Abstract
Rpb5, a subunit shared by the three yeast RNA polymerases, combines a eukaryotic N-terminal module with a globular C-end conserved in all non-bacterial enzymes. Conditional and lethal mutants of the moderately conserved eukaryotic module showed that its large N-terminal helix and a short motif at the end of the module are critical in vivo. Lethal or conditional mutants of the C-terminal globe altered the binding of Rpb5 to Rpb1-β25/26 (prolonging the Bridge helix) and Rpb1-α44/47 (ahead of the Switch 1 loop and binding Rpb5 in a two-hybrid assay). The large intervening segment of Rpb1 is held across the DNA Cleft by Rpb9, consistent with the synergy observed for rpb5 mutants and rpb9Δ or its RNA polymerase I rpa12Δ counterpart. Rpb1-β25/26, Rpb1-α44/45 and the Switch 1 loop were only found in Rpb5-containing polymerases, but the Bridge and Rpb1-α46/47 helix bundle were universally conserved. We conclude that the main function of the dual Rpb5–Rpb1 binding and the Rpb9–Rpb1 interaction is to hold the Bridge helix, the Rpb1-α44/47 helix bundle and the Switch 1 loop into a closely packed DNA-binding fold around the transcription bubble, in an organization shared by the two other nuclear RNA polymerases and by the archaeal and viral enzymes.
INTRODUCTION
Heteromultimeric DNA-dependent RNA polymerases ensure the transcription of all eukaryotic, archaeal, bacterial and chloroplastic genomes, of some mitochondria and of nucleo-cytoplasmic DNA viruses. The bacterial core enzyme, made of the β′, β, ω and α2 subunits, is closely related to the chloroplast-encoded RNA polymerase. The three nuclear RNA polymerases of eukaryotes have a more elaborated core structure of 12 subunits. Five of these subunits (Rpb1, Rpb2, Rpb6 and Rpb3/Rpb11 in RNA polymerase II) correspond to the β′βωα2 bacterial core, six other (Rpb4, Rpb5, Rpb7, Rpb9, Rpb10 and Rpb12) are akin to archaeal polypeptides (1,2) and one, Rpb8, is typically eukaryotic (3). A fourth eukaryotic RNA polymerase, currently defined by its Rpb1-like and Rpb2-like subunits, exists in angiosperm plants (Arabidopsis thaliana and Oryza sativa) and is important for DNA silencing, but its precise subunit composition is not yet known (4).
The acquisition of six additional subunits by the common ancestors of archaea and eukaryotes (or their loss in the bacterial lineage) has been a major twist in the evolutionary history of transcription. However, the biological role(s) of these subunits is not well understood. Their null mutants are lethal in Saccharomyces cerevisiae or, in the case of Rpb4 and Rpb9 (RNA polymerase II) and their RNA polymerase I paralogues Rpa14, Rpa12, are lethal in double-mutant combinations (5,6). Rpb10 and Rpb12 consolidate the enzyme architecture by interacting with the α-like Rpb3 and Rpb11 and their Rpc40 and Rpc19 counterparts in RNA polymerase I and III (7,8). Rpb9 (akin to Rpa12 and Rpc11 in RNA polymerase I and III) contribute to the intrinsic transcript RNase activity of RNA polymerase II and probably control the rate of elongation and/or termination (9–11). Rpb4 and Rpb7 (Rpc17/Rpc25 and Rpa14/Rpa43 in RNA polymerase I and III) form a ‘stalk’ protruding from the RNA polymerase core enzyme (12–14). They are needed for transcript initiation (12,15,16) but may have other functions (17).
The present study deals with the Rpb5 subunit. This polypeptide has a bipartite organization combining a typically eukaryotic N-terminal domain (positions 1–142 in S.cerevisiae) with a C-terminal globe strongly conserved in all non-bacterial enzymes (2). The N-terminal module marks the far end of the DNA channel of the RNA polymerase II crystal (18) and probably accounts for the Rpb5/DNA contacts found some 15–20 nt ahead of the transcription fork in RNA polymerase III (19) and II (20). Its C-terminal part, on the other hand, is tightly connected to the largest subunit Rpb1 (18). Consistent with its location in the periphery of RNA polymerase II, Rpb5 is largely exposed to interactions with general transcription factors or perhaps with specific gene regulators, and previous studies on the human subunit have indeed suggested that it binds specific partners of RNA polymerase II, such as TFIIF, Taf15 (TAFII68) and protein X of the Hepatitis B virus (21–23). However, these interactions do not account for the fact that Rpb5 is shared by all three yeast RNA polymerases (24,25) and is closely related to the archaeal subunit H (2) and to viral Rpb5-like subunits (26,27), which evidently calls for a more general role in transcription.
MATERIALS AND METHODS
Genetic constructions
Yeast strains and plasmids are listed Table 1. Strains YMLF2, YFN25, YFN27, YFN50 and YFN51, depleted of individual subunits of RNA polymerase I, II or III, were constructed by expressing the relevant wild-type gene under the control of a doxycyclin-repressible promoter (28). pAS2Δ plasmids encoding mutant forms of Rpb5 fused to the Gal4BD domain were introduced in strain Y190 to be tested in a two-hybrid assay (29). pACT2 plasmids encoding Rpa190, Rpb1 and Rpc160 fragments fused to the Gal4AD activation domain were isolated from a two-hybrid genomic library (30).
Table 1.
Plasmids and yeast strains
| Name | Yeast markers and promoter | Origin |
|---|---|---|
| Plasmids | ||
| pGEN-RPB5 | ORI (2 μm) TRP1 pPGK1::RPB5 | Cloning of RPB5 between BamHI and KpnI |
| PGEN-RPB5* | ORI (2 μm) TRP1 PPGK1::RPB5 | Two silent mutations create internal cloning sites |
| PGEN-Hs5 | ORI (2 μm) TRP1 pPGK1:: hRPB5 | Human Rpb5 (41) |
| PGEN-Hs5* | ORI (2 μm) TRP1 pPGK1:: hRPB5 | Three silent mutations create internal cloning sites |
| pGEN-rpb5X | ORI (2 μm) TRP1 pPGK1:: rpb5-x | PCR-mediated mutagenesis of pGEN-RPB5 |
| PGEN-ChiX | ORI (2 μm) TRP1 pPGK1::RPB5-chiX | BamHI/KpnI cloning of yeast/human rpb5 chimeras |
| pCM185-RPB10 | CEN TRP1 pTETO7::RPB10 | BamHI/KpnIcloning of RPB10 in pCM185 |
| pCM185-RPA43 | CEN TRP1 pTETO7::RPA43 | StuI/PstI cloning of RPA43 in pCM185 |
| pCM185-RPB11 | CEN TRP1 pTETO7 ::RPB11 | BamHI/PstI cloning of RPB11 in pCM185 |
| pCM185-RPC17 | CEN TRP1 pTETO7::RPC17 | (34) |
| PCM190-RP5Sp | CEN URA3 pTETO7::SpRPB5 | BamHI cloning of the S.pombe RPB5in pCM190 |
| pAS2Δ-Hs5 | ORI (2 μm) TRP1 pADH1::GAL4(1–147): :hRPB5 | XmaI/KpnI cloning of hRPB5 in pAS2Δ |
| PAS2Δ-rpb5x | ORI (2 μm) TRP1 pADH 1::GAL4(1–147): :rpb5 | PCR-mediated mutagenesis of pAS2Δ-RPB5 |
| pACT2-RPA190(1615) | ORI (2 μm) LEU2 pADH1::GAL4(768–881)::RPA190 | pACT2 fusion to a RPA190 fragment (1445–1590 amino acids) |
| pACT2-RPB1(1545) | ORI (2 μm) LEU2 pADH1::GAL4(768–881): :RPB1 | pACT2 fusion to a RPB1 fragment (1169–1406 amino acids) |
| pACT2-RPC160(1594) | ORI (2 μm) LEU2 pADH1::GAL4(768–881)::RPC160 | pACT2 fusion to a RPC160 fragment (1269–1381 amino acids) |
| pSIRT | rDNA 35S (1–685) rDNA 5S | (35) |
| pRS316 (SUP4) | CEN URA3 SUP4ochre | (36) |
| Yeast strains | ||
| Name | Genotype | Origin |
| D471-13B | MATα lys2 ura3-52 rpb1-E1351K ura3-52 trp1-Δ63 leu2-Δ1 rpb5-Δ::ura3::LEU2 // pFL44-RPB5 (URA3 2μ) | YFN2 × GHY530 |
| D481-1B | MATa lys2-801 ade2-1 leu2 his3 ura3 trp1 rpa12-Δ::HIS3 rpb5-Δ::ura3::LEU2 // pFL44-RPB5 (URA3 2μ) | D473-9A × SL35-2C |
| D487-17C | MATα ade2-1 ura3 trp1-Δ63 his3 leu2 dst1-Δ::HIS3 rpb5- Δ::ura3::LEU2 // pFL44-RPB5 (URA3 2μ) | YFN2 |
| GHY530 | MATα his4-912δ lys2-128δ leu2-Δ1 ura3-52 rpb1-E1351K | (46) |
| YCZ44-5B | MATα ade2-1 ura3-52 trp1-Δ63 leu2 lys2 rpb9::KANMX4 rpb5-Δ::ura3::LEU2 // pFL44-RPB5 (URA3 2μ) | YFN2 × Y04437 |
| YMLF2 | MATα ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 RPC17::HIS3 /pCM185::RPC17 | (34) |
| YFN01 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pCM185-RPB5 | This work |
| YFN2 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pFL44-RPB5 | (31) |
| YFN68 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /PGen-RPB5 | This work |
| YFN5 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-RPB5(P151T) | This work |
| YFN6 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-RPB5(H147R) | This work |
| YFN9 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-chi7CAK | This work |
| YFN10 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-chi7K | This work |
| YFN11 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5::URA3::LEU2 /pGEN-chi7CA | This work |
| YFN13 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pCM185-RPB5 | (31) |
| YFN14 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5-Δ::ura3::LEU2 //pCM190-RPB5Sp | This work |
| YFN16 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pCM185-RPB5(H147R) | (31) |
| YFN25 | MATa rpa43Δ::LEU2 ade2-1 ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1 /pCM185-RPA43 | This work |
| YFN27 | MATa ura3-52 his3-Δ200 leu2-3 lys2 ade2-101 trp1-Δ63 rpb11-Δ1::HIS3 /pCM185-RPB11 | This work |
| YFN46 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-RPB5 (P86T, P118T) | This work |
| YFN49 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-RPB5(R200E) | This work |
| YFN50 | MATa ade2-1 ade3 his3 trp1 leu2 ura3 lys2 rpb9-Δ1::HIS3 // pCM185::RPB10 | This work |
| YFN51 | MATa ade2-1 his3-Δ200 trp1-Δ63 leu2 ura3-52 lys2 rpb9- Δ1::HIS3 /pCM185::RPB9 | This work |
| YFN60 | MATa ade2-1 lys2-801 ura3-52 trp1-Δ63 his3-Δ 200 leu2-Δ1 rpb5-Δ::ura3::LEU2 /pGEN-RPB5-Δ14 | This work |
| Y04437 | MATα his3-Δ1 leu2Δ0 met15-Δ0 ura3-Δ0 rpb9Δ::KanMX4 | Euroscarf* |
rpb5 mutants were constructed in plasmid pGEN-RPB5 and tested by plasmid shuffling (31). Amino acid replacements were based on mutant primers amplified with the Pfu DNA polymerase. N-terminal deletions (rpb5-Δ10 to rpb5-Δ38) were made by cloning the appropriate PCR fragment between the BamHI and KpnI sites of pGEN. They retained the first two amino acids of Rpb5 (Met Asp) and eliminated the following amino acids until position 38 inclusively. rpb5-chi alleles resulted from domain swapping with the human subunit. To this end, we introduced two silent mutations in pGEN-RPB5, replacing AGA (position R31) by CGG and GAATTG (E127 and L128) by GAGCTC to generate NgoMI and SacI cloning sites (pGEN-RPB5*). The equivalent mutations were introduced in the human sequence of pGEN-Hs5, together with a third silent mutation replacing (CCAAGT to CCCTCC) at positions P112, S113 and creating a DraIII site naturally present in the yeast RPB5 (pGEN-Hs5*).
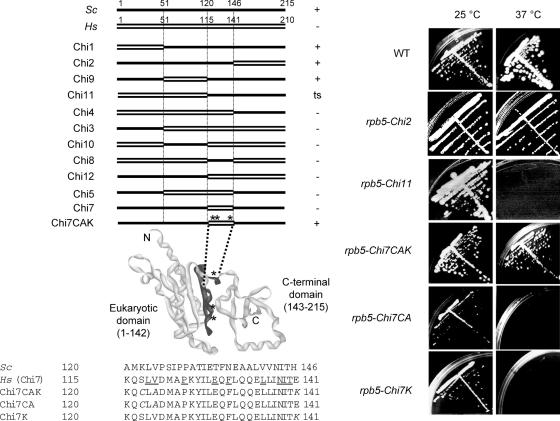
rpb5-chi7 is a lethal mutant where positions 120–146 are replaced by the equivalent human amino acids. The corresponding DNA segment (244–568 nt in the yeast coding sequence) was amplified with Taq DNA polymerase in the presence of 0.1 mM MnCl2 and introduced in strain YFN14 by co-transformation with a linear fragment of pGEN-Chi7 (lacking the SacI/SalI fragment between 373–445 nt), to reconstitute mutant forms of pGEN-Chi7 by recombination in vivo. The host strain used in this experiment, YFN14, bears the RPB5 gene of Schizosaccharomyces pombe (plasmid pCM190-RPB5Sp) that cannot recombine with the rpb5-chi7 fragment, due to a lack of homology (32). One viable clone (rpb5-chi7CAK) was obtained from about 40 000 YFN14 transformants isolated on tryptophan-less medium and cured of the host plasmid by passing them on FOA medium. It resulted from the triple amino acid replacement K115C, S117A, E141K (numbers refer to the human subunit sequence). Further sub-cloning on pGEN resulted in the viable but temperature-sensitive allele rpb5-chi7CA (bearing the K115C and S117A replacements) and rpb5-chi7K, that only differs from the initial lethal allele rpb5-chi7 by the E141K replacement.
RNA analysis
mRNAs were quantified by RT–PCR in conditions generating a linear signal response over a range of 0.1–10 ng RNA. RNAs were extracted from 5 ml cultures using 200 μl of glass beads. A total of 1 μg of RNA was reverse-transcribed for 1 h at 42°C with 100 pmols of appropriate oligonucleotide primers (listed in Supplementary Data S1). The reaction was halted by adding 180 μl water to the 20 μl reaction volume. A total of 10 μl samples were amplified by PCR (15 cycles) in the presence of 25 μCi of [α-32P]dCTP and with 10 pmols of oligonucleotide primers. A total of 5 μl of the reaction mixture were loaded on a 6% RNA polyacrylamide/8 M urea gel and analysed with a Molecular Dynamics PhosphorImager.
Northern blot hybridization was done on 10 μg of RNAs extracted from exponentially growing cells shifted from 25 to 37°C for 7 h in YPD. Cells were harvested at an A600 of 0.4–0.6. The rich (YPD), synthetic complete (SC) and omission media are standard in yeast studies. GAL1 mRNA was measured in cells grown in SC medium with 4% galactose (induction) or 2% raffinose (repression) as sole carbon source, with doxycycline added at doses leading to a 3-fold reduction in growth rate. INO1 mRNA was measured in cells grown in inositol-less medium (33) and in the presence of 0.4 mM inositol.
Protein purification and transcription assays
Transcription was tested on cell-free extracts or on purified RNA polymerase I and III of strains YFN13 (wild-type), YFN6 (rpb5-H147R) and YFN10 (rpb5-chi7K). 0.5 l cultures were harvested during late exponential growth (about 1 g of cell dry weight), re-suspended in 1.5 ml of extraction buffer [0.2 M Tris–Acetate (pH 8.0), 10 mM Mg-Acetate, 10 mM 2-mercaptoethanol, 10% glycerol and 1 mM phenylmethlysulfonyl fluoride (PMSF)] and crushed in an Eaton Press. pSIRT (bearing one copy of the 5S rRNA gene and the 5′ part of the 35S rRNA gene) and pRS316-SUP4 (tRNATyr) were used as RNA polymerase I and III DNA templates, respectively (34,35).
RNA polymerase I was purified to near homogeneity from ten grams of wild-type (YFN13), rpb5-H147R (YFN16) and rpb5-chi7K (YFN10) cells harvested in mid-exponential phase. Cells were washed with extraction buffer [0.2 M Tris–HCl (pH 8), 10 mM 2-mercaptoethanol, 10 mM MgCl2, 1 mM EDTA, 0.3 M ammonium sulphate, 1 mM PMSF and 10% glycerol], re-suspended in 10 ml buffer, frozen at −70°C, broken in an Eaton Press and purified by phosphocellulose and DEAE-cellulose chromatography, followed by a glycerol gradient. Transcription was assayed on poly(dA–dT) (36)
Homology search
Sequence alignments were based on saturating iterative homology search with the standard default Psi-Blast options [(37), see http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi]. In some cases, they were improved by visual inspection, based on the following amino acid conservation groups: AG, ST, CS, DN, DE, EQ, MILV, KR and FWY.
RESULTS
A short domain of the human Rpb5 accounts for its non-complementation in S.cerevisiae
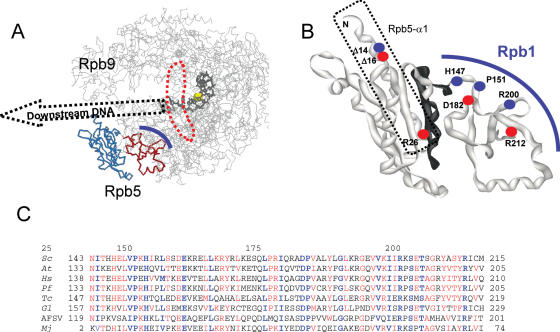
Figure 1 illustrates the bipartite organization of yeast Rpb5, alone or in the crystal structure of RNA polymerase II (18,38). The N-terminal module (positions 1–142 in S.cerevisiae) is only found in eukaryotes. In RNA polymerase II, it occupies the ‘lower’ end of the large DNA Cleft, downstream of the transcription bubble (13,14,18,39). The C-terminal globe, on the other hand, is strongly conserved in all archaea and eukaryotes (2), and in four of the five classes of large nucleo-cytoplasmic DNA viruses (26,27). Poxviral RNA polymerases, on the other hand, have an Rpo22 subunit (40) similar in size to Rpb5, but iterative sequence alignments revealed no significant homology between these two polypeptides.
Figure 1.
Spatial organization and sequence conservation of Rpb5. (A) Spatial organization of yeast RNA polymerase II. The ‘upper’ view using PDB co-ordinates 1I6H from (45) was drawn with Rasmol (http://www.umass.edu/microbio/rasmol/). The RNA polymerase II backbone (without Rpb4/Rpb7) is in grey. The eukaryotic part of Rpb5 (positions 1–142) is shown in blue, and the C-terminal part (positions 143–215) in red. A thick blue line symbolizes the dual binding of Rpb5 to Rpb1. DNA is indicated by a black wire-frame (template strand only) and its hypothetical trajectory downstream of position +4 (where +1 marks the beginning of the DNA–RNA hybrid) is suggested by a dotted arrow. The catalytic Mg(A) atom is shown as a yellow sphere. A doted red line surrounds the invariant Bridge helix. (B) Close-up of the Rpb5 (ribbon structure). Dots indicate the distribution of lethal (red) and conditional (blue) mutants. Positions 121–146, (see Figure 2) are shown in black. A dotted line surrounds the hydrophilic helix Rpb5-α1. (C) Sequence conservation of the C-terminal globe. The amino acid conservation groups considered were: AG, ST, CS, DN, DE, EQ, MILV, KR, FWY, when present in at least half of the compared sequences (red letters). Highly conserved positions present in all amino acid sequences are in blue. Species symbols: S.cerevisiae (Sc), Homo sapiens (Hs), Arabidopsis thaliana (At), Plasmodium falciparum (Pf), Trypanosoma cruzi (Tc) and Giardia lamblia (Gl). These Unikonts (Fungi and Animals, represented here by S.cerevisiae and H.sapiens), Plants (A.thaliana), Alveolates (P.falciparum) or Excavates (T.cruzi) correspond to four of the five main eukaryotic phyla recognized by recent phylogenies (58). One viral (African Swine Fever Virus = AFSV) and one archaeal (Mj = M.jannaschii) subunit are shown for comparison.
The fission yeast Rpb5 is functional in S.cerevisiae, but the human subunit is not (32,41). It has been suggested that the first 139 amino acids of the yeast subunit, practically coinciding with the eukaryotic module, are needed to produce an active yeast-human chimera and that replacing this domain by its human counterpart prevents the activation of some essential yeast gene(s) (42). Given the regulatory role ascribed to Rpb5 by some authors (22,23,42), it was of some interest to know if the non-complementation of the human subunit was due to a specific domain of that subunit. Domain swapping experiments (Figure 2A and B) showed that the first 119 amino acids of the eukaryotic module could be replaced by their human counterpart to form the temperature-sensitive allele rpb5-chi11, and that the yeast and human C-terminal globes were fully interchangeable in vivo (rpb5-chi2). However, lethality was observed for any construction (rpb5-chi3, 4, 5, 7, 8, 10, 12) containing a short domain bearing the human amino acids 115–141 (positions 121–146 in yeast). Moreover, the random mutagenesis of rpb5-chi7 (a lethal allele where yeast positions 121–146 are replaced by the corresponding 115–141 human sequence block), generated the vigorous intragenic suppressor rpb5-chi7CAK, (Figure 2B and C), due to the triple amino acid replacement S117C, V119A, E141K (numbers refer to the human polypeptide).
Figure 2.
In vivo complementation by the human subunit (A) Hybrid constructions based on domain swapping. Black and white boxes correspond to the yeast and human Rpb5, respectively. Symbols on the right denote the mutant growth pattern on complete YPD medium. +: wild- type like growth at 25, 30 and 37°C; ts: no growth at 37°C; −: no growth at all three temperatures. (B) Three viable mutants derived from random mutagenesis of the human domain in the lethal rpb5-chi7 allele (see Materials and Methods for a description of the mutagenesis procedure). A local sequence alignment is shown for the corresponding S.cerevisiae and H.sapiens domain. Amino acid identities (relatively to the S.cerevisiae sequence) are underlined. The amino acids mutated in rpb5-chi7CAK are italicised. The panel above provides a spatial view of Rpb5, with the mutagenized region in black. (C) Growth of the viable rpb5-chi2, rpb5-chi11, rpb5-chi7CAK, rpb5-chi7CA and rpb5-chi7K mutants at 25 and 37°C. Cells were streaked on YPD and examined after five days of incubation.
Positions 120 and 146 mark the end of the eukaryotic module and extend to the first four amino acids of the C-terminal globe shared with archaeal and viral polymerases. Appropriate sub-cloning yielded the rpb5-chi7K and rpb5-chi7CA mutants, viable but with a strong temperature-sensitive defect (Figure 2B), showing that both parts of the yeast central domain are needed to produce a fully functional subunit. Most of the human subunit, on the other hand, is largely (N-part) or fully interchangeable (C-part) from yeast to man. These complementation data extend previous observations showing that the small subunits of RNA polymerases I, II and III are, as a rule, functionally conserved in vivo (7,16,32,41,42,43). However, they are not easily reconciled with a regulatory role of the eukaryotic module of Rpb5 (42).
The N-terminal helix of the eukaryotic domain is critical in vivo
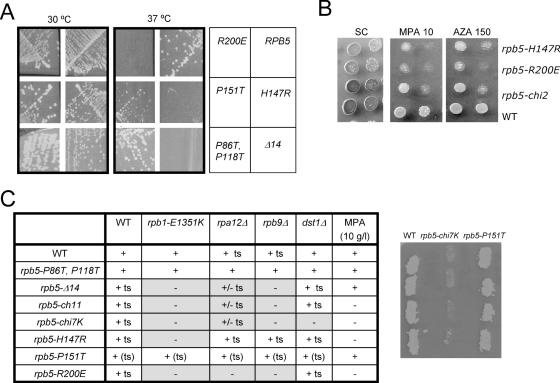
Fifty-five rpb5 mutants (Supplementary Data S2) were generated by domain swapping (see above), N-terminal deletions and site-directed mutagenesis, mostly at highly conserved positions. Rpb5 was fairly resilient to amino acid replacements, even at invariant positions, and most of these mutants had no detectable growth defect. Five lethal (rpb5-R26E, rpb5-D182N, rpb5-R212E, rpb5-Δ16 and rpb5-Δ38) and five fully (rpb5-Δ14, rpb5-H147 and rpb5-R200E) or partly (rpb5-H147Q and rpb5-P151T) temperature-sensitive alleles were nevertheless obtained (Figure 3), adding to the temperature-sensitive rpb5-chi11, rpb5-chi7CA and rpb5-chi7K mutants described above and to a previously described cold-sensitive allele rpb5-V111G (42). Most of these conditional mutants were sensitive to mycophenolate, a drug depleting the cellular pool in NTP (44), with only minor effect of 6-azauracil (Figure 3B). Moreover, rpb5-Δ14, rpb5-chi11, rpb5-chi7K, rpb5-H147R and rpb5-R200E were lethal or had strong synthetic defects with rpb9Δ, lacking the non-essential RNA polymerase II subunit Rpb9 and with the corresponding rpa12Δ mutant in RNA polymerase I. As discussed in more detail below, synthetic lethality also occurred with rpb1-E1351K, at a highly conserved position of Rpb1-α46.
Figure 3.
Conditional mutants of RPB5. (A) Growth pattern of rpb5 mutants. Strains YFN13 (RPB5), YFN6 (rpb5-H147R), YFN5 (rpb5-P151T), YFN49 (rpb5-R200E), YFN60 (rpb5-Δ14) and YFN46 (rpb5-P86T,P118T) were streaked on YPD and tested for growth after three days of incubation at 30 and 37°C. Note the wild-type growth of the rpb5-P86T,P118T double mutant, despite the hypothetical DNA-interacting properties assigned to the corresponding Prolines rings (18,45). (B) Sensitivity to NTP-depleting inhibitors (6-azauracail and mycophenolic acid). Serial dilutions of strains YFN13 (RPB5), YFN6 (rpb5-H147R), YFN49 (rpb5-R200E) and YFN42 (rpb5-chi2) were dropped on SC medium with or without mycophenolate (MPA) or 6-azauracil at the concentration indicated and incubated for three days at 30°C. (C) Summary of the growth defect and synthetic lethality of rpb5 mutants. Strains YFN13 (rpb5Δ/2μ URA3 RPB5), D471-13B (rpb5Δ rpb1-E1351K/2μ URA3 RPB5), YCZ44-5B (rpb5Δ rpb9Δ/2μ URA3 RPB5), D481-1B (rpb5Δ rpa12Δ/2μ URA3 RPB5) and D487-17C (rpb5Δ dst1Δ/2μ URA3 RPB5) were transformed with plasmids of the pGEN-rpb5 and pGEN-Chi series bearing the rpb5-Δ14, rpb5-chi11, rpb5-chi7K, rpb5-H147R, rpb5-P151T or rpb5-R200E alleles, using pGEN-RPB5, pGEN-RPB9 and pGEN-DST1 as positive controls and the void pGEN plasmid as negative control. Four independent transformed clones were re-isolated on selective medium, grown on YPD, replica plated on FOA medium to chase the 2 μ URA3 RPB5 host plasmid and incubated for three days at 30°C. A lack of growth indicated that the transforming plasmid harboured a non-functional rpb5 allele, unable to complement rpb5Δ in the host strain considered. Results are summarized for all four tester strains. The panel on the right illustrates the lethality of rpb5-chi7K and the viability of rpb5-P151T in an rpb9Δ background, as measured after transformation in the YCZ44-5B (rpb5Δ rpb9Δ/2 μ URA3 RPB5) host strain.
The eukaryotic module of Rpb5 is moderately conserved, but two highly conserved sequence blocks are revealed by sequence comparison extending to the entire range of eukaryotic genomes currently available at NCBI (http://www.ncbi.nlm.nih.gov/), as shown in Supplementary Data S3. One conserved block is harboured by the last 12 amino acids of the module and is mutated in the conditional mutants rpb5-chi7CA and rpb5-chi7K described above. The other highly conserved module (positions 11–30) belongs to the long hydrophilic helix Rpb5-α1 and occupies the ‘lower’ far-end of the DNA Cleft (13,14,18), as illustrated in Figure 1A. N-terminal deletions removing the first half of Rpb5-α1 were temperature-sensitive (rpb5-Δ14) or lethal (rpb5-Δ16 and rpb5-Δ38) and an rpb5-R26E charge inversion at its invariant DRGY motif was also lethal (Figure 3 and data not shown). Prolines P86 and P118, shared by the fungal and animal subunits, are exposed to the DNA Cleft, and it has been suggested that they insert their cyclic rings near the deoxyribose moieties of the downstream DNA (39,45). However, they are not (P86) or poorly (P118) conserved in other eukaryotic phyla (Supplementary Data S3), and a double Pro-Thr replacement had no detectable growth defect (Figure 3), suggesting that these DNA contacts, if they occur at all, are of limited biological relevance.
A conserved dual binding of Rpb5 to Rpb1-like subunits of RNA polymerases
In the crystal structure of yeast RNA polymerase II (13,14,18), the C-terminal module of Rpb5 (positions 143–215) binds the largest subunit (Rpb1) at the Rpb1-β24/25 and Rpb1-α44/47-folds. These two binding sites are separated by a large segment of 438 amino acids, containing the ‘lower jaw’ and ‘foot’ modules of RNA polymerase II, with the highly conserved Trigger helix in between (18). As discussed below, the ‘lower jaw’ module also binds subunit Rpb9 (see below, Figure 7A). A weak binding of Rpb5 to Rpb6 is predicted, based on charge interactions involving Rpb5-R169 and Rpb6-D138. These structural data are buttressed by two-hybrid interaction tests and by the genetic characterization of rpb5 mutants detailed below, to the possible exception of the Rpb5–Rpb6 interaction, since an rpb5-R169D charge inversion expected to interfere with this binding was indistinguishable from wild-type (data not shown).
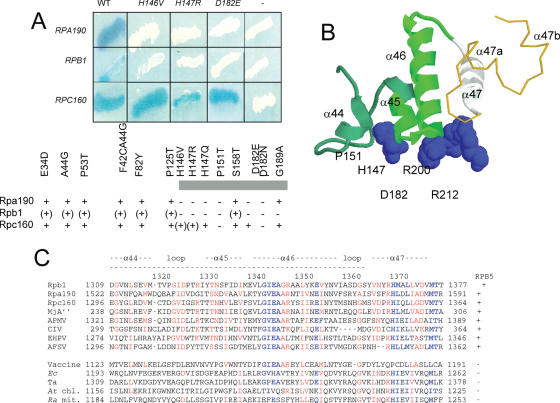
Using a random library of yeast genomic fragments (29,30), 17 distinct Gal4AD fusion clones (10 for Rpa190, 1 for Rpb1 and 6 for Rpc160) were isolated for their ability to specifically interact with a Gal4BD::Rpb5 bait. Amino acid replacements at the C-terminal globe impaired the two-hybrid interaction with Rpb1 and Rpa190, but only had minor effect on the Rpc160 interaction (Figure 4A). Polymerase-specific differences also occurred in two-hybrid interaction tests involving the Rpa190, Rpb1 and Rpc160 domains binding the common subunit Rpb8 (3). They suggest a particularly robust binding of Gal4BD::Rpb5 to its Gal4AD::Rpc160 partner, that may or may not reflect the strength of Rpb5-binding in RNA polymerase III itself. The Rpc160 and Rpa190 inserts were all sequenced and found to overlap between positions 1274–1381 (Rpc160) and 1522–1576 (Rpa190). This Rpa190 segment matched positions 1309–1362 on Rpb1 (1296–1349 on Rpc160), which corresponds to the Rpb1-α44/45/46 helices in the RNA polymerase II crystal (Figure 4B). The single Rpb1 insert was comprised between positions 1170 and 1406, and thus also included the Rpb1-α44/45/46 helices.
Figure 4.
Binding of Rpb5 to the Rpb1-α44/47-fold (A) Two-hybrid interactions between Rpb5 and Rpa190, Rpb1 and Rpc160. Wild-type and mutant forms of Rpb5 fused to Gal4BD in plasmid pAS2Δ were tested against plasmids pACT2-RPA190(1615), pACT2-RPB1(1545) and pACT2-RPC160(1594) listed in table 1 and containing the partner regions of Rpa190, Rpb1 and Rpc160 fused to the Gal4AD domain of pACT2. β-Galactosidase was tested in an overlay assay (29), as shown for the RPB5, rpb5-H146V, rpb5-H147R and rpb5-D182E constructs and as summarized below for the other mutants. +, (+) and − denote positive, reduced and negative responses in the β-galactosidase plate assay, respectively. (B) Spatial organization of H147, P151, D182, R200 and R212 relatively to the Rpb1α44/47-fold in the elongating RNA polymerase II. H147, P151, D182, R200 and R212 positions of Rpb5 are space-filled. The Rpb1-α44/46-fold (positions 1309–1362, corresponding to the minimal two-hybrid domain of Figure 4A is shown in green ribbons. The Rpb1-α47 helix (positions 1363–1377), not comprised in the minimal two-hybrid domain but binding Rpb5, notably through position R212) is shown in white ribbons. A gold thin line corresponding to Switch 1 backbone (positions 1378–1403). (C). Sequence conservation of the Rpb1-α44/47 domains. Highly conserved domain are blue. Species symbols: Sc (S.cerevisiae); Mj (M.jannaschii), Ec (Escherichia coli); Ta (T.aquaticus); At (A.thaliana, chloroplastic), Ra (R.americana, mitochondrial). Viral species: Acanthamoeba Polyphaga Mimivirus (APMV); Chilo Iridescent Virus (CIV); Emiliana Huxleyi PhycoDNA Virus (EHPV); African Swine Fever Virus (AFSV) and Vaccine.
These two-hybrid data are also consistent with the spatial distribution of the five critical amino acids H147, P151, D182, R200 and R212 identified by the lethal (rpb5-D182N, rpb5-R212E) or conditional (rpbH147R, rpb5-H147Q, rpb5-P151T and rpb5-R200E) mutants. These amino acids are quasi invariant in all eukaryotic, archaeal and viral genomes investigated so far (Figure 1). They are close to each other in the yeast RNA polymerase II structure, directly contacting the Rpb1-α45 (H147) and Rpb1-α46 helices (P151, D182, R200 and R212). R200 is also very close one end of the large Rpb1-α47 helix. Finally, rpb5-H147R and rpb5-R200E are synthetic lethal with rpb1-E1351K, a charge inversion at Rpb1-α46 (see Figure 3). rpb1-E1351K alters one of the most strongly conserved positions of Rpb1-α46. This mutant was originally isolated as a suppressor of spt5-242, altering the elongation factor Spt5 (46), which along with the synthetic lethal effects seen here, underscores the importance of the Rpb1-α46 helix.
In summary, the C-terminal globe of Rpb5 binds a conserved fold on Rpa190, Rpb1 and Rpc160, corresponding to a bundle of four helices (Rpb1-α44/47) where α44 to α46 probably contribute to most of this binding. Lethal or conditional rpb5 mutants underscore the importance of this binding. It should be noted that Rpb1-1α44 and α45 are conserved in all Rpb5-containing RNA polymerases but not in the bacterial and chloroplastic enzymes or the mitochondrial polymerase of Reclinomonas americana, that have no Rpb5-like subunit (Figure 4C). The Rpb1-α46/47 sequence, on the other hand, is conserved in all polymerases, with the same spatial organization in yeast and bacteria (see Figure 6 below), showing that their contribution to the RNA polymerase activity is, in part, independent of their binding to Rpb5.
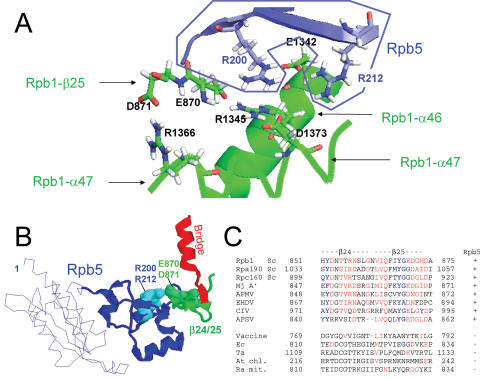
The crystal structure of RNA polymerase II predicts that Rpb5 also binds the Rpb1-β24/25 fold, located immediately after the Rpb1-α25 Bridge helix on Rpb1 (Figures 5–7). These binding sites are separated by <400 amino acids on Rpb1, and are harboured by two distinct polypeptides, A′ and A′′, in archaea and in the chilo iridescent virus (CIV). Rpb1-α44/47, Rpb1-β24/25 and the C-terminal module of Rpb5 are connected by a salt-bridge system (Figure 5B) involving positively and negative charged amino acids of α46 (E1342 and R1345), α47 (D1373), β24 (E870 and D871) and of Rpb5 itself (R200 and R212). These positions are highly conserved in archaeal, eukaryotic and viral RNA polymerases (see the sequence alignments of Figures 1B, 4C and 5C). Charge inversion mutants, such as rpb5-R200E or rpb5-R212E are predicted to impair this salt bridge system, as illustrated in Figure 5B, where position R200 of Rpb5 is seen to be clasped by the negatively charged E870 and E1342 amino acids of Rpb1, whereas R212 binds E1342 on Rpb1-α46. The properties of rpb5-R200E and rpb5-R212E therefore support the idea that the dual binding of Rpb5 to Rpb1-α44/47 and Rpb1-β24/25 is critical in vivo, although Rpb1-β24/25 itself was not isolated as a two-hybrid partner of Rpb5.
Figure 5.
Dual binding of Rpb5 to Rpb1 (A) View of the salt bridge system involving R200 and R212 (Rpb5) and six Rpb1 amino acids: E870, D871 (Rpb1-β25), E1342 and R1345 (Rpb1-α46), R1366 and D1373 (Rpb1-α47). Drawing based on the PDB crystallographic coordinates 1I6H (41), with Hydrogen atoms recalculated by the PDB-viewer programme. The Rpb5 domain is in blue, and the two Rpb1 parts in green. (B) Spatial organization of Rpb5, the Rpb1-β24/25-fold and the Bridge helix (Rpb1-α25), using the same orientation as in Figure 1. Rpb5 is shown in blue, with the eukaryotic backbone domain shown as a thin line. R200 and R212 are space-filled. Rpb1-β24/25 and Rpb1-α25 are shown in green and red, respectively. (C) Conservation of the Rpb1-β24/25 sequence. Species symbols as in Figure 4. Conserved amino acids are in red.
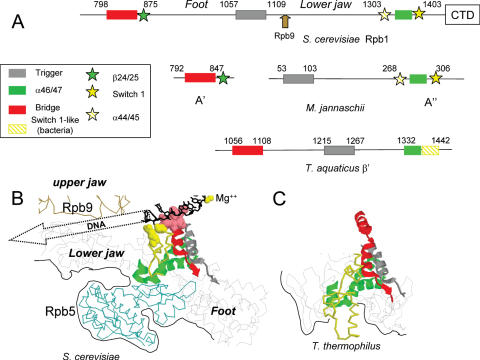
Figure 7.
Spatial organization of Rpb5, Rpb9 and their interacting domains. (A) Distribution of the Bridge, Rpb1-b24/25, Trigger, Rpb1-a44/47 and Switch 1 domains on Rpb1(S.cerevisiae), A′ and A′ (M.jannaschii) and β′ (T.aquaticus). (B) View of the same domains and of Rpb5 and Rpb9 in the elongating RNA polymerase II, based on the PDB crystallographic coordinates 1I6H (45). This view corresponds to the ‘upper’ representation of Figure 1A. The Bridge C-end (positions 831–851), Trigger (1057–1088), α46/47 (1340–1379) and Switch 1 (1380–1406) modules are shown as ribbon structures. DNA-binding amino acids of the Bridge (positions 831–836) and Switch 1 modules (R1386 and E1403) are space-filled. The template strand of DNA (black wire-frame) and the catalytic Mg (yellow sphere) are also indicated. The Rpb9 and Rpb5 backbones are shown in brown and blue, respectively. Black lines correspond to the outer surface of the crystal structure. (C) Equivalent view of the Thermus thermophilus holoenzyme, based on the PDB crystallographic coordinates 1IW7 (47). Note that the amino acid sequences and spatial structures of T.aquaticus and T.thermophilus are practically identical. The T.aquaticus (β′ subunit) equivalent of the yeast bridge (β′F), trigger (β′G) and α46/47 modules correspond to positions 1089–1109, 1215–1246 and 1341–1380, respectively. The yellow backbone (positions 1381–1443 of the β′ subunit) is no related to the Switch 1-fold in sequence (except for its last three positions, see Supplementary Data S3).
β24/25,α44/45 and α47a/b (Switch 1) are conserved in polymerases with Rpb5-like subunits
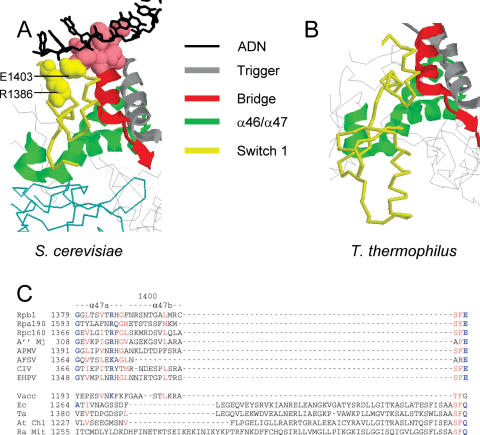
The sequence conservation of Rpb1-β24/25, Rpb1-α44/47 and Rpb1-α47a/b is documented in Figures 4–6. As already mentioned, the α46 and α47 helices are conserved in all polymerases, irrespectively of the presence of an Rpb5-like subunit. In the yeast and bacterial crystal structures (13,14,18,39,45,47,48) they are closely packed with two other strongly conserved folds, the Bridge (Rpb1-α25) and Trigger (Rpb1-α36/37) helices (Figures 6 and 7). Rpb1-β24/25, Rpb1-α44/45 and Rpb1-α47a/b, on the other hand, are only found in Rpb5-containing polymerases, except for the limited conservation of Rpb1-α47a/b seen in poxviruses. The two short helices Rpb1-α47a and α47b immediately follow Rpb1-α46/47 on Rpb1 (Figures 6 and 7), forming the Switch 1 loop that binds the template DNA strand at the downstream fork of the transcription bubble (45). This binding is due to a highly conserved R1386-E1403 dipole located next to the DNA-binding positions 831–836 of the Bridge helix (45) that, again, is only found in Rpb5-containing polymerases. In bacteria, Switch 1 is replaced by a larger fold (positions 1378–1444 of β′ in Thermus aquaticus), with two helices unrelated to Rpb1-α47a and 47b in their amino acid sequences. Unlike the yeast Switch 1 domain, however, this bacterial fold belongs to the outer envelope of the core enzyme and there is so far no evidence that it binds DNA.
Figure 6.
The switch 1 loop in RNA polymerase II. (A) Spatial organization of the Switch 1 loop (1378–1403), Bridge helix (C-end, positions 804–835), trigger helix (positions 1057–1089) and Rpb1-α46/47 domain (1338–1377) in RNA polymerase II, based on the PDB crystallographic coordinates 1I6H (41). (B) Spatial organization of the corresponding bacterial domain, based on the PDB crystallographic coordinates 1IW7 (47). Sequence conservation of the switch 1 loop domain. Species symbols as in Figure 4. (C) Positions R1386 and E1403 are shown in bold characters.
Rpb9 (RNA polymerase II) and Rpa12 (RNA polymerase I) cooperate with Rpb5 in vivo
The large Rpb1 segment (438 amino acids) separating the two Rpb5 binding sites defined above contains the ‘lower jaw’ and ‘foot’ modules of RNA polymerase II (18), with the highly conserved Trigger helix in between (Figure 7A). As illustrated Figure 7B, the ‘lower jaw’ module is projected across the DNA Cleft and binds the Rpb9 upper jaw by a strong Rpb9-β4/Rpb1-β28 β addition motif (18). This organization evidently suggests some cross-talk between the ‘lower’ (Rpb5) and ‘upper’ (Rpb9) parts of the DNA Cleft. This would be consistent with the synthetic lethality mentioned above(Figure 3C) between rpb5 mutants (rpb5-Δ14, rpb5-chi11, rpb5-chi7K and rpb5-R200E) and the rpb9Δ null-mutant or its rpa12Δ counterpart in RNA polymerase I. Since Rpb9 also co-operates with the (non-essential) elongation factor TFIIS factor to activate transcript cleavage in the elongating RNA polymerase II (9), we tested the same rpb5 mutants in a dst1Δ null context. However, rpb5-chi7K was the only rpb5 allele to be affected by the dst1Δ null mutant, indicating that the lethality of the rpb9Δ rpb5 double mutants is not, as a rule, due to a defective transcript cleavage.
rpb5 mutants affect all three nuclear RNA polymerases
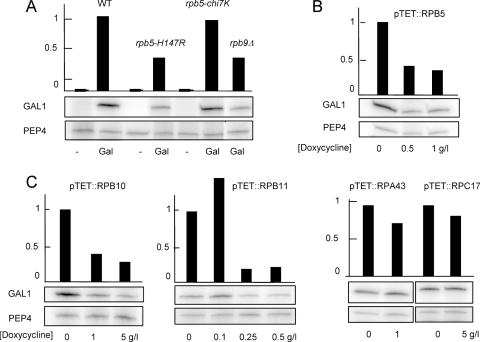
Miyao & Woychick (42) have observed that the cold-sensitive mutant rpb5-V111G partly impairs the gene-specific activation of GAL1 and INO1, with little or no effect on their non-induced level of expression, and have argued that this suggests a specific role of Rpb5 in the gene-specific control of transcription (42). We found a similar gene activation defect with rpb5-H147R and with the slow-growing rpb5-chi6 mutant (shown Figure 7 for rpb5-H147R). Gene-activation defects were also noted for cells partly depleted of the wild-type Rpb2 subunit (49), and we therefore examined GAL1 and INO1 induction in mutants lacking the non-essential subunit Rpb9 (rpb9Δ) or partly depleted for the wild-type Rpb5, Rpb10 or Rpb11 RNA polymerase II subunits. Similar constructions depleted for the Rpa43 or Rpc17 subunits of RNA polymerase I or III served as negative controls. As shown Figure 8 for GAL1, gene activation was distinctly impaired in all strains that were partly depleted in RNA polymerase II. This general effect of RNA polymerase II depletion might well account for the activation defect seen of some rpb5 mutants, thus weakening the current evidence for a direct role of Rpb5 in gene-specific activation.
Figure 8.
Transcriptional activation of GAL1 in RNA polymerase II defective mutants. Strains YFN68 (wild-type), YFN6 (rpb5-H147R), YFN10 (rpb5-chi7K) and YVV9 (rpb9Δ) were grown on raffinose (−) and galactose (Gal). GAL1 and PEP4 mRNAs were assayed by quantitative reverse transcribed PCR (A) as described in Materials and Methods, using the oligonucleotide probes listed in Supplementary Data S1. The same measures are shown for strain YFN13 (pTET::RPB5), YFN50 (pTET::RPB10), YFN27 (pTET::RPB11), YFN25 (pTET::RPA43) and YMLF2 (pTET::RPC17) exponentially grown on Gal medium in the presence of increasing concentrations of doxycycline (B and C). In each case, the higher concentration of doxycycline used reduced growth rate by a factor of three. Note that the depletion of the Rpa43 subunit of RNA polymerase I (YFN25) or the Rpc17 subunit of RNA polymerase III (YMLF2) has no effect on the accumulation of GAL1 mRNA.
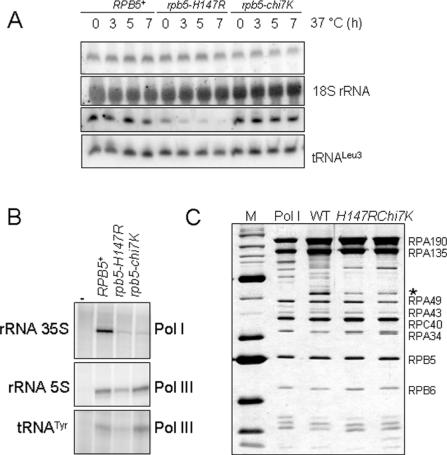
Rpb5 is shared by all three yeast RNA polymerases, and one therefore expects rpb5 mutants to affect RNA polymerase I and III. rpb5-H147R and rpb5-chi7K were thus examined for their influence on RNA polymerase I and III in vivo by comparing the steady-state level of pre-rRNA and pre-tRNALeu to the corresponding mature rRNA (18S) and tRNALeu (50). rpb5-H147R had a strong RNA polymerase III defect whilst rpb5-chi7K had a circa 2-fold reduced pre-rRNA/rRNA ratio, suggesting a partly impaired RNA polymerase I activity (Figure 9A). Cell-free extracts of rpb5-chi7K and rpb5-H147R were also tested for their ability to initiate the transcription of the 35S rDNA template by RNA polymerase I, or to transcribe 5S rDNA and tDNA templates by RNA polymerase III (Figure 9B). rpb5-chi7K mostly affected RNA polymerase I but rpb5-H147R impaired both activities. However, these in vitro transcription defects were not directly related to a catalytic defect since the two mutant RNA polymerase I were comparable to the wild-type control when purified to near homogeneity (Figure 9C) and assayed for their ability to transcribe non-specific poly(dA–dT) templates.
Figure 9.
RNA polymerase I and III defects of rpb5 mutants (A) RNA polymerase I and III activity in vivo. The level of 20S pre-rRNA (compared to the mature 18S rRNA product) and of the pre-tRNALeu3 transcript (compared to the mature tRNA) was measured by northern hybridization using the oligonucleotide probes listed in Supplementary Data S1. Cells were exponentially grown on YPD at 30°C until they reached an O.D. of 0.5 and were then shifted to 37°C for 3, 5 and 7 h. (B) In vitro transcription of 35S rRNA, 5S rRNA and tRNATyr templates. Transcription was assayed on cell-free extract prepared from strains YFN13 (wild-type), YFN6 (rpb5-H147R) and YFN10 (rpb5-chi7K). The templates used were pRS316-SUP4 (tRNATyr) for RNA polymerase III activity (34) and pSIRT (5S rRNA and 35S rRNA) for RNA polymerase I (35). Transcription assays are described in the Materials and Methods section. (C) Subunit composition of the purified RNA polymerase I of strains YFN13, YFN6 and YFN10, using samples with similar specific activities when tested on poly(dA–dT) templates (36). The first two panels, corresponding to standard MW markers and to an RNA polymerase I sample (kindly given by Christophe Carles and Michel Riva) were used to identify individual RNA polymerase I subunits. The star denotes a sub-stoichiometric polypeptide band that was identified by mass spectrometry as being the Rpc53 subunit of RNA polymerase III. However, there is currently no evidence that Rpc53 could be a bona fide subunit of RNA polymerase I.
DISCUSSION
Rpb5 has a bipartite organization combining two globular modules separated by a short hinge (18,38,39). The N-terminal module is only found in eukaryotes, but the C-terminal globe is very closely related to subunit H in archaea (2) and to Rpb5-like subunits of viral RNA polymerases (26,27). Both parts are essential in vivo, and are functionally exchangeable with their human counterpart, except for a small central segment comprised between positions 121–146 in S.cerevisiae. It has been argued that Rpb5 contributes to gene-specific activation, notably during the infectious cycle of the Hepatitis B virus (22), and that this regulatory role accounts for the functional incompatibility of the yeast and human subunits (42). However, our own data make this interpretation rather unlikely, since similar gene-specific defects are obtained by any genetic construction depleting RNA polymerase II in wild-type cells [(49) and this study] and since rpb5 mutants affect all three RNA polymerases.
Rpb5 does not directly belong to the catalytic domain of RNA polymerase II [(13,14,18,45)] and refolding studies in Methanococcus jannaschii have shown that the archaeal enzyme retains some transcriptional activity in the absence of subunit H (1). Moreover, rpb5-chi7K and rpb5-H147R have strong RNA polymerase I defect in vivo or in whole-cell extracts, but remain competent to transcribe a non-specific poly(dA–dT) template. In the RNA polymerase II crystal structure, the C-terminal globe binds the β24/β25 and α44/α47-folds of Rpb1 (18), and the importance of this dual binding fits with the critical Rpb5 positions (H147, P151, D182, R200 and R212) identified in this study. Since Rpb5 is shared by RNA polymerase I, II and III (24,25), the equivalent domains of RNA polymerase I (subunit Rpa190) and III (subunit Rpc160) are expected to also bind Rpb5. This was supported by our two-hybrid data, showing that Rpb5 specifically binds the Rpa190, Rpb1 and Rpc160 domains corresponding to Rpb1-α44/α47 in RNA polymerase II, although some of the two-hybrid clones isolated did not include the α47 helix.
The Rpb1-β24/25 module was not detected in this two-hybrid screening, but sequence comparison show it to be lost in the bacterial, chloroplastic and mitochondrial (R.americana) enzymes that have no Rpb5. It is also lacking in poxviruses, where the Rpo22 subunit is, at best, remotely related to Rpb5. Arginines R200 and R212 of that module are invariant in all Rpb5-like subunits identified so far and belong to an intricate salt bridge system connecting the C-end of Rpb5, the α46 and α47 helices of Rpb1 and E870/D871 (Rpb1-β24). Their charge inversions are lethal (rpb5-R212E)or strongly compromise growth (rpb5-R200E). Intriguingly, R200 may be part of a still poorly defined NTP-binding pocket recently observed in the crystal structure of the free, non-transcribing form of RNA polymerase II (51).
The main function of Rpb5 could therefore be to hold together the Rpb1-β24/25 and Rpb1-α44/47-fold of RNA polymerase II, or their counterparts in the archaeal, viral and RNA polymerase I and III enzymes. Rpb1-β24/25 and Rpb1-α44/47 are widely separated on Rpa190, Rpb1 and Rpc160 and borne by the distinct subunits A′ and A′′ in archaea (1,2) or in the CIV virus (52). They are adjacent to the C-end of the Bridge helix and to the Switch 1 loop, respectively, and their binding to Rpb5 therefore connects the Bridge and Switch 1 domains in the spatial structure of RNA polymerase II, close to the Trigger loop and in direct contact to the template DNA strand at the transcription fork (45). The Bridge (Rpb1-α25), Trigger (Rpb1-α37) and Rpb1-α46/47 helices have the same spatial organization in the yeast and bacterial enzymes (45,47,48). In contrast, Rpb5, Rpb1-α44/45, Rpb1-β24/25 and the Switch 1 loop do not exist in the bacterial enzyme, where the Switch 1 loop is replaced by a larger loop that may or may not bind DNA (47,48).
The strong synergy seen between rpb5 mutants and rpb9Δ or rpa12Δ suggests that Rpb9 and Rpa12 closely cooperate with Rpb5. Like Rpb5, Rpb9 and Rpa12 also belong to a family of subunits or elongation factors conserved in archaea (subunit M/TFS), in the eukaryotic RNA polymerase I (Rpa12), II (Rpb9) and III (Rpc11) and in viral polymerases (9,53). They are also closely related to the C-end of TFIIS, an elongation factor associated to RNA polymerase II (9). In RNA polymerase II, Rpb5 and Rpb9 belong to the ‘upper’ and ‘lower’ jaws of the DNA Cleft, respectively. They are connected by the large Rpb1 segment comprised between the two Rpb5-binding sites, that is projected across the DNA Cleft and binds the N-half of Rpb9 by a strong β-addition motif (18). This Rpb9–Rpb1 interaction and the dual Rpb5–Rpb1-binding may therefore co-ordinate the opening/closing of the DNA Cleft (Rpb9) and the binding of Switch 1 to the transcription fork (Rpb5). Some of the general transcription factors associated with RNA polymerase II may assist in this conformational changes. Thus, the Tfg2 (RAP30) subunit of TFIIF is known to bind Rpb5 in vitro (23), consistent with current structural data on the yeast TFIIF–RNA polymerase II complex (54). In yeast, the binding of TFIIF to RNA polymerase II is hampered in an rpb9Δ mutant (55), perhaps reflecting a defective interaction between Rpb9 and the Tfa1 subunit of TFIIE (56). Finally, the rpb1-E1351K mutant at the Rpb1-α46 helix (found here to be synthetic lethal with rpb5 mutants) suppresses a conditionally defective mutant of the Spt5 elongation factor (46).
Along with Rpb8 (3), the N-terminal module of Rpb5 (positions 1–142) is one of the two structural features that are both typically eukaryotic and shared by all three nuclear RNA polymerases. This module occupies the ‘lower’ jaw of the DNA Cleft (18) and cross-linking data indicate that it contacts the downstream DNA some 15 nt ahead of the transcription bubble (19,20). We were unable to confirm a previous suggestion that its P86 and P118 Prolines are critical for elongation (18). On the other hand, a long and highly conserved Rpb5-α1 helix, formed by the first 30 amino acids of yeast Rpb5, was critical in vivo. This hydrophilic helix occupies the very end of the lower Cleft jaw (18). This part of Rpb5 could therefore directly contact chromatin in elongating RNA polymerases, perhaps through a recently described interaction between Rbp5 and the Rsc4 subunit of the RSC chromatin remodeler (57).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Christophe Carles, Emmanuel Favry, Olivier Gadal, Grant Hartzog, Gérald Peyroche, Michel Riva and Michel Werner for advice and reagents. This work was funded by a European Union grant (FMRX-CT96-0064) and by grants of the Spanish Ministry of Science and Technology (BMMC-07072-C03-03) and of the Junta de Andalucía (CVI258). J.-F.B. was supported by a fellowship from the Fondation pour la Recherche Médicale, C.Z. by a fellowship from the Association pour la Recherche contre le Cancer, M.C.G.-L by a fellowship from University of Jaén-Junta Andalucía and F.N. by a Marie Curie fellowship. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Werner F., Weinzierl R.O. A recombinant RNA polymerase II-like enzyme capable of promoter- specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 2.Langer D., Hain J., Thuriaux P., Zillig W. Transcription in Archaea: similarity to that in Eucarya. Proc. Natl Acad. Sci. USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briand J.F., Navarro F., Rematier P., Boschiero C., Labarre S., Werner M., Shpakovski G.V., Thuriaux P. Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell. Biol. 2001;21:6056–6065. doi: 10.1128/MCB.21.17.6056-6065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wery M., Shematorova E., Van Driessche B., Vandenhaute J., Thuriaux P., Van Mullem V. Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMB0 J. 2004;23:42132–44242. doi: 10.1038/sj.emboj.7600326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadal O., Chédin S., Quémeneur E., Carles C., Sentenac A., Thuriaux P. A34.5, a non-essential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machinery. Mol. Cell. Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadal O., Shpakovski G.V., Thuriaux P. Mutants in ABC10β, a conserved subunit shared by all three yeast RNA polymerases, specifically affect RNA polymerase I assembly. J. Biol. Chem. 1999;274:8421–8427. doi: 10.1074/jbc.274.13.8421. [DOI] [PubMed] [Google Scholar]
- 8.Rubbi L., Labarre-Mariotte S., Chédin S., Thuriaux P. Functional characterization of ABC10α, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem. 1999;274:31485–31492. doi: 10.1074/jbc.274.44.31485. [DOI] [PubMed] [Google Scholar]
- 9.Fish R., Kane C. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 10.Chédin S., Riva M., Schultz P., Sentenac A., Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prescott E.M., Osheim Y.N., Jones H.S., Alen C.M., Roan J.G., Reeder R.H., Beyer A.L., Proudfoot N.J. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl Acad. Sci. USA. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peyroche G., Levillain E., Siaut M., Callebaut I., Schultz P., Sentenac A., Riva M., Carles C. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 RNA polymerase II subunits. Proc. Natl Acad. Sci. USA. 2002;99:14670–14675. doi: 10.1073/pnas.232580799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armache K.J., Mitterweger S., Meinhart A., Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 14.Bushnell D.A., Kornberg R.D. Complete, 12-subunit RNA polymerase II at 4.1 Å resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards A.M., Kane C.M., Young R.A., Kornberg R.D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation at a promoter in vitro. J. Biol. Chem. 1990;266:71–75. [PubMed] [Google Scholar]
- 16.Zaros C., Thuriaux P. Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol. Microbiol. 2005;55:104–114. doi: 10.1111/j.1365-2958.2004.04375.x. [DOI] [PubMed] [Google Scholar]
- 17.Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase at 2.8 A resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 19.Bartholomew B., Durkovich D., Kassavetis G.A., Geiduschek E.P. Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T., Lagrange T., Wang Y.H., Griffith J.D., Reinberg D., Ebright R.H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc. Natl Acad. Sci. USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W., Dorjsuren D., Lin Y., Qin W., Nomura T., Hayashi N., Murakami S. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J. Biol. Chem. 2001;276:12266–12273. doi: 10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y., Nomura T., Cheong J.H., Dorjsuren D., IIda K., Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 23.Le T.T., Zhang S., Hayashi N., Yasukawa M., Delgermaa L., Murakami S. J. Bio. chem. , 138. (Tokyo): 2005. Mutational analysis of human RNA polymerase II subunit 5 (RPB5): the residues critical for interactions with TFIIF subunit RAP30 and hepatitis B virus X protein; pp. 215–224. [DOI] [PubMed] [Google Scholar]
- 24.Buhler J.M., Iborra F., Sentenac A., Fromageot P. Structural studies on yeast RNA polymerases. J. Biol. Chem. 1976;251:1712–1717. [PubMed] [Google Scholar]
- 25.Woychik N.A., Liao S.M., Kolodziej P.A., Young R.A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 26.Iyer L.M., Balaji S., Koonin E.V., Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Raoult D., Audic S., Robert C., Abergel C., Renesto P., Ogata H., La Scola B., Suzan M., Claverie J.M. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 28.Gari E., Piedrafita L., Aldea M., Herrero E. A set of vectors with tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 29.Flores A., Briand J.F., Boschiero C., Gadal O., Andrau J.C., Rubbi L., Van Mullem V., Goussot M., Marck C., Carles C., et al. A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fromont-Racine M., Rain J.C., Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 31.Navarro F., Thuriaux P. In vivo misreading by tRNA overdose. RNA. 2000;6:103–110. doi: 10.1017/s1355838200991714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpakovski G.V., Gadal O., Labarre-Mariotte S., Lebedenko E.N., Miklos I., Sakurai H., Proshkin S.A., Van Mullem V., Ishihama A., Thuriaux P. Functional conservation of RNA polymerase II in fission and budding yeasts. J. Mol. Biol. 2000;295:1119–1127. doi: 10.1006/jmbi.1999.3399. [DOI] [PubMed] [Google Scholar]
- 33.Culbertson M.R., Henry S.A. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975;80:23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siaut M., Zaros C., Levivier E., Ferri M.L., Werner M., Callebaut I., Thuriaux P., Sentenac A., Conesa C. A Rpb4/Rpb7 like complex in yeast RNA polymerase III contains the orthologue of mammalian CGRP-RCP. Mol. Cell. Biol. 2003;23:195–205. doi: 10.1128/MCB.23.1.195-205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musters W., Knol J., Maas P., Dekker A.F., Heerikhuizen H.V., Planta R.J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dieci G., Hermann-Le Denmat S., Lukhtanov E., Thuriaux P., Werner M., Sentenac A. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-Blast protein data searches with composition-based statistics and other refinenements. Nucleics Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todone F., Weinzierl R.O., Brick P., Onesti S. Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target. Proc. Natl Acad. Sci. USA. 2000;97:6306–6310. doi: 10.1073/pnas.97.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cramer P., Bushnell D.A., Fu J., Gnatt A.L., Maier-Davis B., Thompson N.E., Burgess R.R., Edwards A.M., David P.R., Kornberg R.D. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 40.Broyles S.S., Moss B. Homology between RNA polymerases of poxviruses, prokaryotes and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia viruses genes encoding 147 kDa and 22 kDa subunits. Proc. Natl Acad. Sci. USA. 1986;83:3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shpakovski G.V., Acker J., Wintzerith M., Lacroix J.F., Thuriaux P., Vigneron M. Four subunits shared by the three classes of RNA polymerases are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyao T., Woychik N.A. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc. Natl Acad. Sci. USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proshkina G.M., Shematorova E.K., Proshkin S.A., Zaros C., Thuriaux P., Shpakovski G.V. Ancient origin and fast evolution of DNA-dependant RNA polymerase III. Nucleic Acids Res. 2006;34:3615–3624. doi: 10.1093/nar/gkl421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Exinger F., Lacroute F. 6-azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 45.Gnatt A.L., Cramer P., Fu J., Bushnel D.A., Kornberg R.D. Structural basis of transcription: an RNA polymerase elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 46.Hartzog G.A., Wada T., Handa H., Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami K.S., Masuda S., Darst S.A. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 48.Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 49.Archambault J., Jansma D.B., Friesen J.D. Underproduction of the largest subunit of RNA polymerase II causes temperature sensitivity, slow growth and inositol auxotrophy in Saccharomyces cerevisiae. Genetics. 1996;142:737–747. doi: 10.1093/genetics/142.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briand J.F., Navarro F., Gadal O., Thuriaux P. Cross-talk between tRNA and rRNA synthesis. Mol. Cell. Biol. 2001;21:189–195. doi: 10.1128/MCB.21.1.189-195.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westover K.D., Bushnell D.A., Kornberg R.D. Structural basis for transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Schnitzler P., Sonntag K.C., Muller M., Janssen W., Bugert J.J., Koonin E.V., Darai G. Insect iridescent virus type 6 encodes a polypeptide related to the largest subunit of eukaryotic RNA polymerase II. J. Gen. Virol. 1994;75:1557–1567. doi: 10.1099/0022-1317-75-7-1557. [DOI] [PubMed] [Google Scholar]
- 53.Ahn B.Y., Gershon P.D., Kones E.V., Moss B. Identification of Rpo30, a vaccinia virus RNA polymerase gene with structural homology to a eucaryotic transcription elongation factor. Mol. Cell. Biol. 1990;10:5433–5441. doi: 10.1128/mcb.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung W.H., Craighead J.L., Chang W.H., Ezeokonkwo C., Bareket-Samish A., Kornberg R.D., Asturias F.J. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell. 2003;12:1003–1013. doi: 10.1016/s1097-2765(03)00387-3. [DOI] [PubMed] [Google Scholar]
- 55.Ghazy M.A., Brodie S.A., Ammerman M.L., Ziegler L.M., Ponticelli A.S. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol. Cell. Biol. 2004;24:10975–10985. doi: 10.1128/MCB.24.24.10975-10985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Mullem V., Wéry M., Werner M., Vandenhaute J., Thuriaux P. The Rpb9 subunit of RNA polymerase II binds TFIIE and functionally interacts with the SAGA and Elongator Factors. J. Biol. Chem. 2002;277:10220–10225. doi: 10.1074/jbc.M107207200. [DOI] [PubMed] [Google Scholar]
- 57.Soutourina J., Bordas-Le Floch V., Gendrel G., Flores A., Ducrot C., Dumay-Odelot H., Soularue P., Navarro F., Cairns B.R., Lefebvre O., et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.