event is clearly advantageous for the virus. In addition to charged surface residues, a significant alteration concerned the N- and C-terminal segments during evolution of the dUTPase sequence. Co-evolution of the N- and C-termini resulted in (i) an N-terminus pointing out of the compactly folded dUTPase core, stabilized by non-covalent β-strand interaction with the C-terminus, a conformation present also in the absence of the NC domain and (ii) a C-terminus folding back on the active site of the same monomer, with the help of the N-terminal β-strand. This co-folding allows the shortened C-terminus to cope with its handicap and reach to an active site. On this way, betaretroviruses have found a unique economic solution by encoding a shortened though effective dUTPase fused to the NC protein in their very limited genome. The fusion protein may efficiently decrease dUTP pools on the place of reverse transcription without wasting time and energy by hydrolyzing all cellular dUTP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank beamlines and operators at EMBL/DESY, Hamburg, and grants from Hungarian Scientific Research Fund (M27852 to B.G.V., T049073, K61684 to I.S., F046164 to M.F.), Howard Hughes Medical Institutes (#55005628 and #55000342 to B.G.V.), Alexander von Humboldt Foundation (to B.G.V.), Öveges grant from National Office for Research and Technology (to B.G.V.), Varga József Foundation (fellowship to V.N.-P.), Hungarian Economic Competitiveness Operative Programme GVOP-3.2.1.-2004-04-0195/3.0 and GVOP-3.1.1.-2004-05-0412/3.0, Bolyai János Foundation (fellowship to M.F.), Programme 1M6138896301 of the Czech Ministry of Education (to I.P.) and EU FP6 LSHP-CT-2005-012127 (to B.G.V), EU FP6 MRTN-CT-2005-019566 (to M.F), EU FP6 Design Study SAXIER, RIDS 011934 (to D.S. and M.P.). Funding to pay the Open Access publication charges for this article was provided by Howard Hughes Medical Institutes and EU FP6.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pearl L.H., Savva R. The problem with pyrimidines. Nature Struct. Biol. 1996;3:485–487. doi: 10.1038/nsb0696-485. [DOI] [PubMed] [Google Scholar]
- 2.el-Hajj H.H., Zhang H., Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gadsden M.H., McIntosh E.M., Game J.C., Wilson P.J., Haynes R.H. dUtp pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO. J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turelli P., Guiguen F., Mornex J.F., Vigne R., Querat G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-a substitutions. J. Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabas O., Rumlova M., Erdei A., Pongracz V., Pichova I., Vertessy B.G. dUTPase and nucleocapsid polypeptides of the Mason-Pfizer monkey virus form a fusion protein in the virion with homotrimeric organization and low catalytic efficiency. J. Biol. Chem. 2003;278:38803–38812. doi: 10.1074/jbc.M306967200. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Larsson G., Nyman P.O., Kvassman J.O. Kinetic characterization of dUTPase from Escherichia coli. J. Biol. Chem. 1996;271:24010–24016. doi: 10.1074/jbc.271.39.24010. [DOI] [PubMed] [Google Scholar]
- 8.Vertessy B.G. Flexible glycine rich motif of Escherichia coli deoxyuridine triphosphate nucleotidohydrolase is important for functional but not for structural integrity of the enzyme. Proteins. 1997;28:568–579. [PubMed] [Google Scholar]
- 9.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 14.Summers M.F., Henderson L.E., Chance M.R., Bess J.W., Jr, South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C., III, Hare D.R., et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiser A., Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;18:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 16.Barabas O., Nemeth V., Vertessy B.G. Crystallization and preliminary X-ray studies of dUTPase from Mason-Pfizer monkey retrovirus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:399–401. doi: 10.1107/S1744309106008931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CCP4. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Lamzin V.S., Wilson K.S. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–149. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 20.Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53:566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 21.Dosztányi Z., Csizmók V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 22.Fiser A., Vertessy B.G. Altered subunit communication in subfamilies of trimeric dUTPases. Biochem. Biophys. Res. Commun. 2000;279:534–542. doi: 10.1006/bbrc.2000.3994. [DOI] [PubMed] [Google Scholar]
- 23.Larsson G., Svensson L.A., Nyman P.O. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP) Nature Struct. Biol. 1996;3:532–538. doi: 10.1038/nsb0696-532. [DOI] [PubMed] [Google Scholar]
- 24.Mol C.D., Harris J.M., McIntosh E.M., Tainer J.A. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 25.Barabas O., Pongracz V., Kovari J., Wilmanns M., Vertessy B.G. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004;279:42907–42915. doi: 10.1074/jbc.M406135200. [DOI] [PubMed] [Google Scholar]
- 26.Prasad G.S., Stura E.A., Elder J.H., Stout C.D. Structures of feline immunodeficiency virus dUTP pyrophosphatase and its nucleotide complexes in three crystal forms. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1100–1109. doi: 10.1107/s0907444900009197. [DOI] [PubMed] [Google Scholar]
- 27.Vertessy B.G., Larsson G., Persson T., Bergman A.C., Persson R., Nyman P.O. The complete triphosphate moiety of non-hydrolyzable substrate analogues is required for a conformational shift of the flexible C-terminus in E.coli dUTP pyrophosphatase. FEBS Lett. 1998;421:83–88. doi: 10.1016/s0014-5793(97)01545-7. [DOI] [PubMed] [Google Scholar]
- 28.Persson T., Larsson G., Nyman P.O. Synthesis of 2′-deoxyuridine 5′-(alpha,beta-imido) triphosphate: a substrate analogue and potent inhibitor of dUTPase. Bioorg. Med. Chem. 1996;4:553–556. doi: 10.1016/0968-0896(96)00044-2. [DOI] [PubMed] [Google Scholar]
- 29.Mustafi D., Bekesi A., Vertessy B.G., Makinen M.W. Catalytic and structural role of the metal ion in dUTP pyrophosphatase. Proc. Natl Acad. Sci. USA. 2003;100:5670–5675. doi: 10.1073/pnas.1031504100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y., Kaluarachchi K., Giedroc D.P. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein Sci. 1998;7:2265–2280. doi: 10.1002/pro.5560071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huffman J.L., Li H., White R.H., Tainer J.A. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J. Mol. Biol. 2003;331:885–896. doi: 10.1016/s0022-2836(03)00789-7. [DOI] [PubMed] [Google Scholar]
- 32.Whittingham J.L., Leal I., Nguyen C., Kasinatha G., Bell E., Jones A.F., Berry C., Benito A., Turkenburg J.P., Dodson E.J., et al. Dutpase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure. 2005;13:329–338. doi: 10.1016/j.str.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Johansson E., Fano M., Johansson E., Fano M., Bynck J.H., Neuhard J., Larsen S., Sigurskjold B.W., Christensen U., Willemoes M. Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J. Biol. Chem. 2005;208:3051–3059. doi: 10.1074/jbc.M409534200. [DOI] [PubMed] [Google Scholar]
- 34.Diederichs K., Karplus P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature Struct. Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]