Abstract
Riboswitches are highly structured elements in the 5′-untranslated regions (5′-UTRs) of messenger RNA that control gene expression by specifically binding to small metabolite molecules. They consist of an aptamer domain responsible for ligand binding and an expression platform. Ligand binding in the aptamer domain leads to conformational changes in the expression platform that result in transcription termination or abolish ribosome binding. The guanine riboswitch binds with high-specificity to guanine and hypoxanthine and is among the smallest riboswitches described so far. The X-ray-structure of its aptamer domain in complex with guanine/hypoxanthine reveals an intricate RNA-fold consisting of a three-helix junction stabilized by long-range base pairing interactions. We analyzed the conformational transitions of the aptamer domain induced by binding of hypoxanthine using high-resolution NMR-spectroscopy in solution. We found that the long-range base pairing interactions are already present in the free RNA and preorganize its global fold. The ligand binding core region is lacking hydrogen bonding interactions and therefore likely to be unstructured in the absence of ligand. Mg2+-ions are not essential for ligand binding and do not change the structure of the RNA-ligand complex but stabilize the structure at elevated temperatures. We identified a mutant RNA where the long-range base pairing interactions are disrupted in the free form of the RNA but form upon ligand binding in an Mg2+-dependent fashion. The tertiary interaction motif is stable outside the riboswitch context.
INTRODUCTION
RNAs can bind to a large variety of small-molecule ligands with high selectivity and affinity. Initial examples for this include numerous artificially selected RNAs—so-called aptamers—that bind ligands as diverse as nucleotides, amino acids, aromatic dye molecules, coenzymes such as FMN or aminoglycosides. The minimal RNA-motifs required for ligand binding in these aptamers are small and comprise between ∼30 and 50 nt (1). In general, they bind their ligands with affinities in the μM range (1). Structural analysis of many of the aptamer–ligand complexes has shown that the RNAs form intricate binding pockets for their respective ligands involving numerous non-canonical structural elements and combining electrostatic and hydrogen-bonding as well as stacking interactions to recognize their ligands. In the free RNAs, however, the ligand binding region is often completely unstructured and ligand binding is accompanied by an induced fit (2–4).
Recently, naturally occurring RNAs with the capability of binding to small molecule ligands have been discovered and named ‘riboswitches’. Riboswitches are a novel type of genetic control elements that modulate gene expression in bacteria as well as in some plants and fungi (5–7). So far, riboswitches have been reported that bind specifically to essential coenzymes or vitamins, amino acids, glucosamine-6-phosphate and to the purine bases guanine and adenine. They are mostly found in the 5′-untranslated regions (5′-UTRs) of mRNAs and normally consist of two domains, the ligand binding domain—also called the aptamer domain in analogy to the artificially selected RNAs—and an expression platform. Ligand binding to the aptamer domain causes an allosteric structural rearrangement of the expression platform that leads to modulation of gene expression.
Remarkably, the aptamer domains of these naturally occurring riboswitches in general are significantly larger than their artificial counterparts even when they bind to similar ligands and often bind their targets with an affinity in the low nM range. To date, structures are available for the complexes of the aptamer domains of the guanine/adenine-binding riboswitches (8,9), the thiamin pyrophosphate binding riboswitch (10–12) and one of the S-adenosyl methionine binding riboswitches (13) with their respective ligands. Naturally, the details of the RNA–ligand interactions in the three different complexes vary widely due to the chemical diversity of the different ligands. A wide variety of non-canonical structural elements, such as base triples and -quadruples and non-Watson–Crick base pairs are utilized to from the ligand binding pocket and to recognize the ligand similar to the recognition motifs found in the artificial aptamers. However, in contrast to the smaller artificial aptamers the riboswitch structures also revealed numerous long-range RNA–RNA tertiary interactions, such as pseudoknots that are not involved in the formation of the ligand binding pocket but instead stabilize the global fold of the RNA-structure.
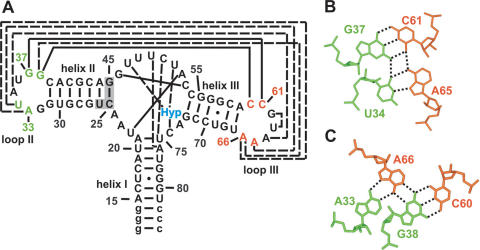
This interplay of tertiary interactions involved in ligand binding and remote to the ligand binding pocket is displayed for instance in the structures of the ligand bound aptamer domains of the guanine/hypoxanthine binding riboswitch and the closely related adenine riboswitch. With only ∼70 nt in length, these aptamer domains are among the smallest naturally occurring riboswitches but still able to adopt an intricate globular 3D fold. Their secondary structure consists of a three-helix junction where the three helices are connected by stretches of single-stranded nucleotides of different length (Figure 1A). These single-stranded nucleotides form the actual ligand binding pocket through the formation of a number of canonical and non-canonical long range base pairing interactions and base triples (Figure 1A). The purine ligand itself is bound through an intermolecular Watson–Crick base pair, a base pairing interaction between the N3/N9-edge of the ligand and an uridine of the RNA (Figure 2B) and a hydrogen bond between an RNA-2′-OH group and the N7-nitrogen of the ligand (8,9,14). In addition to the tertiary RNA-structure elements involved in ligand binding, the two loops capping helices II and III in the secondary structure of the aptamer domain are involved in a tertiary interaction that is remote from the ligand binding site. The two loops interact with each other through the formation of two base quadruples. One consists of a Watson–Crick G:C base pair and a reversed Hoogsteen A:U base pair (Figure 1B), the other one of a Watson–Crick G:C base pair and an A:A base pair (Figure 1C). This loop–loop interaction leads to a tight packing of helices II and III against each other and contributes significantly to the compactness of the aptamer structure in the bound state. The 8 nt contributing to the formation of the two base quadruples in the loop–loop interaction are phylogenetically strictly conserved and replacement of the wild-type loops by stable UUCG-tetraloops (8) abolishes ligand binding to the aptamer domain. Thus, the formation of this additional tertiary interaction that stabilizes the global fold of the RNA is crucial for ligand binding.
Figure 1.
Structure of the complex of the aptamer domain of the guanine riboswitch with hypoxanthine. (A) Secondary structure of the aptamer domain of the guanine binding riboswitch found in the 5′-UTR of the xpt-pbuX mRNA from B.subtilis. Helices I, II and III and the loops II and III are labeled accordingly. Long range base pairing interactions are indicated by solid lines for Watson–Crick base pairs and dashed lines for non-canonical base pairs. Nucleotides added to facilitate in vitro transcription are given in lower case letters and a stabilizing mutation in helix II that has no effect on ligand binding (14) is shaded. (B) Base quadruple formed by nucleotides U34 and G37 of loop II and C61 and A65 of loop III. G37 and C61 are involved in a long-range Watson–Crick base pair, whereas U34 and A65 form a reversed Hoogsteen base pair. Hydrogen bonds are indicated by dashed lines. (C) Base quadruple formed by nucleotides A33 and G38 of loop II and C60 and A66 of loop III. G38 and C60 are involved in a long-range Watson–Crick base pair, whereas A33 and A66 form an A:A base pair. Hydrogen bonds are indicated by dashed lines.
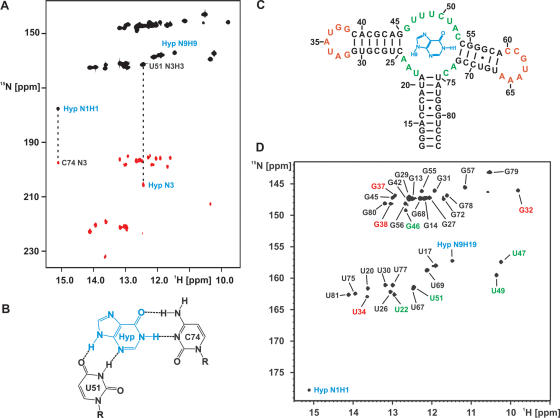
Figure 2.
Hypoxanthine binding to the aptamer domain and NMR resonance assignments for the RNA–hypoxanthine complex. (A) HNN-COSY spectrum for a complex of 15N-labeled RNA and 15N,13C-labeled hypoxanthine at 10°C in 25 mM KPO4-buffer (pH 6.2), 50 mM KCl in the absence of Mg2+-ions. Dashed lines highlight the correlations between hydrogen bond donor imino groups and hydrogen bond acceptor nitrogens due to the intermolecular hydrogen bonds between the N1H1 imino group of hypoxanthine and the C74 N3 nitrogen of the RNA as well as between the N3 of hypoxanthine and the U51 imino group of the RNA. The signal for the N9H9 imino group of bound hypoxanthine that is observable due to hydrogen bonding to a carbonyl group of the RNA is also labeled. (B) Schematic drawing of the intermolecular base pairing between hypoxanthine and the aptamer domain as derived from our NMR-data in the absence of Mg2+ and the X-ray-structure of the hypoxanthine–RNA complex in the presence of Mg2+. (C) Secondary structure and numbering scheme of the hypoxanthine–RNA complex. The ligand is colored blue, nucleotides in the helices are colored black, nucleotides in the ligand binding pockets are shown in green and nucleotides in the loops are highlighted in red. (D) Imino group region of an 1H,15N-HSQC-spectrum of the hypoxanthine–RNA complex at 10°C in the absence of Mg2+. Signals for the imino groups of the ligand are labeled in blue. Signals of the uridine and guanine imino groups are labeled in black for nucleotides in the helices, in green for nucleotides in the ligand binding pockets and in red for nucleotides in the loops.
Interestingly, the ligand is almost completely buried inside the RNA, only 2.2% of its surface are exposed to the solvent (8,9). This has lead to the notion that the ligand binding pocket is most likely in an open, disordered state prior to ligand binding and ligand binding must occur through an induced fit mechanism. In agreement with this hypothesis, in-line probing experiments find the single-stranded nucleotides of the three helix junction susceptible to hydrolytic cleavage in the free RNA but protected in the bound state (5). The induced fit mechanism for ligand binding is further supported by the observation of substantial changes in the NMR-spectra of the RNA upon ligand binding (9,14) and by thermodynamic measurements revealing a large unfavorable entropy contribution to the free energy of ligand binding (15) likely to reflect a local folding event. In contrast, the nucleotides in the loops capping helix II and III are not susceptible to hydrolytic cleavage in the absence of the ligand and show no changes in their protection pattern upon ligand binding. This is suggestive of an ordered structure of the loops already in the free form of the RNA (5).
However, structural information about the free state of the aptamer domain and the conformational transitions taking place upon ligand binding is currently lacking. NMR-spectroscopy in solution is a very valuable technique to probe conformational changes in both proteins and RNA even in cases where the free state is an ensemble of disordered or partially disordered conformers. Here, we use NMR-spectroscopy to compare the ligand free and ligand bound conformation of the guanine riboswitch aptamer domain of the 5′-UTR of the xpt-pbuX mRNA from Bacillus subtilis (5). Due to the size of this RNA (73 nt) that precludes a complete assignment of all the NMR-resonances we used the RNA imino-groups as reporters of conformational changes since these groups display well-resolved chemical shifts and their detectability directly reflects the formation of hydrogen bonds and tertiary structure elements (16). In addition, we probe the binding of divalent cations, such as Mg2+ to the riboswitch aptamer domain in its ligand bound and its free form and report a mutant involving the loop–loop interaction that shows altered folding behavior and Mg2+-requirements for ligand binding. Furthermore, we show that the loop–loop interaction can exist outside the context of the riboswitch and might be used as a RNA-tertiary interaction motif in ‘RNA-tectonics’ (17).
MATERIALS AND METHODS
Synthesis of 15N,13C-labeled hypoxanthine
13C,15N-labeled hypoxanthine was prepared by oxidation of 13C,15N-labeled adenine (14) with sodium nitrite in acidic condition. A total of 4 mg adenine were dissolved by heating in 19 ml of a water/H2SO4 (10:7) mixture. After 5 min the temperature was set to 85°C and 0.01 g NaNO2 were added. Additional H2SO4 and NaNO2 were added every 10 min. The reaction mixture was cooled to room temperature and purified by reversed-phase high-performance liquid chromatography (HPLC) as described in (14).
RNA preparation
15N- and 15N,13C-labeled nucleotides were purchased from Silantes (Munich). Labeled RNAs were prepared by in vitro transcription with T7 RNA polymerase from linearized plasmid DNA templates and purified as described (18). The unlabeled RNA-constructs containing only helices II and III of the aptamer domain were chemically synthesized and purchased from Dharmacon (Boulder, CO). They were deprotected and lyophilized as described by the protocol supplied by the manufacturers. Concentrations were measured by ultraviolet (UV) absorption at 260 nm. All RNAs were folded into a monomeric form by denaturing at 368 K at a concentration of 0.25 mM and subsequent dilution to 0.05 mM with ice-cold water and finally exchanged into NMR buffer [25 mM KPO4 (pH 6.2) and 50 mM KCl] using Centricon-10 or Centricon-3 microconcentrators (Amicon, Inc.).
NMR-spectroscopy
NMR experiments were performed on Bruker AV 700, AV 800 and AV 900 MHz spectrometers equipped with 5 mm TXI-HCN cryogenic probes and z-axis gradients. All spectra were processed and analyzed using the Bruker NMRsuite (XwinNMR 3.5) and Xeasy (19). NMR spectra were recorded in 90% H2O/10% D2O at a temperature of 283 K using the WATERGATE (20) water suppression scheme including water flip-back pulses (21). 1H,15N-HSQC-, 1H,13C-HSQC-, 2JHN-1H,15N-HSQC- (22) and 2D- or 15N-edited 3D-NOESY-experiments were carried out using standard pulse sequences (23). The 2hJNN-HNN-COSY experiments (24) were performed as described (14).
Native gel electrophoresis
RNA was loaded in a buffer containing 50 mM Tris–acetate, 100 mM sodium acetate (pH 8.0) (running buffer). Electrophoresis was performed in 10% acrylamide:bisacrylamide (37.5:1) gels in running buffer at room temperature at 150 V for 8 h. The gel was stained with ethidium bromide.
RESULTS
Hypoxanthine binding to the G-switch aptamer domain in solution
The guanine riboswitch aptamer domain binds to both guanine and hypoxanthine albeit with a 10-fold lower affinity for the latter (5). X-ray structures were determined for the aptamer domain in complex with both compounds and revealed an identical mode of ligand recognition (8,9). Due to the absence of the amino group in the 2-position of the ligand, hypoxanthine forms two intermolecular hydrogen bonds less than guanine. Due to its superior solubility we used hypoxanthine as the RNA–ligand in our NMR studies and tested the binding mode of hypoxanthine in solution [25 mM KPO4-buffer (pH 6.2), 50 mM KCl, no Mg2+ and 10°C] by NMR-spectroscopy. First, addition of 15N,13C-labeled hypoxanthine to a sample of 15N-labeled RNA resulted in the appearance of numerous new signals with narrow line widths in the imino-group region of a 1H,15N-HSQC-spectrum. Most importantly, signals for the N1H1 and the N9H9 imino groups of hypoxanthine could be observed (Figure 2A and B) and unambiguously identified in a H(N)CO-experiment employing the selective 15N,13C-labeling of the ligand (data not shown). These signals are not observable for the free ligand due to fast exchange of the imino protons with the bulk solvent. The appearance of new signals for RNA imino groups upon hypoxanthine addition and the appearance of signals for the hypoxanthine imino groups show that hypoxanthine binds to the aptamer RNA in solution even in the absence of Mg2+. The line widths for the hypoxanthine imino group signals as well as for the signals of the complexed RNA are in agreement with the formation of a high-affinity complex, which is in slow exchange on the NMR-timescale indicating a dissociation constant smaller then 1 μM.
A HNN-COSY-experiment was used to identify NH•••N-type hydrogen bonds between the bound hypoxanthine and the RNA. The N1H1 imino group of bound hypoxanthine showed a correlation to a cytidine N3 nitrogen (Figure 2A) indicating the formation of a Watson–Crick base pair between the ligand and the RNA (Figure 2B). In addition, an uridine imino group of the RNA is correlated with the N3 nitrogen of the bound hypoxanthine (Figure 2A) in agreement with a base pairing interaction of this uridine with the N3/N9-edge of the bound ligand (Figure 2B). The N9H9-imino group of hypoxanthine, which is involved in a hydrogen bond and therefore protected against exchange with the solvent can be detected but shows no correlation to a nitrogen in the HNN-COSY-experiment. This is expected when the hydrogen bond acceptor is an oxygen (Figure 2A and B). The observed hydrogen bonding pattern between the bound hypoxanthine and the RNA in solution is in full agreement with that observed in the X-ray structure of the hypoxanthine-RNA complex reported by Batey et al. (8) (Figure 2B). The addition of up to 2 mM EDTA to remove traces of divalent cations did not lead to changes in the NMR-spectra of the complex (data not shown).
NMR-assignments and solution structure of the ligand bound aptamer domain
The imino region of the 1H,15N-HSQC-spectrum of the aptamer domain bound to hypoxanthine is exceptionally well resolved for an RNA of this size and displays numerous signals with narrow line widths and homogeneous intensity (Figure 2D). The aptamer domain contains 21 guanine nucleotides and 17 uridine nucleotides in its sequence. In the 1H,15N-HSQC-spectrum 19 signals are observed in the region of the guanine imino groups and 14 signals are observed in the region of the uridine imino groups (Figure 2D) along with signals for the two imino groups of the bound ligand (see above). The observable signals were completely assigned using a combination of 2D-1H,1H-NOESY-, 3D-1H,1H,15N-NOESY-HSQC and 2D-1H,1H,(13C)-NOESY-HSQC-experiments (data not shown). The assignments were confirmed by comparing the spectra to those of a guanine-RNA and a 7-deaza-guanine-RNA complex (14) and to those of a hypoxanthine complex with a mutant RNA (see below). The two guanine imino signals that are not observable belong to G12 in the 5′-terminal closing base pair of helix I and G62 in the loop III region. The three uridine signals that are missing are U36, U48 and U63. The imino groups of U36 and U48 are exposed to the solvent in the X-ray structure of the hypoxanthine–RNA complex. G62 and U63 are located at the tip of loop III, which is also directly exposed to the solvent. The number and the location of the observed imino group signals for the hypoxanthine-RNA complex in solution agree remarkably well with the X-ray structure (8,9). In addition, the observed intra- and intermolecular NOEs between the RNA imino groups and the protons of the bound hypoxanthine, respectively, can all be predicted using the X-ray structure of the complex. Furthermore, the observed chemical shift changes upon modifying the ligand from hypoxanthine to guanine to 7-deaza-guanine (data not shown) and the observed intermolecular (see above) and intramolecular hydrogen bonding patterns in the HNN-COSY experiment (Figure 2A and see below) all indicate that the structure of the hypoxanthine–RNA complex in solution is virtually identical to the structure in the crystal.
Effects of Mg2+ on the ligand bound aptamer domain
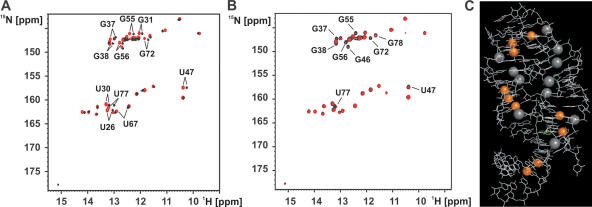
To gain insight into the possible role of divalent cations for the structure and stability of the hypoxanthine–RNA complex we titrated the 1:1 complex with MgCl2 up to a final Mg2+-concentration of 5 mM and followed the chemical shift changes induced by the presence of the divalent metal ion in 1H,15N-HSQC-spectra. In general, Mg2+ induced only very limited chemical shift changes and no new imino group signals appeared in the spectra. This indicates that the presence of Mg2+ does not change the structure of the hypoxanthine–RNA complex and does not promote the formation of additional structure elements. The following imino groups display chemical shift changes by more then three line widths (>66 Hz) in either the 1H- or the 15N-dimension in the presence of Mg2+: U26, U30, G31, G37, G38, U47, G55, G56, U67, G72 and U77 (Figure 3A). Mapping of these residues onto the X-ray structure of the complex reveals five spatially distinct regions that are affected by the presence of Mg2+-ions. G37 and G38 are part of the loop–loop interaction between loops II and III and U67 is part of the closing base pair of helix III right next to G38. U47 is part of the ligand binding core. U30 and G31 are located at the tip of helix II facing loop II. G55, G56 and G72 are located at the 5′ end of helix III and close in space to U26 of helix II whereas U77 is in the center of helix I. Thus, the observed chemical shift changes indicate the presence of up to five putative Mg2+-binding sites in the hypoxanthine–RNA complex.
Figure 3.
Divalent cation binding to the hypoxanthine–RNA complex. (A) Overlay of the imino group regions of 1H,15N-HSQC-spectra for the hypoxanthine–RNA complex at 10°C in the absence of Mg2+-ions (black) and the presence of 5 mM MgCl2 (red). Only small chemical shift changes and no new signals are observed upon addition of Mg2+. Resonances shifting by more then three line widths are labeled. (B) Overlay of the imino group regions of 1H,15N-HSQC-spectra for the hypoxanthine–RNA complex at 10°C in the presence of 5 mM Mg2+ (black) and in the presence of 5 mM Mg2+ and 5 μM Mn2+ (red). Resonances that are strongly broadened or disappear due to paramagnetic line broadening in the presence of 8 μM Mn2+ are labeled. (C) Mapping of the position of the imino groups experiencing line broadening upon Mn2+-addition (8 μM) on the 3D structure of the hypoxanthine–RNA complex. The affected imino groups are highlighted as orange spheres. The RNA is shown as white lines. Four well-defined divalent cation binding sites are observed. The position of [Co(NH3)6]3+-ions of cobalt hexamine groups found in the X-ray structure of the complex is indicated by gray spheres.
In addition, we titrated the hypoxanthine–RNA complexe in the presence of 5 mM MgCl2 with 5 and 8 μM MnCl2. Mn2+ is a paramagnetic analog of Mg2+. Its binding should induce paramagnetic line broadening in the NMR-signals of nucleotides in spatial proximity to the binding site (16). The results of this titration experiment with 5 μM Mn2+ are shown in Figure 3B. The imino group signals of G37, G38, G46, U47, G55, G56, G72, U77 and G78 either disappear completely or are very strongly broadened. At higher Mn2+-concentrations (8 μM) U34 is also affected (data not shown). U34 is close in space to G37 and G38 involved in the loop–loop interaction. These residues are similar to the ones that experience significant chemical shift changes upon addition of Mg2+. The exceptions are U30 and G31 where no line broadening is observed. Therefore, the paramagnetic line broadening data support the presence of four of the divalent cation binding sites mapped by chemical shift perturbation experiments using Mg2+. The spatial location of these four divalent metal-binding sites is mapped on the 3D structure of the hypoxanthine–RNA complex in Figure 3C. Also indicated is the location of [Co(NH2)6]3+-ions found in the X-ray structure of the complex. The location of the [Co(NH2)6]3+-ions found in the crystal coincide with three of the divalent cation binding sites identified in our NMR-experiments although sometimes more then one [Co(NH2)6]3+-ion is found per binding site. One exception is the region around nucleotides U77 and G78 in helix I where we find both chemical shift changes induced by Mg2+ and line broadening upon addition of Mn2+ but where no [Co(NH2)6]3+-ion is observed in the X-ray structure. In this region the RNA used for NMR contains the wild-type U17:G79 wobble base pair, which differs from the RNA used in the X-ray study where a canonical Watson–Crick base pair is introduced. The other exception is helix II where [Co(NH2)6]3+-ions are found in the X-ray structure (8) but no paramagnetic line broadening is observed in the presence of Mn2+. We find, however, chemical shift changes induced by Mg2+ in this area (G31 and U30).
Most importantly with respect to our further findings all three experimental approaches (Mg2+-induced chemical shift perturbations, paramagnetic line broadening with Mn2+ and X-ray crystallography) consistently find divalent-metal ion binding sites in the vicinity of the two base quadruples forming the loop–loop interaction.
Whereas Mg2+ is not changing the structure of the complex it influences its stability. 1H,15N-HSQC-spectra recorded at different temperatures show that many imino group signals including those of the bound ligand broaden at ∼25°C and disappear entirely at higher temperature in the absence of Mg2+ indicating the melting of the complex structure whereas these signals are still observable at 42°C in the presence of 5 mM Mg2+ (data not shown).
Residual structure of the free aptamer domain
To characterize the structural transitions in the RNA induced by ligand binding in more detail we compared the imino regions of 1H,15N-HSQC-spectra of the RNA in its free and hypoxanthine bound states in the absence of Mg2+. The imino group signals of guanine and uridine nucleotides in RNA are well-suited reporters of structural transitions since they are only observable when the imino group is involved in a hydrogen bond due to a base pairing interaction or sterically protected against fast exchange with the bulk solvent. In addition, the chemical shifts of imino groups are sensitive to the type of interactions and due to the large aromatic ring current effects also sensitive to changes in aromatic stacking interactions.
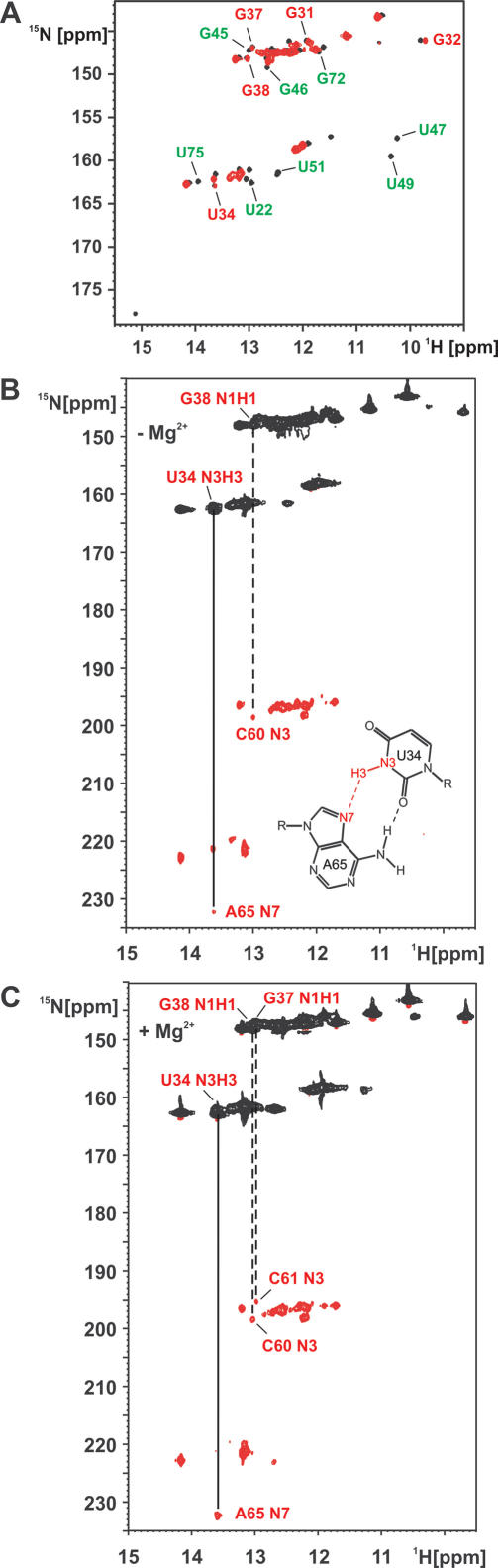
The 1H,15N-HSQC-spectrum of the free RNA contains significantly fewer signals compared to the one of the hypoxanthine bound state (Figure 4A). In addition, signal line widths are less homogeneous in the free state. This indicates that the free state of the RNA is less structured and resembles an ensemble of structures with intermediate exchange kinetics on the NMR time scale. However, many signals can be assigned by comparing the two spectra of the free and the bound states. These assignments were confirmed and extended by 2D-1H,1H-NOESY- and 3D-1H,1H,15N-NOESY-HSQC-experiments on the free RNA (data not shown). Comparison of the two spectra reveals that the secondary structure of the RNA is very similar in the free and the bound state, e.g. Watson–Crick base pairing in helices I, II and III is similar in both states. In addition, the G:U base pairs U17:G79 in helix I and G57:U69 in helix III are observed in both the free and the bound state.
Figure 4.
Conformational changes of the RNA upon hypoxanthine binding and the conformation of the free RNA. (A) Overlay of the imino group regions of 1H,15N-HSQC-spectra for the hypoxanthine–RNA complex (black) and the free RNA (red) at 10°C. Signals of imino groups are labeled in green for nucleotides in the ligand binding core and in red for nucleotides of loop II and III. The signals for the core nucleotides, such as U22, G45, G46, U47, U49, U51, G72 and U75 are only present in the bound state. In contrast, the imino groups of G32, U34, G37 and G38 in loop II as well as those for the closing base pairs of helix II G31 are observable in both the free and the bound form and have virtually identical chemical shifts. (B) HNN-COSY spectrum for the free RNA at 10°C in 25 mM KPO4-buffer, 50 mM KCl in the absence of Mg2+-ions. A solid line indicates a correlation corresponding to a hydrogen bond between an uridine imino group and an adenine N7-nitrogen due to the presence of the reversed Hoogsteen base pair formed by U34 in loop II and A65 in loop III as shown schematically in the inset. A dashed line indicates the hydrogen bond between the imino group of G38 in loop II and the N3 nitrogen of C60 in loop III as expected in a canonical Watson–Crick G:C base pair. The presence of these hydrogen bonds indicates that the two base quadruples that stabilize the loop–loop interaction are present already in the free RNA. (C) HNN-COSY spectrum for the free RNA at 10°C in 25 mM KPO4-buffer, 50 mM KCl in the presence of 5 mM magnesium. The cross peak corresponding to the hydrogen bond in the reversed Hoogsteen base pair formed by U34 in loop II and A65 in loop III is stronger as in the absence of Mg2+ and the correlation between the imino group of G37 and the N3 of C61 due to the presence of the G37:C61 Watson–Crick base pair becomes detectable indicating that these base pairs are further stabilized in the presence of divalent cations.
In contrast, the signals for nucleotides in the ligand binding core of the RNA are completely absent in the 1H,15N-HSQC-spectrum of the free RNA as are the signals corresponding to nucleotides of the closing base pairs of the three helices facing the ligand binding core. No signals can be found for the imino groups of U22, G45, G46, U47, U49, U51, G72 and U75 (Figure 4A). The nucleotides in the ligand binding core are apparently not involved in any stabilizing hydrogen bonding interactions in the free state of the RNA. Therefore, the ligand binding core is likely to be flexible in the absence of ligand.
However, the spectrum contains more signals then just those due to the Watson–Crick and G:U base pairs of the three helices. The remaining signals can be assigned to the imino groups of G32, U34, G37 and G38 that correspond to nucleotides involved in the loop–loop interaction between loops II and III. In addition, the signal for the imino group of G31 in the closing base pair of helix II facing loop II can be found. All these signals have very similar shifts in the free and the bound state of the RNA indicating that they are involved in similar base pairing interactions. G37 and G38 in loop II are part of long range Watson–Crick base pairs with C61 and C60 in loop III, respectively. U34 forms a reversed Hoogsteen base pair with A65. The G37:C61 base pair and the U34:A65 base pair constitute a base quadruple that is further stabilized by hydrogen bonding interactions between G37 and U34 as well as between C61 and A65 (Figure 1B). Similarly the G38:C60 base pair forms a base quadruple with an unusual A35:A66 base pair (Figure 1C). The imino group of G32 is not hydrogen bonded but sterically protected from exchange with the solvent due to stacking interactions (Figure 4A and B). To further probe the hydrogen bonding interactions of these bases, we recorded an HNN-COSY spectrum of the free RNA. The imino group of G38 showed the expected correlations with the C60 N3 nitrogen due to the formation of a Watson–Crick G:C base pair. Moreover, the U34 imino group showed a correlation to an adenine N7-nitrogen as expected for a reversed Hoogsteen A:U base pair (Figure 4B). The same correlations have been observed for these nucleotides in the HNN-COSY experiment with the hypoxanthine bound RNA. Together with the very similar chemical shifts in both states these data demonstrate that the long range base pairs between loop II and loop III are already present in the free RNA and that the geometry of the base quadruples and the conformation of the adjacent nucleotides is the same in the free and the hypoxanthine bound RNA. The virtually identical chemical shifts for the imino protons of nucleotides involved in the loop–loop interaction in the free and the ligand bound RNA furthermore indicate the free RNA is not in fast equilibrium between a conformation with the loop–loop interaction formed and an open conformation. In this case significant changes in the chemical shifts for those nucleotides would be observed upon ligand binding since the chemical shifts in the free form would be a population average between the open and the closed form. A slow equilibrium in the free RNA between a conformation with a loop–loop interaction and an open conformation would likely result in additional signals for hydrogen-bonded imino protons of nucleotides in stable Watson–Crick base pairs in helices II and III, which are in close proximity to loops II and III, respectively. Such additional signals are not observed.
However, the line widths of the imino group signals of G37 and U34 in the free RNA are larger then those in the hypoxanthine bound RNA indicating increased dynamics and a reduced stability of the G37:C61 and the U34:A65 long range base pairs in the free state of the RNA. The 1H,15N-HSQC-spectra of the free RNA are not concentration dependent in a range of RNA-concentrations from 150 to 750 μM indicating that the formation of the loop–loop interaction is not due to dimerization at the high-RNA concentrations typically used for NMR-spectroscopy.
The addition of Mg2+ to the free RNA results in only minor chemical shift changes and does not lead to the appearance of new signals e.g. for nucleotides of the ligand binding core region. A HNN-COSY experiment in the presence of Mg2+, however, reveals that the cross peak corresponding to the correlation between U34 NH and the A65 N7 of the reversed Hoogsteen U34:A65 base pair becomes stronger and the correlation between the imino group of G37 and the N3 nitrogen of C61 is now readily detectable (Figure 4C). Therefore, Mg2+ apparently stabilizes the loop–loop interaction.
Finally, it should be noted, that small chemical shift differences are observed between the free and the bound RNA also for helical regions, e.g. in helices II and III. These chemical shift changes indicate subtle rearrangements in the structure upon ligand binding that do not change the secondary structure, such as rearrangements in helix–helix packing interactions.
A mutant destabilizing the loop–loop interaction
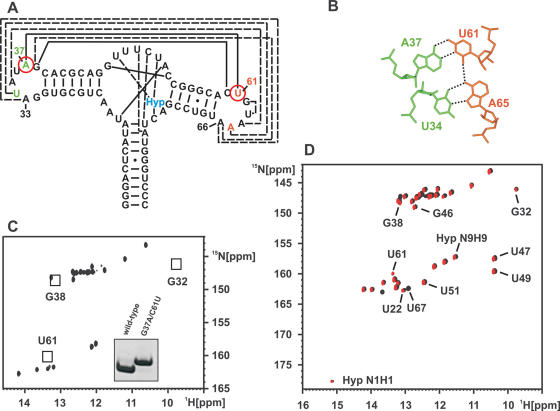
To further probe the role of the loop–loop interaction between loop II and III we introduced a mutation in one of the two base quadruples that hold the loops together. Specifically, we altered the G37:C61 Watson–Crick base pair that forms a base quadruple with the U34:A65 reversed Hoogsteen base pair into an A37:U61 Watson–Crick base pair (Figure 5A). Formally, this mutation eliminates two hydrogen bonds from the base quadruple, one in the 37:61 base pair and one between positions 37 and 34 (Figure 5B). The 1H,15N-HSQC-spectrum of this RNA in its free form is shown in Figure 5C. Comparison with the spectrum of the free wild-type RNA reveals that the signal of the G37 imino group is absent as expected due to its mutation to an adenine. Furthermore, the signals for the imino groups of G32 and G38 are absent in the spectrum of the mutant RNA both in the absence (data not shown) and in the presence of Mg2+ (Figure 5C). Therefore, the mutation disrupts the loop–loop interaction in the free RNA.
Figure 5.
Ligand binding and conformational changes of the G37A/C61U-mutant. (A) Location of the G37A/C61U-mutations in the secondary structure of the aptamer domain. It replaces a long-range Watson–Crick G:C base pair with a weaker A:U Watson–Crick base pair. (B) Consequences of the double mutation on hydrogen bonding of the base quadruple formed between loop II and III. The mutation results in the formal loss of two hydrogen bonds in the base quadruple. (C) Imino group region of an 1H,15N-HSQC-spectrum of the free G37A/C61U-RNA in the presence of Mg2+. The imino group signals for nucleotides G32 and G38 are missing in the mutant due to the absence of a loop–loop interaction. They are observable only in the free wild-type RNA due to the presence of the loop–loop interaction there. In addition, no signal is observed for U61 in the free mutant RNA. The introduction of the G37A-mutation also eliminates the signal for G37 imino group. Inset: Native gel electrophoresis comparing the wild-type RNA (left) and the G37A/C61U-mutant (right). The lower electrophoretic mobility of the mutant indicates that the mutant is less compact compared to the wild-type in agreement with the absence of a loop–loop interaction. (D) Overlay of the imino group regions of 1H,15N-HSQC-spectra for the hypoxanthine–RNA complexes at 10°C for the G37A/C61U-mutant RNA (red) and the wild-type RNA (black) in the presence of 5 mM Mg2+. In both cases, signals for the bound hypoxanthine are observable (labeled Hyp N1H1 and Hyp N9H9) as well as signals for nucleotides in the ligand binding core. This demonstrates that both RNAs are capable of ligand binding under these conditions. In addition, G32 and G38 are now observable in both RNA's indicating that upon ligand binding the loop–loop interaction forms also in the G37A/C61U-mutant. In addition, an imino group signal is observable for U61 in the mutant due to the formation of the A37:U61 Watson–Crick base pair instead of the G37:C61 Watson–Crick base pair in the wild-type RNA.
The absence of the loop–loop interaction in the G37A/C61U-mutant RNA was further corroborated by native gel electrophoresis. Compared to the wild-type RNA the mutant migrates slower in the gel (Figure 5C, Inset) indicating a less compact conformation.
Addition of hypoxanthine to the free RNA in the absence of Mg2+ does not lead to notable changes in the spectrum of the mutant RNA. No signals for bound hypoxanthine are observable. Thus, the mutant RNA does not bind hypoxanthine in the absence of Mg2+ (data not shown). However, in the presence of 5 mM Mg2+ the addition of equimolar amounts of hypoxanthine to the mutant RNA results in a 1H,15N-HSQC-spectrum that is very similar to that of the complex of hypoxanthine with the wild-type RNA (Figure 5D). As observed for the hypoxanthine complex with the wild-type RNA the complex with the mutant RNA is in slow exchange on the NMR time scale indicating a similar stability. Specifically, we do not only observe two signals for the N1H1 and N9H9 imino groups of the bound hypoxanthine but also the signals typical for the nucleotides in the ligand binding core of the RNA as well as signals for G32 and G38 with chemical shifts very close to those found in the wild-type RNA. In addition, we find a new uridine imino signal that corresponds to the U61 in the A37:U61 Watson–Crick base pair introduced by the mutation. The presence of this A37:U61 base pair was further confirmed in a HNN-COSY experiment and the assignments were established using 2D-1H,1H-NOESY- and 3D-1H,1H,15N-NOESY-HSQC-experiments (data not shown). This indicates that ligand binding in the presence of Mg2+ in the G37A/C61U-mutant RNA not only induces the folding of the ligand binding core but in addition the formation of the loop–loop interaction stabilized by the two base quadruples. Chemical shift comparisons and a HNN-COSY experiment with the hypoxanthine bound mutant confirmed the similarity in ligand binding modes of the mutant and the wild-type RNA. Furthermore, titration with 5 μM Mn2+ in the presence of 5 mM Mg2+ established the presence of binding sites for divalent cations in the mutant at the same locations as in the wild-type RNA. The only notable difference is the absence of an imino group signal for U34 involved in the U34:A65 reversed Hoogsteen base pair. Apparently, the missing hydrogen bond between A37 and U34 in the mutated base quadruple destabilizes this base pair resulting in fast exchange of the U34 imino group with the solvent.
The loop–loop interaction is stable outside the riboswitch context
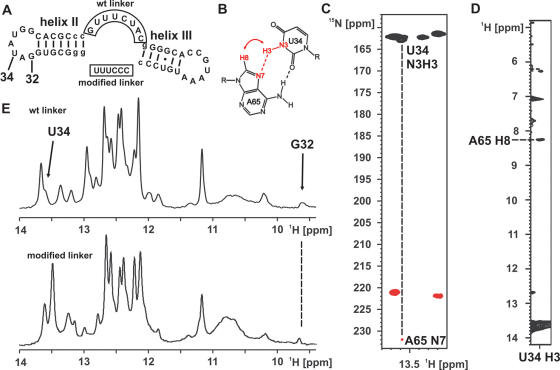
Since the loop–loop interaction between loops II and III did not require the presence of the ligand or the folding of the core region we wanted to know whether this interaction is stable outside the context of the riboswitch sequence. We therefore synthesized two RNA constructs that contain the helices II and III with altered sequences, the loops II and III in the wild-type sequence and a linker of different sequence and length (Figure 6A). The imino proton signals of G32 and U34 as well as the presence of a reversed Hoogsteen A:U base pair were used as reporters for the presence of the loop–loop interaction. The expected NOE-pattern for such a base pair and the expected HNN-COSY correlation is shown schematically in Figure 6B. Therefore, the construct containing the wild-type linker sequence was synthesized both in unlabeled and 15N-G,U,C, 13C,15N-A-labeled form. An imino proton spectrum of an RNA corresponding to helix II alone did not show any evidence for a structure in the isolated loop II and only imino proton resonances and NOEs compatible with Watson–Crick base pairs (data not shown). Figure 6C shows the result of a HNN-COSY experiment for the construct with the wild-type linker. The correlation between the U34 NH and an adenine N7 corresponding to the expected hydrogen bond in a reversed Hoogsteen A:U base pair is clearly visible. In addition, we observed the expected NOE-cross peak between the U34 NH and the H8 of the adenine (Figure 6D). The identity of the H8 proton was further confirmed in a 1H,13C-HSQC with the isotopically labeled construct with the wild-type linker where the adenine H2 and H8 resonances can be distinguished by the chemical shift of their attached carbon (data not shown). Furthermore, 1D-proton spectra of the imino proton region revealed a signal in both constructs with a chemical shift very similar to G32 (Figure 6E). The identity of this imino group as belonging to a G was established for the construct with the wild-type linker in a 1H,15N-HSQC (data not shown). Thus, even in the absence of helix I and altered linker sequences the two loops interact and the presence of the G32 imino resonance as well as the reversed Hoogsteen A:U base pair in both constructs indicate a very similar conformation compared to the loop–loop interaction in the full-length aptamer domain.
Figure 6.
A loop–loop interaction outside the riboswitch context. (A) RNA-constructs containing only helices II and III with the wild-type and a modified (boxed) linker sequence. Nucleotides that correspond to sequences different from the natural riboswitch are given in lower-case letters. The numbering is analogous to the aptamer domain. (B) Schematic drawing of a reversed Hoogsteen A:U base pair with the uridine N3H3 adenine N7 hydrogen bond (dashed red line) and the expected NOE contact between uridine H3 and adenine H8 (red curved arrow) indicated. (C) Detail of the HNN-COSY spectrum for the RNA at 10°C at the position of the U34 imino group showing the correlation expected due to the hydrogen bond to the adenine N7-nitrogen. (D) Detail of a 2D-Watergate-1H,1H-NOESY-spectrum at 10°C at the position of the U34 imino group showing the NOE-cross peak to the adenine H8 in agreement with the formation of a reversed Hoogsteen A:U base pair. (E) 1D-1H-imino proton spectra of the RNAs shown in (A) at 10°C reveal the presence of a signal for the G32 imino proton in both RNAs.
DISCUSSION
Many RNA molecules bind their ligands through an ‘induced fit’-mechanism. The ligand binding pocket in the free state of the RNA is often unstructured and dynamically disordered. Such a binding mechanism carries a large entropic penalty that contributes unfavorably to the free energy of binding but has the advantage that the ligand can be enveloped by the RNA thereby utilizing as many functional groups in the ligand for intermolecular interactions as possible. Other RNAs avoid the entropic costs to the free energy of binding by presenting a preformed interaction interface to the ligand (18,25–27). The ligand binding domains of naturally occurring riboswitches often contain conserved sequence elements that are not directly involved in ligand recognition. X-ray structure analysis of RNA–ligand complexes of these riboswitches has revealed that these elements are involved in RNA–RNA-tertiary interactions that stabilize the global fold of these RNAs (8–13). In some cases it has been shown that the destruction of these structure elements abolishes ligand binding (8,28) and is necessary for the function of the riboswitches. Similarly, recent studies on the naturally occurring forms of both the hammerhead (29,30) and the hairpin ribozyme (31) have revealed that these ribozymes contain tertiary structure elements remote from the catalytic core that preorganize the global fold of the ribozyme, increase its catalytic efficiency and render it less dependent on the presence of divalent cations.
In the present study we used NMR-spectroscopy to investigate the binding of hypoxanthine to the aptamer domain of the guanine riboswitch and to characterize the conformational transitions of the RNA upon ligand binding in solution. We found that the ligand binds with a binding mode identical to that found in the X-ray structure of the complex in a magnesium-independent manner to an RNA with a ligand binding pocket that lacks stabilizing hydrogen bonding interactions and is likely to be disordered and unstructured but with a preorganized tertiary interaction between loop II and loop III in its free state. The loop–loop interaction is mediated by two base quadruples that adopt the same conformation and base pairing pattern in the free and the bound state of the RNA. A Mg2+-binding site that is located in close proximity to these base quadruples is involved in additional stabilization of the two base quadruples but nonessential for its formation.
The presence of the loop–loop interaction with its two base quadruples fully formed already in the absence of ligand explains the absence of hydrolytic cleavage in this region observed in in-line probing experiments with the free RNA as well as the lack of change in protection patterns in this region upon ligand binding (5). The small chemical shift changes observed for residues in the three helices upon ligand binding suggest that the loop–loop interaction organizes the global fold of the RNA prior to ligand binding in such a way that only small changes in inter-helical packing are necessary to accommodate the ligand. We found evidence of increased dynamics in the region of the loop–loop interaction for the free RNA compared to the ligand bound state, such as increased line-widths of NMR-signals for loop nucleotides in the free RNA. This, however, does not indicate that the loop–loop interaction is only transiently formed. Instead the NMR-data in conjunction with the native gel electrophoresis results and the in-line probing data (5) suggest that the loop–loop interaction is stable in the free RNA and not in equilibrium with an open form. Increased line widths are also observed for NMR-signals of nucleotides in the core of the three helices suggesting increased dynamics in inter-helical packing in the free RNA. Changes in the relative orientations of helix II and III with respect to each other will lead to transient deformations in the base pair geometries of the base quadruples in the loop–loop interactions and therefore cause the observed line-broadening for the nucleotides involved in the loop–loop interactions.
No evidence for an ordered structure is found in the ligand binding core region of the RNA in analogy to what is observed in artificial aptamers. The combination of the preorganized global fold of the RNA with a dynamic and unstructured binding pocket might contribute to lower the entropic cost for ligand binding while still allowing to completely envelope the ligand and thereby maximizing intermolecular interactions.
However, a preorganized global fold is not an absolute requirement for ligand binding as exemplified by the behavior of the G37A/C61U-mutant. This RNA is still capable of high-affinity ligand binding despite the absence of the loop–loop interaction in the free form of the RNA. The ligand binds to this RNA in an induced fit mechanism that includes both folding of the core region as well as the formation of the base quadruples in the loop–loop contact. Here it seems plausible that the favorable enthalpic contributions of base pair formation at least partially compensate for the higher entropic costs. Furthermore, in contrast to the wild-type RNA Mg2+-binding is now required to drive the reaction towards complex formation. However, the complete elimination of loop–loop interactions as described by Batey et al. (8) abolishes ligand binding.
It will be interesting to compare the thermodynamics and the kinetics of ligand binding to the wild-type and the mutant RNA in detail to gain further insight into the role of preformed tertiary interaction elements for ligand binding. In addition, it will be revealing to investigate other riboswitch systems, such as the thiamine riboswitch (10,11), the SAM-riboswitches (13,28,32), the glmS-ribozyme (27,33–35) and the lysine-riboswitch (36) where extensive tertiary interaction elements not directly involved in ligand binding have been revealed by X-ray structure analysis of the RNA–ligand complexes or proposed based on biochemical data with regard to the presence and stability of such stabilizing elements prior to ligand binding. In the light of our results described above it is conceivable that a wide range of behavior can be expected ranging from totally preorganized 3D folds to simultaneous induced folding of ligand binding and tertiary structure elements. This is illustrated by recent single-molecule FRET-experiments (37) on the aptamer domain of the adenine dependent riboswitch, which is highly similar to the guanine riboswitch studied here but for instance contains a loop II extended by a non-canonical U:U base pair. In the adenine riboswitch the loop–loop interaction can form in the absence of the ligand but depends on the presence of Mg2+-ions and is only transiently stable. In this regard the aptamer domain of the adenine riboswitch represents an intermediate compared to the wild-type guanine riboswitch where the loop–loop interaction is stable and Mg2+-independent and its G37A/C61U-mutant where the ligand and Mg2+-ions must bind cooperatively to induce the formation of the loop–loop interaction.
Finally, the stability of the wild-type loop–loop interaction motif even outside the context of the riboswitch contexts leads us to belief that it constitutes an autonomous tertiary interaction motif similar to the tetraloop–tetraloop receptor interaction (38) or the A-minor motif (39). It might be present in other RNA folds and is probably a useful building block in designing 3D RNA-objects in ‘RNA-tectonics’ (17).
Acknowledgments
The authors thank C. Richter for continuous support with the NMR-spectrometers. The authors are grateful to M. Görlach, E. Duchardt and K. Abarca Heidemann for critical reading of the manuscript. The authors thank E. Stirnal for excellent technical support. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through the SFB 579 ‘RNA–ligand interactions’ (A9, A10), the Center of Biomolecular Magnetic Resonance, Johann-Wolfgang-Goethe-University, Frankfurt, Germany and by start-up funding from the Department of Biochemistry, University of Texas Health Science Center, San Antonio (USA). J.N. was supported by a fellowship of the Fonds der Chemischen Industrie. Funding to pay the Open Access publication charges for this article was provided by the Department of Biochemistry, University of Texas Health Science Center, San Antonio.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 2.Nonin S., Jiang F., Patel D.J. Imino proton exchange and base-pair kinetics in the AMP–RNA aptamer complex. J. Mol. Biol. 1997;268:359–374. doi: 10.1006/jmbi.1997.0986. [DOI] [PubMed] [Google Scholar]
- 3.Jucker F.M., Phillips R.M., McCallum S.A., Pardi A. Role of a heterogeneous free state in the formation of a specific RNA–theophylline complex. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 4.Patel D.J., Suri A.K., Jiang F., Jiang L., Fan P., Kumar R.A., Nonin S. Structure, recognition and adaptive binding in RNA aptamer complexes. J. Mol. Biol. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 5.Mandal M., Boese B., Barrick J.E., Winkler W.C., Breaker R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 6.Sudarsan N., Barrick J.E., Breaker R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubodera T., Watanabe M., Yoshiuchi K., Yamashita N., Nishimura A., Nakai S., Gomi K., Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 8.Batey R.T., Gilbert S.D., Montange R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 9.Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thore S., Leibundgut M., Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A., Polonskaia A., Phan A.T., Breaker R.R., Patel D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards T.E., Ferre-D'Amare A.R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Montange R.K., Batey R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 14.Noeske J., Richter C., Grundl M.A., Nasiri H.R., Schwalbe H., Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Furtig B., Richter C., Wohnert J., Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 17.Chworos A., Severcan I., Koyfman A.Y., Weinkam P., Oroudjev E., Hansma H.G., Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 18.Stoldt M., Wohnert J., Ohlenschlager O., Gorlach M., Brown L.R. The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition. EMBO J. 1999;18:6508–6521. doi: 10.1093/emboj/18.22.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartels C., Xia T.-H., Billeter M., Güntert P., Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 20.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 21.Grzesiek S., Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 22.Sklenar V., Peterson R.D., Rejante M.R., Feigon J. Correlation of nucleotide base and sugar protons in a 15N-labeled HIV-1 RNA oligonucleotide by 1H-15N HSQC experiments. J. Biomol. NMR. 1994;4:117–122. doi: 10.1007/BF00178339. [DOI] [PubMed] [Google Scholar]
- 23.Wijmenga S.S., van Buuren B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. NMR Spectroscopy. 1998;32:287–387. [Google Scholar]
- 24.Dingley A.J., Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 25.Ohlenschlager O., Wohnert J., Bucci E., Seitz S., Hafner S., Ramachandran R., Zell R., Gorlach M. The structure of the stem–loop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Jansen J.A., McCarthy T.J., Soukup G.A., Soukup J.K. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nature Struct. Mol. Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- 27.Hampel K.J., Tinsley M.M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel B.A., Grundy F.J., Henkin T.M. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol. Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- 29.Khvorova A., Lescoute A., Westhof E., Jayasena S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 30.De la Pena M., Gago S., Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murchie A.I., Thomson J.B., Walter F., Lilley D.M. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol. Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- 32.Corbino K.A., Barrick J.E., Lim J., Welz R., Tucker B.J., Puskarz I., Mandal M., Rudnick N.D., Breaker R.R. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth A., Nahvi A., Lee M., Jona I., Breaker R.R. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson S.R., Been M.D. A pseudoknot in the 3′ non-core region of the glmS ribozyme enhances self-cleavage activity. RNA. 2005;11:1788–1794. doi: 10.1261/rna.2203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soukup G.A. Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure. Nucleic Acids Res. 2006;34:968–975. doi: 10.1093/nar/gkj497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grundy F.J., Lehman S.C., Henkin T.M. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl Acad. Sci. USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeMay J.F., Penedo J.C., Tremblay R., Lilley D.M., Lafontaine D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 39.Nissen P., Ippolito J.A., Ban N., Moore P.B., Steitz T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]