Abstract
Alternative splicing has been associated with increased evolutionary changes and with recent exon creation or loss. The addition of a new exon can be explained by its inclusion in only a fraction of the transcripts leaving the original form intact and giving to the new form the possibility to evolve independently but the exon loss phenomenon is less clear. To explore the mechanism that could be involved in CFTR exon 12 lower splicing efficiency in primates, we have analyzed the effect of multiple synonymous variations. Random patterns of synonymous variations were created in CFTR exon12 and the majority of them induced exon inclusion, suggesting a suboptimal splicing efficiency of the human gene. In addition, the effect of each single synonymous substitution on splicing is strongly dependent on the exonic context and does not correlate with available in silico exon splicing prediction programs. We propose that casual synonymous substitutions may lead to a reduced splicing efficiency that can result in a variable proportion of exon loss. If this phenomenon happens in in-frame exons and to an extent tolerated by the cells it can have an important evolutionary effect since it may generate a substrate for natural selection of new splicing isoforms.
INTRODUCTION
Pre-mRNA splicing is a complex mechanism that relies on the correct identification of protein-coding sequences (exons), on the transcribed RNA, from the more abundant, non-coding sequences (introns). This identification requires not only the presence of the ‘core’ splicing recognition features such as the 5′- and 3′-splice sites, branch-point sequences, and polypyrimidine tracts but is also modulated by additional cis-acting elements (1). These elements are localized either within the coding sequence or close to introns and may enhance or antagonize RNA processing. (2–4). Alternative pre-mRNA splicing, a pervasive feature in mammalian genomes, generates multiple transcripts from a single pre-mRNA and is a fundamental mechanism for the regulation of gene expression and the generation of proteome diversity. From an evolutionary point of view, alternative splicing is associated with a large increase in the frequency of recent creation and/or loss of exons, and it has the potential ability to generate minor, species-specific alternative spliced exons (5). In the case of the generation of new alternative spliced forms the new exon has been viewed as an internal paralog to its own gene in which a new function can potentially evolve without disrupting the original function of the gene. In humans one possible mechanism to generate new exons is the inclusion of parts of Alu sequences into mature mRNAs. These mobile elements can provide preformed splice sites that can be tested during the evolution (6,7) and in fact internal exons that contain an Alu sequence are predominantly, if not exclusively, alternatively spliced (8). On the other hand the mechanism that leads to a reduced splicing efficiency in constitutively included exons and the significance of the exon loss is less clear.
Exonic splicing regulatory elements contribute significantly to constitutive and alternative splicing regulation (1,2,9). These are highly degenerated RNA sequences that interact with several classes of positive and negative splicing trans-acting regulatory factors. Exonic splicing regulatory elements overlap with the amino acid code and are found in both pre-mRNAs that are constitutively (10,11) and alternatively spliced (1,12). Their importance in pre-mRNA splicing is relevant in fields as diverse as clinical genetics and molecular evolution. In fact, extensive evidence indicates that exonic point mutations may affect pre-mRNA splicing in several human disease genes (2,4,13–23). In particular, synonymous sites represent an interesting feature of exonic splicing regulatory elements because they may affect the splicing efficiency without theoretically changing the protein sequence. In several cases they may have pathological consequences (2,4,24). To identify the ‘splicing code’ hidden in exonic sequence and thus to predict the splicing phenotype of exonic mutations in human genes, several in silico programs have been developed (3,9,25,26), but few studies have systematically evaluated their reliability in clinical genetics. On the other hand, exonic splicing enhancers are widely distributed among metazoans from flies to humans (1), they have been reported also in yeast (27) and suggested to play a role in species-specific alternative splicing regulation (5). However, the effect of evolutionary related exonic nucleotide substitutions on the splicing efficiency and on the generation of new alternative splicing events is largely unexplored.
The CFTR exon 12 show reduced splicing efficiency in the primates (28–30) and its length being multiple of three, its skipping maintains in-frame the final protein. The alternative spliced form has up to now not been ascribed any functional role. Even if complete skipping causes severe classical cystic fibrosis (30,31), the functional significance and the mechanism that have generated this alternative splicing with partial skipping in the human lineage (between 5 and 30% in humans in vivo) (30,32) is unknown. Interestingly, the evolutionary differences in CFTR exon 12 in most mammals are mainly restricted to synonymous substitutions which are distributed in the entire exon length at 14 different sites (28). The only exception is found in the rodent lineage that has additional four non-synonymous substitutions (28). In the human CFTR exon 12, we have previously analyzed the exon splicing efficiency of several single synonymous changes in the central region of the exon (28). As ∼25% of them negatively affect the splicing process inducing exon skipping, these sites cannot evolve freely and are significantly constrained by splicing requirements, a fact that is supported by recent statistical and large scale genome comparative evidences (24,33). Synonymous substitutions in CFTR exon 12 might affect RNA secondary structure as previously suggested by statistical analysis (34), a possibility that needs additional experimental validation.
In this paper we have evaluated the functional role of multiple synonymous substitutions in order to understand why the CFTR exon 12 has a lineage-specific alternative splicing and to get more insight on the underlying regulatory mechanism. The effect of multiple random synonymous substitutions on the human CFTR exon 12 splicing pattern indicate that its splicing efficiency is the combinatorial result of single mutations and that the synonymous sites in the human gene have unexpectedly evolved toward a reduced splicing efficiency. This reduced splicing efficiency may be simply a background noise level tolerated by the cells but in turn generates a substrate for natural selection of new splicing isoforms.
MATERIALS AND METHODS
Hybrid minigene constructs
By PCR-mediated site-directed mutagenesis with ex12-Xba Rev 5′-TTCTGTTAAAACATCTAGATATCC-3′ and exon 12-Xba Dir 5′-ACTCTCCTTTTGGATATCTAGATG-3′ we introduced in the WTB minigene (22) a single 52T substitution that creates a unique XbaI restriction site. The resulting minigene (pTBCFex12 52T) was digested with AccI and XbaI, dephosphorylated and the corresponding exonic sequence between positions 13 and 52 replaced with two pairs of degenerated single stranded 5′-phosphorylated oligomers that contained a randomized 13-degenerated codons core (differing at conserved Leu and Ser codons for I and II pairs, respectively).
I-dir ATACAARGAYGCNGAYTTRTAYTTRTTRGAYTCNCCNTTYGGNTAT
I-rev CTGATANCCRAANGGNGARTCYAAYAARTAYAARTCNGCRTCYTTGT
II-dir ATACAARGAYGCNGAYCTNTAYCTNCTNGAYAGYCCNTTYGGNTAT
II-rev CTGATANCCRAANGGNGARTCYAAYAARTAYAARTCNGCRTCYTTGT
Each annealed oligonucleotide duplex (10 μg each) was ligated into the pTBCFex12 52T transformed into the DH5α strain of Escherichia coli. Random transformed colonies were collected, sequenced and then individually analyzed for splicing efficiency. Out of 22 single synonymous substitutions between positions 13 and 52 in the CFTR exon 12, 19 have been previously reported (28). The missing three mutations (31g, 46tc and 49c) were introduced in the previously described WTB minigene (22) between the AccI and BamHI sites, which were substituted with the appropriate AccI–BamHI cassettes created by PCR-mediated site-directed mutagenesis. The same strategy was used for the preparation of the minigenes reported in Figure 4. The oligonucleotides used for PCR-mediated mutagenesis are available upon request. All mutations were confirmed by sequencing.
Analysis of the hybrid minigene expression
Transient transfection of Hep3B cells, RNA extraction, reverse transcription RT–PCR, and quantitation of PCR products were done as described previously (22). PCRs were optimized to remain in the exponential range of amplification and products were routinely fractionated in 1.5% (w/v) agarose gel. For quantitation of the PCR, 32dCTP was included in the PCR mixture, the products loaded on 5% denaturing polyacrylamide–8 M urea gel, dried and exposed to a Cyclone Instant Imager. The counts of each splicing band were corrected by the number of C/G present in the PCR-product sequence.
Statistical analysis and in silico predictions
Statistical analysis was performed with StatView program and data were evaluated with nonparametric Kruskal Wallis and Mann Whitney tests. In silico analysis was performed using the following web-based resources, ESEfinder (http://rulai.cshl.edu/tools/ESE/) (25), RESCUE-ESE (http://genes.mit.edu/burgelab/rescue-ese/) (3), and PESX (http://cubweb.biology.columbia.edu/pesx/) (26). The threshold score for the enhancer or silencer motifs were set to the values suggested by the programs. The relationship between the number of splicing regulatory motifs and the percentage of exon inclusion was evaluated with linear regression using StatView program.
RESULTS
The composition of synonymous site in human CFTR exon 12 is suboptimal for splicing efficiency
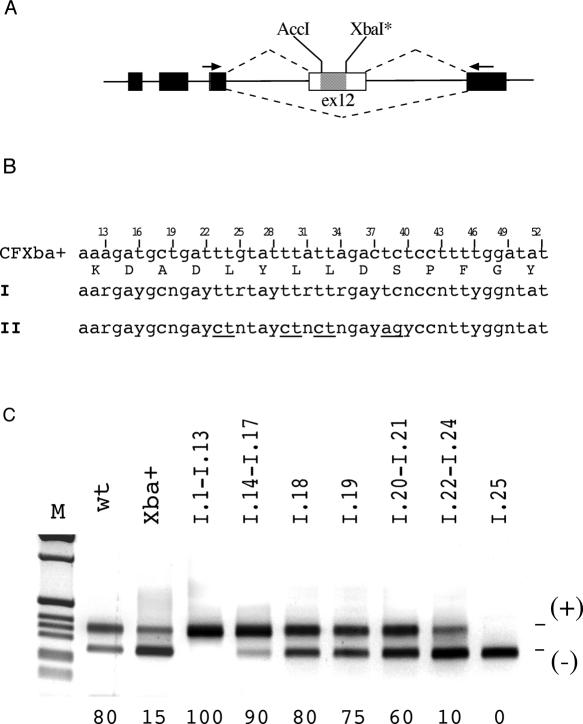
We have followed our earlier observations of the effect of site-directed mutants selected from the evolutionary divergences in mammals with random mutagenesis to explore the limits of the exon sequence variability. We have replaced the (wild-type) WT CFTR exon 12 sequence between positions 13 and 52 with two degenerated oligonucleotides pairs that differ at conserved Leu and Ser codons (Figure 1B). To facilitate cloning procedures the minigene contained a XbaI site that was inserted in position 52 changing the C with a T (Figure 1A) and the I and II oligomers ligated between the AccI and XbaI sites. Single clones derived from the ligations were isolated, sequenced and analyzed for splicing efficiency. We have evaluated a total number of 25 and 17 variants randomly selected from the I and II sequences, respectively. Compared with WT the I and II mutants showed a mean of 7.6 and 15.6 synonymous substitutions, respectively. Among the total number of 42 random sequences, 22 (53%) showed complete exon inclusion, 4 (9%) severe exon skipping (<15% of exon inclusion) and intermediate levels were observed in 16 (38%) sequences. Considering the 25 clones derived for the synonymous changes generated with the I oligonucleotide, 13 (52%), 8 (32%) and 4 (16%) variants showed complete, intermediate and low exon inclusion levels, respectively. Interestingly, only one clone showed complete exon skipping (I.25), whereas no minigene variants with low percentages of exon inclusion (<15%) were observed in the II group (Figure 2). The creation of the XbaI site used to facilitate cloning of the oligos had a conspicuous negative effect on the splicing pattern as expected from our previous work (28). On the other hand most of synonymous changes produce a significant improvement not only of the defective splicing caused by the 52 C to T change (XbaI site creation) but also relative to the WT pattern (Figure 1C). To further explore its potential confounding effect we restored the WT 52C in selected clones derived from the I oligo. Eight clones, indicated with a (C) in Figure 2, from the group with high, intermediate and low exon inclusion were mutated to 52C and analyzed for splicing efficiency in transfection experiments. This analysis revealed that the 52 T to C mutation did not affect the splicing pattern in clones with complete exon inclusion (I.1, I.5, I.8 and I.13) and with complete exon skipping (I.25) (Figure 2). On the other three intermediate clones (I.18, I.19 and I.23) this T to C substitution increased the percentage of exon inclusion. Overall the results indicate that the 52T variant used to facilitate cloning does not affect the final results and that the majority of random multiple synonymous changes enhance CFTR exon 12 inclusion.
Figure 1.
Combined effect of multiple random synonymous substitutions on CFTR exon 12 splicing pattern. (A) Schematic representation of the CFTR hybrid minigene used in transient transfection assay showing the location of the two restriction sites in exon 12 that were used for cloning the I and II degenerated oligonucleotides. Exonic and intronic sequences are shown as boxes and lines, respectively, the hatched box corresponds to the substituted sequence and the arrows show the specific oligonucleotides used in RT–PCR analysis. The two alternative splicing possibilities are indicated as dotted lines. The XbaI site was created by site-directed mutagenesis and corresponds to 52T synonymous substitution. (B) Nucleotide composition of part of the human CFTR exon 12 between the AccI and XbaI sites. Third nucleotide positions are numbered according to their location in the exon. The two 14-degenerated codons oligonucleotides I and II (II differs to I at conserved Leu and Ser codons, which are underlined) are indicated. These oligomers were cloned in the CFTR exon 12 to create random changes at synonymous sites between the exonic positions 13 and 52. (C) Effect of multiple random synonymous substitutions on splicing efficiency. Representative samples of the CFTR exon 12 minigenes derived from the degenerated oligonucleotides I are shown and their sequences are represented in Figure 2. Minigenes were transfected in Hep3B cells and analyzed for splicing efficiency. The percentages of exon inclusion are reported at the bottom of each lane and are the mean of three independent experiments done in duplicate. The effect of the multiple random synonymous substitutions is compared with the WT and the CFXba+ minigenes. The exon 12 inclusion and skipping forms are indicated.
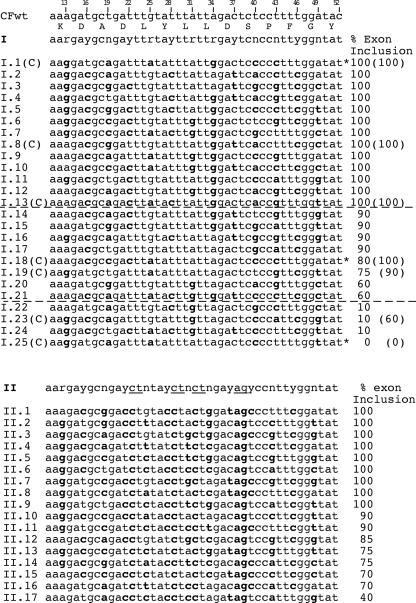
Figure 2.
Effect of CFTR exon 12 random multiple synonymous substitutions on the splicing efficiency. The two 13-degenerated codons oligonucleotides I and II were cloned in the CFTR exon 12 to create random changes at synonymous sites between the exonic positions 13 and 52. Resulting hybrid minigenes (I.1 to I.25 and II.1 to II.17) were then isolated and individually analyzed for splicing efficiency. The position of the mutations relative to the wild-type (CFWT) exon is shown at the top and synonymous mutations are in bold in comparison to human sequence. Number on the right of each sequence indicates the percentage of exon inclusion as determined by minigene splicing assay and is the mean of three independent experiments done in duplicate. In some clones derived from degenerated oligonucleotide I (indicated with a additional (C)) we changed the T52 to the wild-type C52 and the resulting percentage of exon inclusion is indicated in parenthesis on the right. The clones with the asterisks I.1, I.18 and I.25 were selected for subsequent experiments shown in Figure 4. Compared with WT sequence the I mutants showed a mean of 7.6 synonymous substitutions, the II mutants 15.6.
Correlation between single and multiple synonymous substitutions on the splicing efficiency
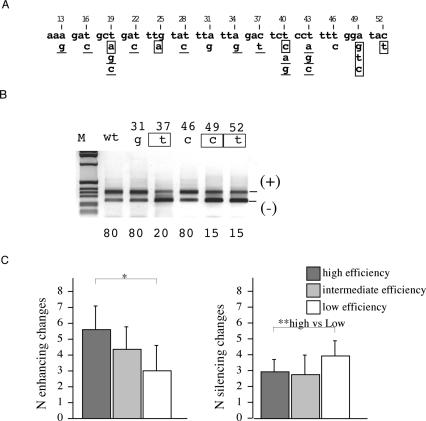
We have previously reported the effect of 19 out of 22 single synonymous substitutions between positions 13 and 52 on the exon 12 splicing efficiency (28). To compare the relationship between the multiple at the single synonymous substitutions we now have evaluated the three lacking variants and the complete analysis is shown in Figure 3A and B. Compared with the WT basal splicing efficiency each synonymous substitution can induce exon skipping, exon inclusion or have no effect. Out of 22 single synonymous substitutions, the majority (13) have a positive effect on splicing-inducing exon inclusion, 7 induce exon skipping and 7 did not affect the splicing pattern (Figure 3A). As the majority of substitutions preferentially induce exon inclusion we tested if the splicing pattern observed in multiple substitutions can be the result of the effect of each single substitution. To analyze the relationship between the single and multiple synonymous substitutions each change was classified as positive, negative or neutral according to its effect on splicing; the I minigenes with multiple changes were also separated in three groups according to their splicing efficiency: high, intermediate and low that correspond to 100, 99–50 and 50–0%, respectively. In each group the number of synonymous differences that induce exon inclusion or exclusion were recorded and analyzed for statistical significance. The number of enhancing changes decreased progressively with the reduction of the efficiency of splicing (Figure 3C). This difference was statistically significant between the three groups and between the high and low efficiency groups. On the other hand, a higher number of silencing changes were found in the low efficiency group and in this case a significant correlation was evident between the high and low efficiency groups (Figure 3C).
Figure 3.
The effect of multiple synonymous substitutions on splicing correlates with the sum of the effect of each single substitution. (A) Nucleotide composition of part of the human CFTR exon 12 between the AccI and XbaI sites. Third nucleotide positions are numbered according to their location in the exon. The 22 possible single synonymous changes are shown. According to their individual effect on splicing in WT context, the synonymous substitutions were classified as positive, (mutations that induce exon inclusion, underlined) negative (exon skipping, boxed ) or neutral (no effect in black). (B) Representative RT–PCR products from transfection experiments showing the effect of single synonymous substitutions. The 31g, 46c and 49c mutations, not tested previously (28), are compared with WT and other selected mutants. The exon 12 inclusion and skipping forms are indicated. The percentages of exon inclusion are reported at the bottom of each lane and are the mean of three independent experiments done in duplicate. (C) Correlation between splicing efficiency of single and multiple synonymous substitutions. The clones shown in Figure 2 with random multiple substitutions originated from the I oligonucleotides were grouped according to their splicing efficiency: high, intermediate and low corresponding to 100, 99–50 and 50–0% of splicing, respectively. For each group the number of positive or negative synonymous substitutions were calculated (y axis) and analyzed for statistical significance (*, P < 0.05 between the three different groups), (**, P < 0.05 between the high ad low efficiency groups).
The context determines the exon splicing efficiency of synonymous substitutions
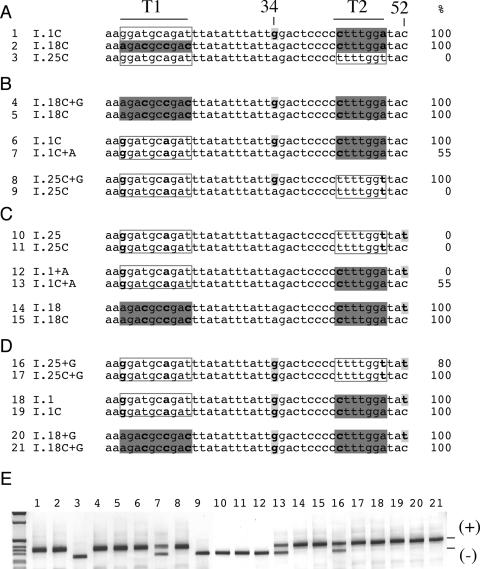
To test the importance of the context in determining the effect on splicing of single nucleotide substitutions we focus our attention on the G to A substitutions at position 34 and the T to C mutation at position 52, two mutations that induce significant exon skipping in the WT minigene (Figure 3A). The effect of these substitution on splicing was analyzed in three different contexts derived from the I.1, I.18 and I.25 minigenes with random CFTR exon 12 multiple synonymous substitutions (Figure 4). Strikingly, the 34G to A substitution may have no effect on exon inclusion (compare lanes 4 and 5), may cause partial exon skipping (compare lanes 6 and 7) or complete exon skipping (compare lanes 8 and 9) (Figure 4B). The 52T to C substitution did not affect the complete exon inclusion in three contexts: I.18 (lanes 14 and 15), I.1C (lane 18 and 19) and I.18+G (lanes 20 and 21) but increase the exon splicing efficiency in I.25+G (lanes 16 and 17, from 100 to 80%) and in I.1+A (lanes 12 and 13, from 55 to 0%). In the context of the exon already completely excluded (I.25), this substitution did not affect splicing (compare lanes 10 and 11). These results clearly show that the context where a synonymous mutation occur determine its effect on the splicing pattern.
Figure 4.
The context determines the exon splicing efficiency of synonymous substitutions. Significant sequence differences among the minigene constructs are highlighted Percentages of exon inclusion, determined by splicing assay, are indicated on the right. (A) Sequence comparison among I.1C, I.18C and I.25C minigenes. Significant sequence differences among the minigenes are highlighted and correspond to the T1 tract (A13, C16, C19 and C22), to position 34 (G or A) and at the T2 tract (C43 and A49). Compared with I.25C, I.18C had changes at T1 tract sequence (A13, C16, C19 and C22) and T2 tract (C43 and A49) whereas A1C showed differences at G34 and T2 (shown in bold and highlighted). (B) Splicing effect of the G to A substitution in position 34 in the three different contexts derived from I.1C, I.18C and I.25C. The G to A substitution at position 34 did not affect splicing in I.18C (compare I.18C+G with I.18C), caused partial exon skipping in I.1C (compare I.1C with I.1C+A) and complete exon skipping in I.25 (compare I.25C+G with I.25C). (C) Splicing effect of the T to C substitution in position 52 in three different contexts derived from I.1C, I.18C and I.25C. The T to C substitution in position 52 did not affect splicing in I.25 and I.18 context but induce partial exon inclusion in the A1 context. (D) Splicing effect of the T to C substitution in position 52 in three different contexts in the presence of the G to A substitution in position 34. In this case, the T to C substitution in position 52 did not affect splicing in I.1 and I.18 context but induce partial exon inclusion in the I.25 context. (E) RT–PCR products of transfection experiments. The different minigenes were transfected in Hep3B cells and analyzed for splicing efficiency. The exon 12 inclusion and skipping forms are indicated.
Correlation between the splicing efficiency of synonymous substitutions and the in silico predictions
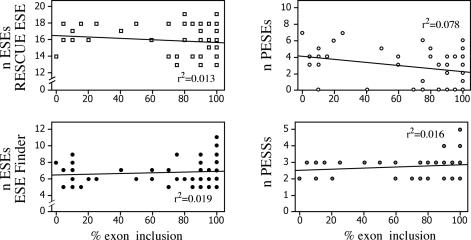
We tested if the exon inclusion efficiency resulting from 22 single and 55 multiple synonymous substitutions is dependent on predictable Exonic Splicing regulatory sequences. The number of predicted sites using ESE finder (25), RESCUE-ESE (3) and PESX (26) programs were plotted against the percentage of exon inclusion and the results are shown in Figure 5. No significant correlation was observed between the number of predicted sites and the splicing efficiency for both the single and double mutants. In addition, the combined evaluation of the effect of both putative exonic splicing silencer (PESS) and putative exonic splicing enhancers (PESE), calculated as the difference between them, did not show significant correlation (data not shown).
Figure 5.
Number of predicted exonic splicing enhancers motifs do not correlate with splicing efficiency. The percentages of exon inclusion detected by splicing assay were compared with the number of predicted exonic splicing regulatory motifs. The analysis includes 22 minigenes with a single substitution, 6 with multiple synonymous substitutions (shown in Figure 4) and 49 minigenes derived from random multiple synonymous substitutions (shown in Figure 2). Splicing regulatory motifs were predicted using RESCUE-ESE (3), ESE finder (19) and PEXS (26) programs.
DISCUSSION
Alternative splicing has been associated with increased evolutionary changes and in particular with a large increase in frequency of recent exon creation and/or loss (5). The addition of a new exon to a gene, the so called exonization process, might result in its inclusion in only a fraction of the transcripts, leaving the original form intact and giving to the new form the possibility to evolve independently. In humans intronic Alu repeats can turn into new alternative exons by providing potential splice sites (6,7). In this paper, we explore possible mechanisms of the exon loss process focusing on the variability of exonic splicing regulatory elements in the CFTR exon 12 at synonymous sites. As per definition, synonymous mutations do not change the protein, but they may affect splicing regulatory elements and modify the percentage of exon inclusion, hence having an important effect on human disease as well as in the creation of new alternative splicing isoforms. We have previously observed that single nucleotide substitutions at synonymous sites of the human exon can significantly affect its splicing pattern inducing both exon skipping or inclusion (28). We show here that multiple random synonymous substitutions preferentially increase the exon inclusion indicating that the composition of the human CFTR exon 12 is suboptimal for splicing efficiency (Figure 2). This effect on splicing was studied further by analyzing the correlation between single and multiple synonymous substitutions. The analysis of each single nucleotide variation showed that the majority of them preferentially induce exon inclusion than skipping (13 and 7, respectively) (Figure 3) indicating that the final exon splicing efficiency is due to some extent to the combined additive effect of the single synonymous changes. Since the alternatively spliced CFTR exon 12 minus form does not seem to have a clear functional role, its exon skipping might be simply ascribed to aberrant splicing generated by the ‘noise’ of the splicing machinery induced fortuitously by the synonymous substitutions (35). However, the aberrant splicing ‘noise’ might play a role in evolution by generating new alternative protein isoforms to be eventually selected. In fact our results provide a possible explanation for the previously reported (5) large increase in frequency of recent exon loss during evolution. Successive multiple synonymous mutations can induce partial skipping of a constitutively included in-frame exon. This will result in a slightly reduced production of the original protein and in the generation of a new alternatively spliced isoform. This situation ensures preservation of function and the new isoform without the exon can be evolutionary selected. Interestingly, new minor and not evolutionary conserved alternatively spliced exons are frequently reported in several disease-causing genes like NF1, CFTR and BRCA1. These events may result from the fortuitous insertion of negative splicing regulatory elements due to retrotransposition, as previously found in the CFTR exon 9 (36), or due to changes in exonic splicing regulatory elements. It remains to be determined how frequently species-specific new alternative splicing forms generated by substitutions at synonymous site are present in the human genome.
To explore the underlying splicing regulatory mechanism affected by multiple synonymous substitutions we have used available in silico programs that identify exonic splicing regulatory elements. With ∼70 mutants tested this is to our knowledge the largest analysis on a single exon that compares the splicing effect of multiple mutations in their original context with in silico predictions. The fact that the number of predicted exonic splicing enhancers and silencer motifs does not correlate with the splicing efficiency (Figure 5) indicates that the in silico programs are not useful to predict the effect of multiple mutations at least in the case of CFTR exon 12. This can be due to the fact that the available prediction programs deal with just a subgroup of splicing regulatory elements, which are not well represented in the CFTR exon 12. Since these programs are widely used in clinical genetics, it seems useful that their predictions should be experimentally validated in appropriate splicing assays that take into account the original gene context as far as possible. Furthermore, discrete sequences may have a strong context-dependent effect and can function as enhancer or silencer depending on the exonic context (9,12). On the other hand, the correlation between the effect of single and multiple synonymous substitutions and their context-dependent effect (Figure 3) suggests the presence of multiple overlapping splicing regulatory elements distributed along the entire CFTR exon 12, with both enhancer and silencer functions that are not really considered in the currently available prediction programs. At present their properties are difficult to explore by means of in silico analysis. It is possible that the entire sequence of the CFTR exon 12 is entirely made up by Composite Exonic Regulatory Element of Splicing (CERES) as previously suggested (22). The composite nature of these elements may result from the binding of multiple antagonistic splicing regulatory factors with different affinity that may be also modulated by RNA secondary structure.
Acknowledgments
We thank Rodolfo Garcia for critical reading of the manuscript and Cristiana Stuani for technical assistance. This work is supported by the Associazione Ricerca sul Cancro, Telethon Onlus Foundation (Italy) and the Italian Cystic Fibrosis Research Foundation. Funding to pay the Open Access publication charges for this article was provided by ICGEB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Ann. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 3.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 4.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 5.Modrek B., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 6.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 7.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 8.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goren A., Ram O., Amit M., Keren H., Lev-Maor G., Vig I., Pupko T., Ast G. Comparative analysis identifies exonic splicing regulatory sequences-the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Mayeda A., Screaton G.R., Chandler S.D., Fu X.D., Krainer A.R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaal T.D., Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muro A.F., Caputi M., Pariyarath R., Pagani F., Buratti E., Baralle F.E. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 15.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 17.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 18.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 19.Liu H.X., Cartegni L., Zhang M.Q., Krainer A.R. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nature Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- 20.Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 21.Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense and neutral mutations define juxtaposed regulatory elements of splicing in CFTR Exon 9. J. Biol. Chem. 2003;278:2680–2688. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 22.Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 23.Aznarez I., Chan E.M., Zielenski J., Blencowe B.J., Tsui L.C. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mol. Genet. 2003;12:2031–2040. doi: 10.1093/hmg/ddg215. [DOI] [PubMed] [Google Scholar]
- 24.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nature Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 25.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb C.J., Romfo C.M., van Heeckeren W.J., Wise J.A. Exonic splicing enhancers in fission yeast: functional conservation demonstrates an early evolutionary origin. Genes Dev. 2005;19:242–254. doi: 10.1101/gad.1265905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagani F., Raponi M., Baralle F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slomski R., Schloesser M., Berg L.P., Wagner M., Kakkar V.V., Cooper D.N., Reiss J. Omission of exon 12 in cystic fibrosis transmembrane conductance regulator (CFTR) gene transcripts. Hum. Genet. 1992;89:615–619. doi: 10.1007/BF00221949. [DOI] [PubMed] [Google Scholar]
- 30.Hull J., Shackleton S., Harris A. Analysis of mutations and alternative splicing patterns in the CFTR gene using mRNA derived from nasal epithelial cells. Hum Mol Genet. 1994;3:1141–1146. doi: 10.1093/hmg/3.7.1141. [DOI] [PubMed] [Google Scholar]
- 31.Zielenski J., Markiewicz D., Lin S.P., Huang F.Y., Yang-Feng T.L., Tsui L.C. Skipping of exon 12 as a consequence of a point mutation (1898 + 5G→T) in the cystic fibrosis transmembrane conductance regulator gene found in a consanguineous Chinese family. Clin. Genet. 1995;47:125–132. doi: 10.1111/j.1399-0004.1995.tb03944.x. [DOI] [PubMed] [Google Scholar]
- 32.Bremer S., Hoof T., Wilke M., Busche R., Scholte B., Riordan J.R., Maass G., Tummler B. Quantitative expression patterns of multidrug-resistance P-glycoprotein (MDR1) and differentially spliced cystic-fibrosis transmembrane-conductance regulator mRNA transcripts in human epithelia. Eur. J. Biochem. 1992;206:137–149. doi: 10.1111/j.1432-1033.1992.tb16911.x. [DOI] [PubMed] [Google Scholar]
- 33.Parmley J.L., Chamary J.V., Hurst L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 34.Meyer I.M., Miklos I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorek R., Shamir R., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]