Abstract
Despite the fact that cold shock domain proteins (CSDPs) and glycine-rich RNA-binding proteins (GRPs) have been implicated to play a role during the cold adaptation process, their importance and function in eukaryotes, including plants, are largely unknown. To understand the functional role of plant CSDPs and GRPs in the cold response, two CSDPs (CSDP1 and CSDP2) and three GRPs (GRP2, GRP4 and GRP7) from Arabidopsis thaliana were investigated. Heterologous expression of CSDP1 or GRP7 complemented the cold sensitivity of BX04 mutant Escherichia coli that lack four cold shock proteins (CSPs) and is highly sensitive to cold stress, and resulted in better survival rate than control cells during incubation at low temperature. In contrast, CSDP2 and GRP4 had very little ability. Selective evolution of ligand by exponential enrichment (SELEX) revealed that GRP7 does not recognize specific RNAs but binds preferentially to G-rich RNA sequences. CSDP1 and GRP7 had DNA melting activity, and enhanced RNase activity. In contrast, CSDP2 and GRP4 had no DNA melting activity and did not enhance RNAase activity. Together, these results indicate that CSDPs and GRPs help E.coli grow and survive better during cold shock, and strongly imply that CSDP1 and GRP7 exhibit RNA chaperone activity during the cold adaptation process.
INTRODUCTION
The freezing tolerance of plants increases after a period of exposure to low temperatures that remain above freezing; this process is known as cold acclimation. A variety of genes induced during cold acclimation have been identified from numerous plant species (1–7). In prokaryotes, a similar acclimation process, termed the ‘cold shock response’, has been extensively characterized in Escherichia coli (8,9), and it has been shown that the cold shock protein (CSP) family of E.coli was induced at high levels during the cold acclimation phase (10). CSPs contain a domain with high similarity to the cold shock domain (CSD), which is present in eukaryotic Y-box proteins. These proteins are involved in the regulation of gene expression at the transcription or translation level (11). The CSD comprises ∼65–75 amino acid residues and is capable of binding RNA, single-stranded DNA and double-stranded DNA (12). It has been suggested that E.coli CspA functions as an RNA chaperone that facilitates translation at low temperature by blocking the formation of secondary structures in mRNA (13). CSPs in prokaryotes are small in size (∼7–10 kDa), and this small size CSP has been shown to be sufficient for nucleic acid-binding and the cold shock response by functioning as an RNA chaperone (14).
Several cold shock domain proteins (CSDPs) have been documented in plants, including tobacco (15), wheat (16) and Arabidopsis (17). Plant CSDPs differ from those found in prokaryotes because they contain additional glycine-rich regions interspersed with CCHC-type zinc fingers at the C-terminal half (Figure 1A). Although the nucleic acid-binding property and function of CSD have been well described, functional roles of the glycine-rich region with CCHC-type zinc finger motifs have not been established. It has been determined that the Arabidopsis genome contains four CSDPs, the functions of which are largely unknown (17). Although plant CSDPs are suggested to play similar roles as CSPs in prokaryotes during the cold acclimation process, the information on their roles is severely limited. In recent, a CSDP isolated from winter wheat was shown to function as an RNA chaperone (18).
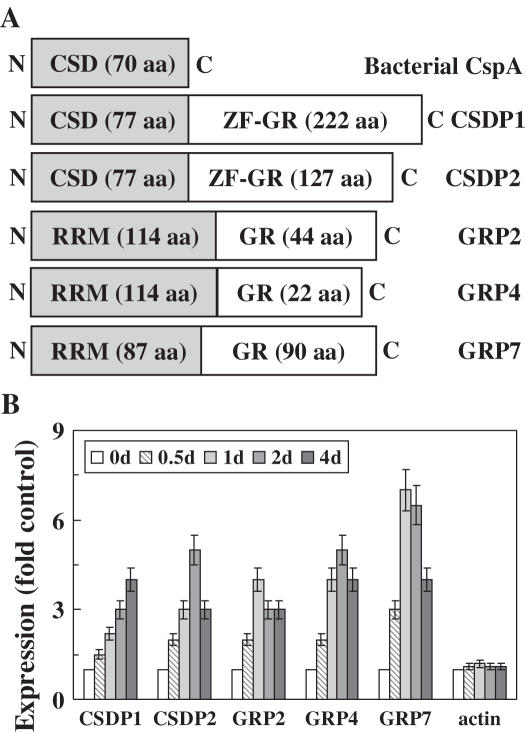
Figure 1.
The structure and cold-regulated expression of Arabidopsis CSDPs and GRPs investigated in this study. (A) Schematic representation of the structures of CSDPs and GRPs; CSD, cold shock domain; GR, glycine-rich; ZF-GR, zinc finger glycine-rich. (B) Effect of cold stress on the expression of CSDPs and GRPs was investigated in A.thaliana subjected to cold stress at 4°C for the indicated time.
During the last two decades, glycine-rich RNA-binding proteins (GRPs) that contain one or more RNA-recognition motifs (RRMs) at the N-terminus and a glycine-rich region at the C-terminus have been identified in a variety of plant species (19,20). Although the function of GRPs was not characterized in detail, it has been suggested that some may play a role in stress responses, as their mRNA levels increased following exposure to cold, wounding, water stress, plant hormones or viral infection [(19) and references therein]. More specifically, the role of plant GRPs during cold acclimation has been implicated by the fact that they were highly induced by cold temperature. Since it was found that cyanobacteria lack CSPs, instead containing a cold-induced RRM protein (Figure 1A), it was hypothesized that the function of RRM proteins may substitute for the function of CSPs in cyanobacteria (14,21). It is likely that plant RRM-type GRPs play a similar role to bacterial CSPs and/or plant CSDPs during cold stress. Therefore, it is of interest to investigate CSDPs and GRPs together as a regulator of gene expression upon a downshift in temperature. However, our understanding of the function and importance of GRPs and CSDPs in the cold stress response are quite limited due to the lack of clear in vivo functional data.
We recently showed that an Arabidopsis GRP, designated as at RZ-1a, which contains N-terminal RRM and C-terminal glycine-rich domains interspersed by CCHC-type zinc fingers, plays a role in the enhancement of freezing tolerance of Arabidopsis plants (22). As a step toward understanding the function of plant GRPs and CSDPs during the cold adaptation process, we aimed to test whether plant GRPs and CSDPs exhibit growth-stimulating activity of E.coli during cold shock. GRP2 (accession no. At4g13850), GRP4 (accession no. At3g23830) and GRP7 (accession no. At2g21660) investigated in this study belong to the group of GRP family members found in Arabidopsis thaliana (12). The three GRPs are selected for investigation, because they are markedly up-regulated by cold stress (23–25). GRP2 and GRP7 have been suggested to play roles in cold stress responses (23,24), and GRP4, which contains a shorter glycine-rich region compared to GRP2 and GRP7, had no impact on cold stress resistance of Arabidopsis plants (25). CSDP1 (accession no. At4g36020) and CSDP2 (accession no. At4g38680) investigated in this study are members of the Arabidopsis CSDP family (17). CSDP1 and CSDP2 differ from each other in that CSDP1 comprising 299 amino acids contains seven CCHC-type zinc fingers at the C-terminus, whereas CSDP2 comprising 204 amino acids has two CCHC-type zinc fingers at the C-terminus. The primary structures of GRPs and CSDPs are schematically represented in Figure 1A. Here, GRPs and CSDPs from Arabidopsis were investigated for their growth-stimulating abilities under cold stress of E.coli BX04 mutant, which lacks four CSPs (CspA, CspB, CspE and CspG) and is highly sensitive to cold stress. This article provides new evidence to indicate that GRPs and CSDPs from A.thaliana help E.coli grow and survive better during cold shock, implying that GRP7 and CSDP1 exhibit RNA chaperone activity during the cold adaptation process.
MATERIALS AND METHODS
Plant growth and stress treatment
A.thaliana ecotype Col-0 used in this study was grown at 23 ± 2°C under long day conditions (16 h-light/8 h-dark cycle) as described previously (22). To test the effect of cold stresses on gene expression, plants that had been grown in pots were placed at 4°C for 1–4 days under a 16 h photoperiod. The samples were collected at indicated time intervals, frozen immediately into liquid nitrogen, and used for RNA extraction and subsequent analysis. The entire experiment was repeated at least three times.
RNA extraction and quantitative RT–PCR
Total RNA was extracted from the frozen samples using the Plant RNeasy extraction kit (Qiagen, USA). The concentration of RNA was quantified by spectrophotometric measurements, and 5 μg of total RNA was separated in 1% formaldehyde agarose gel to determine the concentration and to monitor the integrity of the samples. For detection of the RNA transcripts in cold stress-treated samples, real-time quantification of RNA targets was performed in the Rotor-Gene 2000 real-time thermal cycling system (Corbett Research, Australia) using QuantiTect SYBR Green RT–PCR kit (Qiagen) as described previously (26). The gene-specific primers used in real-time RT–PCR are listed in Supplementary Table 1. A control RT–PCR was performed with the same amount of total RNA using the primer pair specific to Actin gene.
Vector construction
The pINIII vector was kindly provided by Dr M. Inouye. The coding region of CSDP and GRP cDNAs was prepared by PCR, and was subcloned into the NdeI/BamHI site of pINIII vector (27). The constructs were designated pINIII-CSDP and pINIII-GRP. To construct the truncated version of the expression vector for GRP7 and CSDP1, the cDNA containing the N-terminal region (from 1 to 87 amino acid of GRP7 and from 1 to 99 amino acid of CSDP1) and the cDNA containing the C-terminal region (from 88 to 177 amino acid of GRP7 and from 100 to 299 amino acid of CSDP1) were subcloned into pINIII, and the constructs were designated as pINIII-GRP7N, pINIII-CSDP1N, pINIII-GRP7C and pINIII-CSDP1C, respectively. All DNA manipulations were performed according to standard procedures (28), and GRP and CSDP coding regions and junction sequences were confirmed by DNA sequencing.
Cold shock test in E.coli
pINIII expression vectors containing either full-length CSDPs, full-length GRPs, the N-terminal or C-terminal region of CSDP1, the N-terminal or C-terminal region of GRP7, or the pINIII vector without a DNA insert as a control were used for the cold shock test. E.coli BX04 mutant cells (29) that lacked four CSPs and are highly sensitive to cold stress were obtained from Dr M. Inouye. The BX04 mutant cells transformed with each vector were grown in Luria–Bertani (LB) medium containing ampicillin and kanamycin, and the overnight cultures of the mutant cells containing each construct were inoculated into new LB medium. When the optical density at 600 nm reached ∼0.8, the cells were streaked on LB-agar plates containing 0.2 mM isopropyl-d-thiogalactopyranoside (IPTG) and incubated under low temperatures. The growth of the cells was inspected every day. In separate experiments, the BX04 mutant cells transformed with each vector were grown in LB medium, the cells were subjected to cold shock at 17°C, and the growth rate was monitored by measuring the optical density at 600 nm.
Cell viability test
E.coli BX04 mutant cells containing pINIII-CSDP, pINIII-GRP or pINIII vector were grown to exponential phase, and were then transferred to 4°C. Cultures were removed from low temperature at the time points indicated in Figure 4 and plated on LB-agar plates, which were then incubated at 37°C. Counts of viable colonies were converted into percentages, taking the colony counts at zero time point as 100%. The entire experiment was repeated three times.
Nucleic acid-binding assay and selective evolution of ligand by exponential enrichment
The DNA sequence encoding CSDP1 and GRP7 proteins was cloned into the pET-22b(+) vector (Invitrogen), and the labeled proteins were synthesized by coupled transcription-translation (Promega) in the presence of [35S]methionine. All experimental conditions were maintained as described previously (22). The oligonucleotide template and primers for selective evolution of ligand by exponential enrichment (SELEX) were as follows: T7PRO5, 5′-TAATACGACTCACTATAGGGATCCAAGATGCCGAC-3′; T7TEMP, 5′-GTTCCAATAGAGATGTAACG(N)65GTCGGCATCTTGGATCCCTATAGTGAGTCGTATTA-3′; T7PRO3, 5′-GTTCCAATAGAGATGTAACG-3′. The in vitro-transcribed RNAs were incubated with the GST–GRP7 fusion proteins bound to glutathione sepharose 4B resin. All experimental conditions were maintained as described previously (30). This process was repeated for five to eight cycles, and the final products were gel-purified, ligated into pGEM-T Easy vector (Promega) and sequenced.
Nucleic acid melting assay
The molecular beacon used in this study was a 78 nt-long, 9 bp-containing, hairpin-shaped molecule labeled with a fluorophore (tetramethyl rhodamine) and quencher (dabcyl) as described by Phadtare et al. (31). For the expression and purification of recombinant CSDP–GST and GRP–GST-fusion proteins in E.coli, the coding regions of each CSDP and GRP were cloned into pGEX-5X-2 vector (Amersham Pharmacia Biosciences), and the constructs were transformed into BL21 DE3 competent cells (Promega). Transformants were cultured and induced by the addition of IPTG (0.4 mM), and the recombinant proteins were purified with glutathione sepharose 4B resin. Fluorescence measurements were performed on a Spectra Max GeminiXS spectrofluorometer (Molecular Devices) with excitation and emission wavelengths of 555 and 575 nm, respectively. All experimental conditions were maintained as described previously (31).
Ribonuclease assay
RNA substrate was prepared by transcribing the pET-22b(+) plasmid cut with BamH1 using T7 RNA polymerase, which results in 175 nt-long RNA. 32P-labeled RNA substrates were incubated with the purified GST-fusion proteins in the binding buffer for 15 min on ice. The reaction mixture was loaded onto an 8% acrylamide gel. All experimental conditions were maintained essentially as described previously (13).
RESULTS
Cold-regulated expression patterns of CSDPs and GRPs in A.thaliana
The transcript levels of CSDPs and GRPs in A.thaliana subjected to cold stress treatment were measured by quantitative real-time RT–PCR. The histograms in Figure 1B represent the mean values of different experiments conducted with different RNA preparations. The expression of GRP2 whose transcript level is not affected noticeably by drought, abscisic acid, or wound treatment (24) was markedly induced by cold stress. GRP4 and GRP7, the expression of which decreased by drought or salt stress (25), were up-regulated by cold stress. Cold-regulated expression of GRP2, GRP4 and GRP7 was also reported by microarray analysis (32). Additionally, the expression of CSDP1 and CSDP2 was markedly up-regulated by cold treatment such that their expression levels increased up to 4- to 5-fold during the 4 days of cold treatment (Figure 1B). It was observed that the transcript levels of CSDP1 and CSDP2 were down-regulated by dehydration stress, and the expression of CSDP1 decreased by salt stress (data not shown). Cold-regulated expression of CSDP1 and CSDP2 was also reported by microarray analysis (32). It is apparent that cold treatment greatly increased the expression of GRPs and CSDPs, as reported previously (17,23–25,32). The expression of RD29A cold stress response marker was increased to 18-fold by cold treatment (data not shown), and no significant changes in the transcript level of Actin was observed (Figure 1B), indicating that our experimental conditions and real-time RT–PCR analysis were valid by means of which to follow the changes in transcript levels in stress-treated samples.
Arabidopsis CSDPs and GRPs help E.coli grow better during cold shock
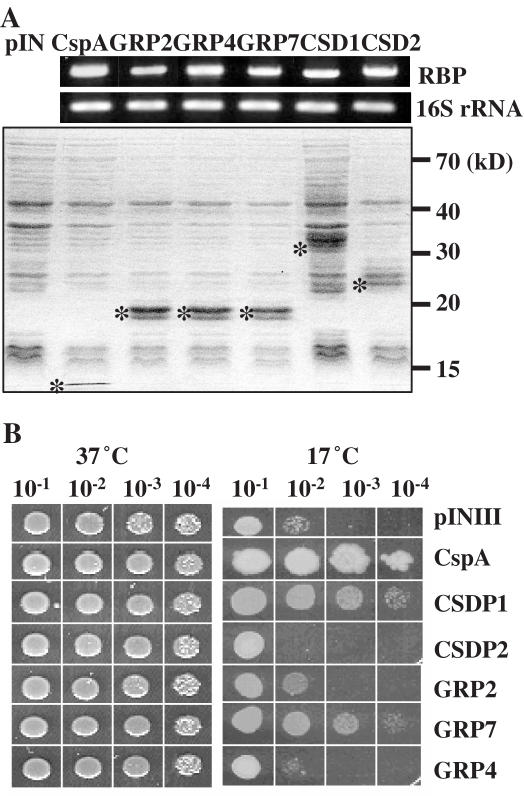
CSPs have been implicated to function as RNA chaperones and confer cold tolerance in prokaryotes (8,9,13). To understand the functional role of CSDPs and GRPs in cold stress, we examined each CSDP and GRP gene for its ability to complement defects in the growth of BX04 cells at low temperature. The coding sequences of CSDP and GRP genes, as well as the E.coli CspA gene as a positive control, were inserted into a pINIII vector (27), and the colony-forming abilities of BX04 cells transformed with these clones were examined on LB plates at 17°C in the absence and the presence of IPTG. We first confirmed by RT–PCR and SDS–PAGE analyses that CSDPs and GRPs are expressed quite equally in BX04 cells (Figure 2A). When the BX04 cells harboring each construct were incubated at 37°C, all cells grew well without any noticeable difference (Figure 2B). In contrast, when the cells were subjected to cold shock at 17°C, the growth of BX04 cells was quite different in that the growth rate of BX04 expressing either CSDP1 or GRP7 was much higher than that of the control cells expressing the vector only (Figure 2B). GRP2 was able to partially complement the cold sensitivity of BX04 when incubated at 17°C. In comparison, the BX04 cells expressing either CSDP2 or GRP4 did not grow well when incubated at 17°C. As shown in the previous report (29), CspA used as a positive control successfully complemented the cold-sensitive phenotype of BX04. These growth-stimulating abilities of CSDPs and GRPs were observed only when the BX04 cells were cultured in the presence of IPTG. These results demonstrate that plant CSDPs and GRPs are able to suppress the cold sensitivity of E.coli cells at low temperature, although the ability to complement is not the same for all CSDPs and GRPs. It appears that the degree of complementation exists in the order of CSDP1 > GRP7 > GRP2.
Figure 2.
Effect of CSDPs and GRPs on the growth of E.coli during cold shock. (A) The expression of GRP2, GRP4, GRP7, CSDP1, CSDP2 or CspA in BX04 cells was verified by RT–PCR and SDS–PAGE analyses. The bands corresponding to each protein were indicated by asterisks. (B) The growth of the BX04 cells expressing each gene was measured by spotting the diluted cultures (10−1 to 10−4 serial dilution from the cultures at OD600 = 1.0) on LB-agar plates, and the cells were incubated at 37 and 17°C. The pictures were taken after 1 day for cells incubated at 37°C and 5 days for cells incubated at 17°C.
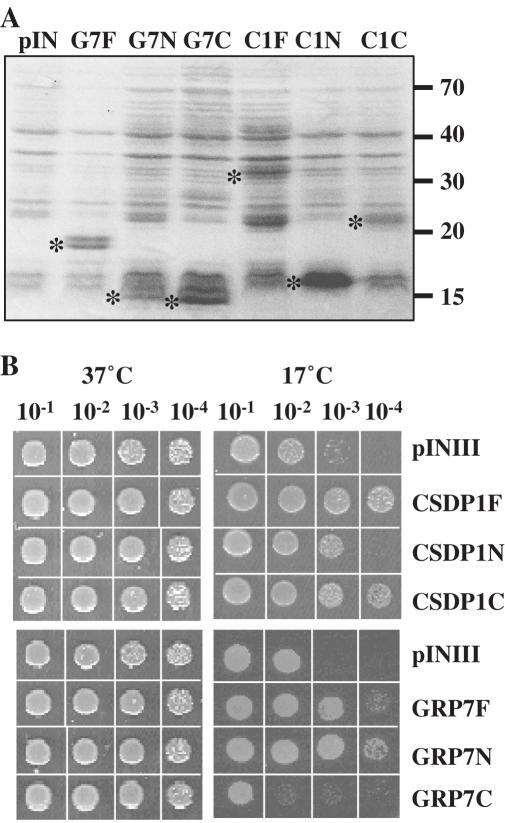
Since it is apparent that CSDP1 and GRP7 complement the cold sensitivity of BX04 cells, we next wanted to test whether the N-terminal region containing RRM or the C-terminal region containing the glycine-rich domain of GRP7 and the N-terminal region containing CSD or the C-terminal region containing the zinc finger glycine-rich domain of CDSP1 (Figure 1A) are involved in this process. The N-terminal or C-terminal regions of each gene were inserted into pINIII expression vector, and the constructs containing either the N-terminal region (pINIII-GRP7N and pINIII-CSDP1N) or C-terminal region (pINIII-GRP7C and pINIII-CSDP1C) were tested for their abilities to suppress the cold sensitivity of BX04 cells. The expression of N-terminal and C-terminal regions of CSDP1 and GRP7 was also confirmed by SDS–PAGE analysis (Figure 3A). When incubated at 17°C, BX04 cells harboring pINIII-CSDP1C grew better than cells containing pINIII-CSDP1N or pINIII vector (Figure 3B), although pINIII-CSDP1C expression was lower than pINIII-CSDP1N expression in BX04 cells (Figure 3A). In comparison, BX04 cells harboring pINIII-GRP7N grew better than cells containing pINIII-GRP7C or pINIII vector (Figure 3B). These results revealed that the N-terminal CSD alone in CSDP1 is not enough for the activity, but the C-terminal region containing seven zinc fingers has the growth-stimulating activity during the cold adaptation process. It is also apparent that the N-terminal RRM in GRP7 is sufficient for the growth-stimulating activity of GRP7 during the cold adaptation process.
Figure 3.
Effect of N-terminal and C-terminal regions of CSDP1 and GRP7 on the growth of E.coli during cold shock. The expression of the full-length (C1F and G7F), N-terminal (C1N and G7N), or C-terminal (C1C and G7C) CSDP1 and GRP7 in BX04 cells was verified by SDS–PAGE analyses. The bands corresponding to each protein were indicated by asterisks. The BX04 cells harboring each construct were analyzed as described in Figure 2.
Arabidopsis CSDPs and GRPs increase the viability of E.coli during cold shock
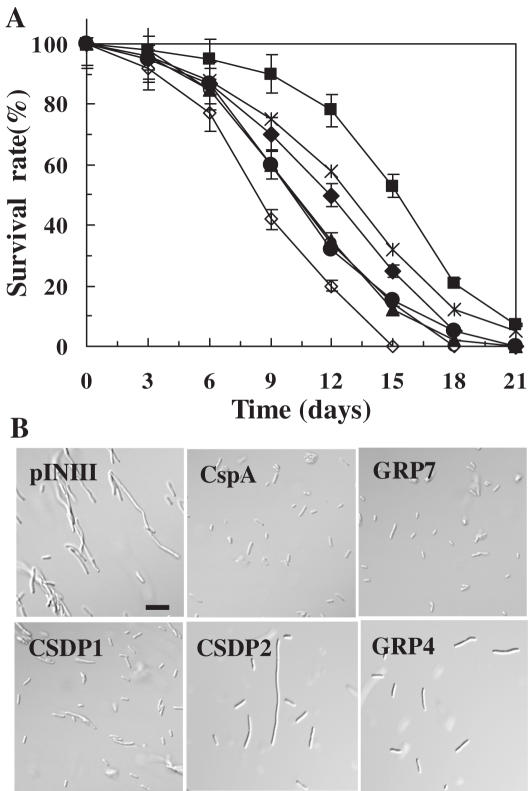
To further confirm that CSDPs and GRPs play roles as suppressors of cold sensitivity of BX04 cells during cold shock, the BX04 cells expressing either CSDP or GRP were subjected to cold shock at 4°C, and the survival rates of the cells were measured at the indicated time intervals (Figure 4A). After 6 days of incubation at 4°C, ∼77% of the E.coli harboring pINIII vector survived, whereas ∼83 to 88% of BX04 cells containing GRP or CSDP survived. In comparison, ∼95% of the BX04 cells containing pINIII-CspA survived. After 12 days of incubation at 4°C, ∼20% of the E.coli harboring pINIII vector survived, whereas ∼45 and 58% of pINIII-GRP7 and pINIII-CSDP1 cells survived, respectively. In comparison, ∼78% of pINIII-CspA cells and 32 to 35% of BX04 cells containing either pINIII-GRP2, pINIII-GRP4 or pINIII-CSDP2 survived. E.coli cells harboring pINIII vector all died 15 days after incubation at 4°C, whereas ∼20% of pINIII-GRP7 cells and 32% of pINIII-CSDP1 cells survived. BX04 cells harboring pINIII-CSDP1 and pINIII-CspA maintained viability 21 days after incubation at 4°C, whereas the rest of the cells died. These results indicate that, during the incubation at 4°C, the viability of BX04 cells depends heavily on the presence of CSDP1 or GRP7.
Figure 4.
Effect of CSDPs and GRPs on the viability of E.coli and suppression of the cell-division defect of BX04 by CSDP1 and GRP7 during cold shock. (A) The BX04 cells harboring pINIII-GRP2 (black triangle), pINIII-GRP4 (open circle), pINIII-GRP7 (black diamond), pINIII-CSDP1 (cross), pINIII-CSDP2 (black circle), pINIII-CspA (black square), or pINIII (open diamond) vector were subjected to cold shock at 4°C, and the survival rates of the cells were measured at the indicated time intervals. (B) The BX04 cells expressing GRP4, GRP7, CSDP1, CSDP2, CspA or pINIII were grown in LB medium at 15°C for 2 days, and were then observed under a light microscope. The bar corresponds to 10 μm in length.
Arabidopsis CSDPs and GRPs suppress the cell-division defect of BX04
Since it has been reported that BX04 cells incubated at 15°C for 72 h were all elongated and formed filamentous cells that resulted from the impairment of septum formation (29), we next wanted to test whether the Arabidopsis CSDP and GRP genes suppress the cell-division defect of BX04. As shown in Figure 4B, it was apparent that BX04 cells expressing pINIII control vector were elongated and formed filamentous cells when incubated at 15°C for 2 days, and this cell morphology was completely recovered to normal shape by the expression of CspA as observed previously (29). It was also evident that no filamentous cells were observed for BX04 cells containing either CSDP1 or GRP7 gene when grown at 15°C for 2 days. In contrast, the BX04 cells expressing CSDP2 or GRP4 that does not show the ability to suppress the cold sensitivity of the cells were elongated and formed filamentous cells at low temperature (Figure 4B). These results demonstrate that CSDP1 and GRP7 genes suppress the cell-division defect of BX04, implying that Arabidopsis CSDP, GRP and E.coli CspA share common basic activity that enables them to protect E.coli cells against cold shock stress.
DNA and RNA-binding properties of CSDP1 and GRP7
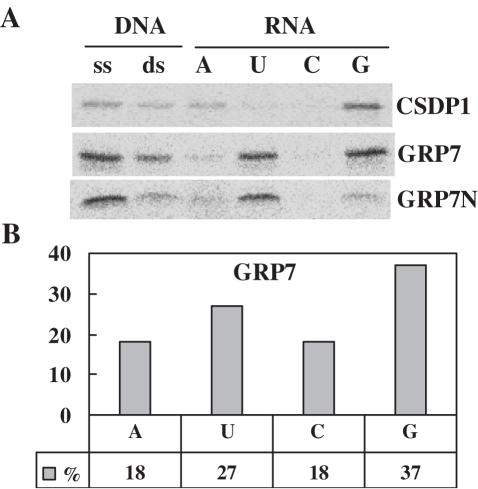
Since RNA chaperones have been known to bind RNA sequences nonspecifically, the nucleic acid-binding capability of CSDP1 and GRP7 was investigated. When the in vitro-synthesized 35S-labeled proteins were mixed with ssDNA, dsDNA (calf thymus) and four kinds of ribonucleotide homopolymers bound to agarose beads at different NaCl concentrations, it was apparent that CSDP1 bound preferentially to ssDNA and poly(G), and GRP7 bound preferentially to ssDNA, and poly(G) and poly(U) sequences (Figure 5A). It was observed that the binding property of the N-terminal region of GRP7 was quite similar to that of the full-length GRP7 (Figure 5A). To further determine whether GRP7 binds equally to all G- or U-rich RNAs, or whether it favors specific primary sequences for binding, SELEX was performed using recombinant GRP7-GST protein and in vitro-transcribed RNAs with 65 nt-long randomized regions. Sequence analysis of the RNAs enriched after eight rounds of SELEX revealed that GRP7 binds G- or U-rich RNAs but the primary sequence of the RNA is not important (Figure 5B and Supplementary Table 2).
Figure 5.
Nucleic acid-binding property of CSDP1 and GRP7. (A) The in vitro-synthesized 35S-labeled proteins were mixed with ssDNA, dsDNA (calf thymus) and four kinds of ribonucleotide homopolymers bound to agarose beads at 250 mM NaCl. Bound proteins were separated by SDS–PAGE, and the protein bands were visualized by an Image analyzer. (B) SELEX was performed using recombinant GRP7–GST protein and in vitro-transcribed RNAs with 65 nt-long randomized regions. The average occurrence of each nucleotide in 30 different RNA sequences is represented.
DNA melting activities of CSDP1 and GRP7
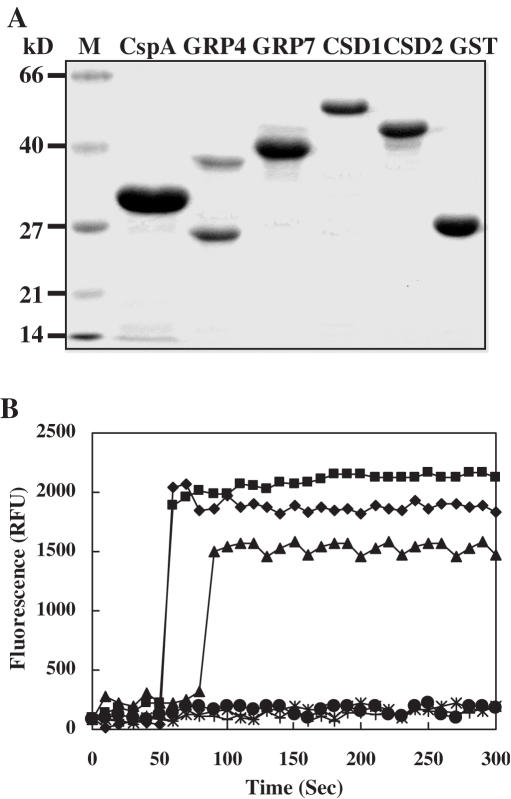
To better understand whether CSDP1 and GRP7 exert their role by RNA chaperone activity, DNA melting activities of the recombinant proteins were measured by using the 78 nt-long, 9 bp-containing, hairpin-shaped molecular beacon. The total bacterial extracts expressing CSDP1, CSDP2, GRP4, GRP7, CspA (positive control) or pINIII vector (negative control) as well as the purified CSDP1-GST, CSDP2-GST, GRP4-GST, GRP7-GST or CspA-GST recombinant proteins (Figure 6A) were tested for DNA melting activities. It was apparent that the addition of the lysates of the cells expressing CSDP1, GRP7 or CspA as a positive control resulted in stably increased beacon fluorescence. In contrast, the cells expressing CSDP2, GRP4 or empty pINIII vector showed no DNA melting activity (data not shown). It is noteworthy that these DNA melting activities were detected only when the E.coli cells were induced by the addition of IPTG. We also performed DNA melting assay with the purified CSDP1-GST, CSDP2-GST, GRP4-GST and GRP7-GST recombinant proteins. For unknown reasons, in the purification of GRP4, both GRP4–GST-fusion protein and GST protein were co-purified (Figure 6A). As shown in Figure 6B, the addition of CspA-GST, CSDP1-GST or GRP7-GST proteins resulted in greatly increased fluorescence, whereas the addition of CSDP2-GST, GRP4-GST or GST alone did not result in any fluorescence. There was a lag of ∼30 s in the appearance of fluorescence in GRP7 sample compared with CSDP1 and CspA samples. This pattern of delayed appearance in fluorescence was always observed irrespective of the amounts (5–20 μg) of CSDP and GRP used for the analysis (data not shown). We do not know at present whether the delayed appearance of fluorescence represents the intrinsic property of GRP7 or results from other experimental conditions. These results demonstrate that CspA, CSDP1 and GRP7 contain DNA melting activity, implying their roles as RNA chaperones during the cold adaptation process.
Figure 6.
Nucleic acid melting activity of CSDP1 and GRP7. (A) SDS–PAGE gel showing the expression and purification of recombinant proteins in E.coli; M, MW marker. (B) The fluorescence of a 78 nt-long molecular beacon was monitored as 10 μg of CSDP1-GST (square), 7 μg of GRP7-GST (triangle), 5 μg of CspA-GST (diamond), 7 μg of GRP4-GST (asterisk), 10 μg of CSDP2-GST (circle) or 5 μg of GST (plus) were added.
CSDP1 and GRP7 function as RNA chaperones
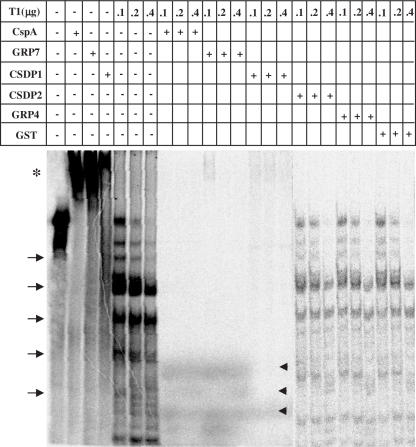
Based on the observations that CSDP1 and GRP7 complement successfully the cold sensitivity of Csp-deficient mutant, bind RNAs with low sequence-specificity, and contain DNA melting activity, it is speculated that CSDP1 and GRP7 may function as an RNA chaperone. We attempted to further examine, by the ribonuclease assay, whether CSDP1 or GRP7 binding to RNA secondary structure facilitates the RNA susceptibility to ribonuclease T1. The RNA substrate used in this study was prepared by transcribing the pET-22b(+) plasmid. When CSDP1, GRP7 or CspA (as a positive control) was added, the RNA was supershifted to upper positions indicated by asterisks (Figure 7). Binding of CSDP1 and GRP7 to this functionally un-related RNA sequence further indicates that CSDP1 and GRP7 bind to RNA with a sequence in an independent manner. It was noted that GST itself did not bind to this RNA (data not shown). When RNase T1 was added to the RNA and the mixture was incubated for 15 min on ice, several RNase T1-resistant bands appeared at lower positions indicated by arrows. However, when CSDP1, GRP7 or CspA was added before the addition of RNase T1, RNase-resistant bands disappeared and the new cleavage products appeared at lower positions indicated by arrowheads (Figure 7). At the lowest RNase T1 concentration in the presence of CSDP1 and GRP7, the broad, supershifted bands were observed. In contrast, no new cleavage products of RNA substrate appeared by the addition of CSDP2, GRP4 or GST that does not show the ability to suppress the cold sensitivity of the BX04 cells (Figure 2). The cleavage of RNA by RNase T1 was dependent on the concentration of CSDP1 and GRP7 in that more severe cleavage occurred by increasing the amount of proteins in the reaction mixture (Supplementary Figure 1). Similar results were obtained with RNase A; in the absence of CSDP1 and GRP7, several RNase A-resistant bands appeared at lower positions indicated by arrows. However, when CSDP1, GRP7 or CspA was added before the addition of RNase A, RNase-resistant bands disappeared and the new cleavage products appeared at lower positions indicated by arrowheads (Supplementary Figure 2). The fact that the original bands disappeared and the new cleavage products appeared in the presence of CSDP1 or GRP7 indicates that CSDP1 and GRP7 destabilize the secondary structures existing in RNA molecules to further digest the RNAs into smaller fragments. The results further support the proposition that CSDP1 and GRP7 function as RNA chaperones during the cold adaptation process, as does bacterial CspA.
Figure 7.
Enhancement of RNase activity by CSDP1 and GRP7. The [α-32P]-labeled RNA substrate was prepared by transcribing pET-22b(+) plasmid with T7 RNA polymerase, and the RNase assays were carried out for 15 min on ice. The amounts of RNase T1 used are shown at the top in micrograms. When the protein was used (indicated by +), 5 μg of CspA-GST, 7 μg of CSDP1-GST, 7 μg of CSDP2-GST, 5 μg of GRP4-GST, 5 μg of GRP7-GST and 3 μg of GST were added. The asterisk indicates supershifted band, the arrows indicate several RNase T1-resistant bands, and the arrowheads indicate new cleavage products appeared in the presence of CspA, CSDP1 or GRP7.
DISCUSSION
CSDP1 and GRP7 exhibit RNA chaperone activity during the cold adaptation process
Plant CSDPs and GRPs have been implicated to play roles in the cold adaptation process because they were strongly induced by cold temperatures. However, their functional roles and importance in the cold stress response are largely unknown. The present data revealed that Arabidopsis CSDPs and GRPs help E.coli grow and survive better during cold shock, which implies that CSDP1 and GRP7 play a positive role during the process of cold adaptation. In E.coli, downshifts in temperature during growth induce cellular adaptation to low temperature (8); cold-induced proteins, including members of the CspA family, are induced and play roles in survival at low temperature (8,9). In animal and bacterial cells, CSPs were induced in response to cold stress, and either overexpression or deletion of specific CSPs in target cells resulted in altered resistance of the cells to low temperature (13,29,33,34). In a recent study, a plant homology of CSD protein was identified in winter wheat (16). The CSD proteins from winter wheat and Arabidopsis CSDPs investigated in this study have high sequence similarity with the bacterial CSP family in the region of N-terminal CSD. In addition to this CSD, plant CSD proteins contain additional C-terminal glycine-rich domains interspersed with CCHC zinc fingers [(16,17), Figure 1A]. Although the roles and importance of these additional motifs found in plant CSDPs are largely unknown in terms of the stress response, the present data appear to indicate that plant CSDPs play similar roles as positive regulators of cell growth at low temperature, as do bacterial CSPs. It should be noted that CSDP1 has growth-stimulating activity at low temperature but CSDP2 does not have the activity. Since CSDP1 and CSDP2 differ from each other in that CSDP1 contains seven CCHC-type zinc fingers at the C-terminus, whereas CSDP2 has two CCHC-types zinc fingers at the C-terminus, it appears that the number and a proper arrangement of C-terminal CCHC-type zinc fingers are important for activity.
It has been suggested that bacterial CSPs function as mRNA chaperones by destabilizing the over-stabilized secondary structures in mRNAs for efficient translation at low temperatures (9,13,14). GRPs are also shown to play a role in the cold adaptation process. With the observation that cyanobacteria do not have any CSD proteins but do contain a cold-induced RRM protein instead, it was proposed that the function of RRM proteins may substitute for the function of CSD proteins in cyanobacteria (14). The GRPs investigated in this study do not have the same CSD found in bacteria; these GRPs contain similar structural features to the RRM protein found in cyanobacteria. Although not proven conclusively in this study, it is therefore proposed that the higher growth of E.coli cells expressing CSDPs or GRPs during cold shock resulted from the RNA chaperone activity of CSDPs and GRPs. The findings that CSDP1 and GRP7 complement successfully the cold sensitivity of Csp-deficient mutant, contain DNA melting activity, and enhance RNase cleavage activity further support the proposition that CSDP1 and GRP7 have RNA chaperone activity. This consideration is supported by our recent findings that demonstrate that atRZ-1a, an Arabidopsis GRP that contains an N-terminal RRM and C-terminal glycine-rich domains, interspersed by CCHC-type zinc fingers, plays a role in the enhancement of the freezing tolerance of Arabidopsis plants (22). In the analysis of the roles of GRP2 and GRP7 in the plant response to cold stress, overexpression of GRP2 and GRP7 also increased the cold and freezing tolerances of Arabidopsis plants (J. S. Kim and H. Kang, unpublished data), further supporting the proposition that GRPs exert RNA chaperone activity during the cold adaptation process. It is of worth to note that overexpression of GRP4, which shows negligible growth-stimulating ability in E.coli during cold stress (Figure 2) has no DNA and RNA melting activities (Figures 6 and 7), does not increase the cold or freezing resistance of Arabidopsis plants (25). Because GRP4 has a shorter glycine-rich region compared to GRP7, and it appears that the size of glycine-rich region at the C-terminus is important for activity.
Structural features important for nucleic-aid binding and chaperone activity
Investigation of the structural motifs necessary for binding target RNAs or DNAs and exerting biological function is important to allow for better understanding of the action mechanism of CSDPs and GRPs in the cold adaptation process. It has been reported that the S1 domain of polynucleotide phosphorylase (PNPase), which has a 3D structure quite similar to that of CspA although the primary structures differ substantially, complements the cold sensitivity of BX04 mutant (29). This finding suggests that the β-barrel structures found in CspA and in the S1 domain of PNPase, rather than their amino acid sequences themselves, are important for the growth-stimulating activity of E.coli during cold stress. Because GRPs contain RRM at the N-terminus and a glycine-rich region at the C-terminus, and CSDPs contain CSD at the N-terminus and glycine-rich domains interspersed with CCHC zinc fingers at the C-terminus, it is of interest to know whether these structural motifs are important for the growth-stimulating activity of E.coli during cold stress. The RRM is one of the best studied RNA-binding motifs and plays a role in binding the target RNA sequence, and the glycine-rich domain has been suggested to be involved in sequence-specific binding, as well as in interaction with other protein or ligand molecules (35). GRP2 binds preferentially to U-rich RNA sequences (24), and GRP7 prefers G- and U-rich RNA sequences to A- and C-rich sequences for their binding (36,37). We have recently shown that GRP4 binds sequence nonspecifically to RNA homopolymers (25). It was observed that the binding specificity of the truncated GRP7N containing only the N-terminal RRM is similar to that of the full-length GRP7 (Figure 5A). It is interesting to note that the growth-stimulating activity of the N-terminal region of GRP7 is higher than that of the C-terminal region of GRP7 in E.coli under cold stress (Figure 3). Although the importance of N-terminal RRM and the C-terminal glycine-rich region in the process of cold adaptation is not clear, the present study demonstrates that the N-terminal RRM is important for nucleic acid-binding and RNA chaperone activity, and the C-terminal glycine-rich region contributes to attain full activity.
It has been observed that CspA from E.coli binds single-stranded DNA and RNA sequences nonspecifically (13), and CspB, CspC and CspE from E.coli bind to specific RNA/single-stranded DNA sequences (38). It was suggested that non-specific RNA-binding of CspA is important for its role as an RNA chaperone (13,38). As revealed by in vitro nucleic acid-binding assay and SELEX, GRP7 binds G- or U-rich RNAs but the primary sequence of the RNA is not important (Figure 5 and Supplementary Table 2). It is interesting to note that CspB from E.coli, cold-induced RNA-binding protein from cyanobacteria, and GRPs from Arabidopsis prefer the poly(U) sequence. Although the primary sequence of the RNA is not important for GRP7 binding, it is possible that GRP7 recognizes specific secondary structural elements in the RNA. We examined this possibility by predicting the secondary structures of the RNAs listed in Supplementary Table 2 using Zuker's MFOLD program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold.html). Two folding patterns of the RNA sequences were observed; one- or two-hairpin structure with bulges and internal loops (data not shown). Although no common secondary structural elements were found in the RNAs, it is of interest to examine whether the one- or two-hairpin structure is important for GRP7 binding.
The in vitro nucleic acid-binding assay revealed that CSDP1 binds preferentially to single-stranded DNA and G-rich RNA (Figure 5A). It was apparent that the N-terminal region of CSDP1, which lacked all C-terminal zinc fingers, did not recover successfully growth of the mutant (Figure 3). This result is in line with a recent report demonstrating that deletion of all C-terminal zinc fingers in wheat WCSP1 abolished the growth-stimulating activity during cold stress (18). It is of interest to note that the C-terminal region of CSDP1 comprising seven zinc fingers has a much higher growth-stimulating activity than the N-terminal region of CSDP1 in E.coli under cold stress (Figure 3). Loss of the growth-stimulating activities in WCSP1 lacking three zinc fingers (18), in CSDP1 lacking seven zinc fingers (Figure 3), and in CSDP2 containing two zinc fingers (Figure 2), together with the finding that C-terminal region of CSDP1 comprising seven zinc fingers has a higher growth-stimulating activity (Figure 3), strongly suggest that the C-terminal CCHC-type zinc fingers in CSDPs are important determinants for this activity. Because plant CSDPs containing various copies of zinc fingers display different growth-stimulating activity of BX04 cells under cold stress, it is proposed that both CSD and zinc finger glycine-rich domains are important for nucleic acid-binding and RNA chaperone activity. In particular, the number of zinc fingers and/or a proper arrangement of CSD and zinc finger domains are very important for the activity of CSDPs during the cold adaptation processes.
Putative target genes regulated by CSDP1 or GRP7 in E.coli during cold stress
With the observation that CSDPs and GRPs display growth-stimulating activity in E.coli under cold stress, and bind RNAs with low sequence-specificity, the next important question is to determine how CSDPs and GRPs exert their roles during downshifts in temperature. RNA-binding proteins interact with the target RNAs and regulate RNA processing and/or translation, leading to ultimate modulation of protein synthesis of the target genes. Because GRP7 binds G- or U-rich RNAs but the primary sequence of the RNA is not important, it is difficult to search for target RNAs. In order to discover some clues regarding the molecular mechanism underlying the roles of CSDP1 and GRP7 on the cold stress adaptation process, we used proteome analysis to investigate what kinds of proteins are modulated in E.coli that expressed CSDP1 or GRP7 and showed enhanced cold tolerance compared to the control cells. Our preliminary proteome analysis demonstrated that several proteins are either up- or down-regulated by the expression of CSDP1 or GRP7 in E.coli under cold stress (Supplementary Table 3). In the analysis of primary sequences of the mRNAs encoding the up- or down-regulated proteins, no significant sequence similarity was found (data not shown).
To determine whether the changes in protein accumulation resulted from variation in the transcription of corresponding target genes, the transcript levels of these genes were investigated by RT–PCR analysis. No significant changes in the levels of RNA transcripts were observed between the cells expressing CSDP1, GRP7 or pINIII during cold shock at 15°C for 12–24 h, indicating that CSDP1 or GRP7 did not influence the transcription of these putative target genes (Supplementary Figure 3). Given that CSDP1 and GRP7 appear to bind the mRNAs sequence nonspecifically, it is likely that they recognize and bind to the secondary structural elements found in the mRNAs, leading to the altered expression levels of the corresponding proteins. Although we do not presently know whether all of these genes contribute to the enhancement of cold tolerance in BX04 cells that express CSDP1 or GRP7, it is tempting to speculate that CSDP1 and GRP7 modulated the expressions of these genes at posttranscriptional level, which resulted in increased growth and survival rates of E.coli under cold stress.
In conclusion, the present work provided novel information to increase our knowledge of the roles of CSDPs and GRPs in response to cold stress. The presence of an RRM similar to cold-regulated RRM in other bacteria and animal cells, the cold-dependent induction pattern, efficient complementation of the cold sensitivity of BX04 mutant cells, DNA melting activity and the enhancement of RNase activity suggest that GRPs and CSDPs both display RNA chaperone activities during the cold acclimation process. It appears that CSDPs and GRPs exert their functions by modulating the expression of several genes at a posttranscriptional level. Because our knowledge regarding the molecular mechanism underlying CSDPs or GRPs-mediated gene regulation under cold stress is far from sufficient, further analysis including identification of the RNA targets and any protein factors interacting with CSDPs and GRPs will be of great importance to discover a comprehensive picture of the functional roles of CSDPs and GRPs as regulators of gene expression under cold stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Dr M. Inouye and Dr S. Phadtare for BX04 mutant cell and pINIII vector. This work was supported by the SRC program of MOST/KOSEF (R11-2001-092-02007-0) to the Agricultural Plant Stress Research Center of Chonnam National University. Funding to pay the Open Access publication charges for this article was provided by MOST/KOSEF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crespi M.D., Zabaleta E.J., Pontis H.G., Salerno G.L. Sucrose synthase expression during cold acclimation in wheat. Plant Physiol. 1991;96:887–891. doi: 10.1104/pp.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn M.A., Hughes M.A., Zhang L., Pearce R.S., Quigley A.S., Jack P.L. Nucleotide sequence and molecular analysis of the low temperature induced cereal gene, BLT4. Mol. Gen. Genet. 1991;229:389–394. doi: 10.1007/BF00267460. [DOI] [PubMed] [Google Scholar]
- 3.Gilmour S.J., Artus N.N., Thomashow M.F. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 1992;18:13–21. doi: 10.1007/BF00018452. [DOI] [PubMed] [Google Scholar]
- 4.Houde M., Danyluk J., Laliberte J.F., Rassart E., Dhindsa R.S., Sarhan F. Cloning, characterization, and expression of a cDNA encoding a 50-kilodalton protein specifically induced by cold acclimation in wheat. Plant Physiol. 1992;99:1381–1387. doi: 10.1104/pp.99.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monroy A.F., Castonguay Y., Laberge S., Sarhan F., Vezina L.P., Dhindsa R.S. A new cold-induced alfalfa gene is associated with enhanced hardening at subzero temperature. Plant Physiol. 1993;102:873–879. doi: 10.1104/pp.102.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarillo J.A., Capel J., Leyva A., Martinez-Zapater J.M., Salinas J. Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol. Biol. 1994;25:693–704. doi: 10.1007/BF00029607. [DOI] [PubMed] [Google Scholar]
- 7.Ferullo J.M., Vezina L.P., Rail J., Laberge S., Nadeau P., Castonguay Y. Differential accumulation of two glycine-rich proteins during cold-acclimation alfalfa. Plant Mol. Biol. 1997;33:625–633. doi: 10.1023/a:1005781301718. [DOI] [PubMed] [Google Scholar]
- 8.Thieringer H.A., Jones P.G., Inouye M. Cold shock and adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 9.Phadtare S., Alsina J., Inouye M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999;2:175–180. doi: 10.1016/S1369-5274(99)80031-9. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein J., Pollitt N.S., Inouye M. Major cold shock protein of Escherichia coli. Proc. Natl Acad. Sci. USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolffe A.P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioassays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 12.Lorković Z.J., Barta A. Genomic analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W., Hon Y., Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 14.Graumann P.L., Marahiel M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 15.Kingsley P.D., Palis J. GRP2 proteins contain both CCHC zinc fingers and a cold shock domain. Plant Cell. 1994;6:1522–1523. doi: 10.1105/tpc.6.11.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlson D., Nakaminami K., Toyomasu T., Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- 17.Karlson D., Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakaminami K., Karlson D., Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl Acad. Sci. USA. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachetto-Martins G., Franco L.O., de Oliveira D.E. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim. Biophys. Acta. 2000;1492:1–14. doi: 10.1016/s0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 20.Albà M.M., Pagès M. Plant proteins containing the RNA-recognition motif. Trends Plant Sci. 1998;3:15–21. [Google Scholar]
- 21.Maruyama K., Sato N., Ohta N. Conservation of structure and cold-regulation of RNA-binding proteis in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acid Res. 1999;27:2029–2036. doi: 10.1093/nar/27.9.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y.O., Kim J.S., Kang H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005;42:890–900. doi: 10.1111/j.1365-313X.2005.02420.x. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter C.D., Kreps J.A., Simon A.E. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermel M., Guermann B., Delage L., Grienenberger J.-M., Maréchal-Drouard L., Gualberto J.M. A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl Acad. Sci. USA. 2002;99:5866–5871. doi: 10.1073/pnas.092019599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak K.J., Kim Y.O., Kang H. Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J. Exp. Bot. 2005;56:3007–3016. doi: 10.1093/jxb/eri298. [DOI] [PubMed] [Google Scholar]
- 26.Kim J.S., Kim Y.O., Ryu H.J., Kwak Y.S., Lee J.Y., Kang H. Isolation of stress-related genes of rubber particles and latex in fig tree (Ficus carica) and their expressions by abiotic stress or plant hormone treatments. Plant Cell. Physiol. 2003;44:412–419. doi: 10.1093/pcp/pcg058. [DOI] [PubMed] [Google Scholar]
- 27.Masui Y., Coleman J., Inouye M. Multipurpose expression cloning vehicles in Escherichia coli. In: Inouye M., editor. Experimental Manipulation of Gene Expression. NY: Academic Press; 1983. pp. 15–32. [Google Scholar]
- 28.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Xia B., Ke H., Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 30.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 31.Phadtare S., Inouye M., Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 32.Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 33.Danno S., Nishiyama H., Higashitsuji H., Yokoi H., Xue J.-H., Itoh K., Matsuda T., Fujita J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka K., Fang L., Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Micobiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 35.Hanano S., Sugita M., Sugiura M. RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol. 1996;31:57–68. doi: 10.1007/BF00020606. [DOI] [PubMed] [Google Scholar]
- 36.Ludevid M.D., Freire M.A., Gómez J., Burd C.G., Albericio F., Giralt E., Dreyfuss G., Pagès M. RNA binding characteristics of a 16 kDa glycine-rich protein in maize. Plant J. 1992;2:999–1003. [PubMed] [Google Scholar]
- 37.Hirose T., Sugita M., Sugiura M. Characterization of a cDNA encoding a novel type of RNA-binding protein in tobacco: its expression and nucleic acid-binding properties. Mol. Gen. Genet. 1994;244:360–366. doi: 10.1007/BF00286687. [DOI] [PubMed] [Google Scholar]
- 38.Phadtare S., Inouye M. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 1999;33:1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x. [DOI] [PubMed] [Google Scholar]