Abstract
Switching from a replicative to a translesion polymerase is an important step to further continue on replication at the site of DNA lesion. Recently, RAD18 (a ubiquitin ligase) was shown to monoubiquitinate proliferating cell nuclear antigen (PCNA) in cooperation with RAD6 (a ubiquitin-conjugating enzyme) at the replication-stalled sites, causing the polymerase switch. Analyzing RAD18-knockout (RAD18−/−) cells generated from human HCT116 cells, in addition to the polymerase switch, we found a new function of RAD18 for S phase-specific DNA single-strand break repair (SSBR). Unlike the case with polymerase switching, PCNA monoubiquitination was not necessary for the SSBR. When compared with wild-type HCT116 cells, RAD18−/− cells, defective in the repair of X-ray-induced chromosomal aberrations, were significantly hypersensitive to X-ray-irradiation and also to the topoisomerase I inhibitor camptothecin (CPT) capable of inducing single-strand breaks but were not so sensitive to the topoisomerase II inhibitor etoposide capable of inducing double-strand breaks. However, such hypersensitivity to CPT observed with RAD18−/− cells was limited to only the S phase due to the absence of the RAD18 S phase-specific function. Furthermore, the defective SSBR observed in S phase of RAD18−/− cells was also demonstrated by alkaline comet assay.
INTRODUCTION
Various types of DNA lesions generated by endogenous and environmental factors in living organisms can be repaired by various repair mechanisms (1). However, if a lesion is not yet removed because of a limited repair capacity of cells and if the DNA replication machinery encounters this lesion before repair, then the replication machinery stalls at this lesion, causing a gap in the newly synthesized strand opposite to the damaged site. Further the gap could cause a secondary damage, much severer than the first one, such as DNA double-strand break (DSB), and thus could cause cell death unless the gap is filled. This gap filling is called as postreplication repair (PRR) and such gap filling function is conserved from bacteria through to human cells (2,3).
In the budding yeast Saccharomyces cerevisiae, genes belonging to the RAD6 epistasis group are involved in the PRR pathway (4). Among these genes, those encoding Rad6 (a ubiquitin-conjugating enzyme, E2) and Rad18 (a ubiquitin ligase, E3) play crucial roles in this pathway (5,6). Yeast Rad18 was shown to bind to single-stranded DNA and also to form a tight complex with Rad6 (5,7) whereas Rad6 does not have any DNA-binding activity. Thus it was proposed that such Rad18 bound to the gap regions would recruit Rad6 to the replication-stalled sites, where Rad6 modulates the function of a stalled DNA replication machinery with its ubiquitin-conjugating activity (5). Recently, in yeast cells, the proliferating cell nuclear antigen (PCNA), a DNA polymerase sliding clamp involved in DNA replication and repair, was shown to be monoubiquitinated at the lysine-164 site in a Rad18/Rad6-dependent manner, which was necessary for carrying out translesion synthesis (TLS) by Rad30, a yeast homolog of polymerase η (poly η), which is a member of the RAD6 epistasis group (8–11). Similarly, monoubiquitination of human and mouse PCNAs was observed in a RAD18/RAD6-dependent manner at the sites of replication forks stalled by ultraviolet (UV) light-induced lesions, and caused a polymerase switch from a replicative to a translesion polymerase, pol η (12,13). Thus, the mechanism of polymerase switching through the monoubiquitination of PCNA is conserved in various species.
The mutation of either RAD18 or RAD6 gene in yeast cells resulted in hypersensitivity to various DNA-damaging agents including UV light and methylmethanesulfonate (MMS) (14), and caused the attenuation of the mutation-inducing ability following such exposure (15). In mammals, only a single homolog of yeast RAD18 has been identified (16). The human and mouse RAD18 proteins can interact with two forms of homologs of yeast Rad6, RAD6A and RAD6B, both in vitro and in vivo (16–18). Either overexpression of a dominant-negative RAD18 mutant protein in human cells or targeted disruption of RAD18 in mouse embryonic stem cells resulted in an increased sensitivity to various DNA-damaging agents and also enhanced the genomic instability as determined by increases in sister-chromatic exchange (SCE) and also by stable transformation (16,17). These phenotypes were also observed in the RAD18-knockout chicken DT40 cells (19).
In the present study, to understand the role of RAD18 in human cells, we have generated RAD18-knockout (RAD18−/−) cells from wild-type human HCT116 cells, and investigated how RAD18 was involved in the repair mechanism of DNA DSB or single-strand break (SSB) in human cells inducing DSBs and SSBs by X-ray irradiation and also with topoisomerase inhibitors in RAD18−/− cells. As described above, RAD18 has been shown to function at the polymerase switching for TLS through PCNA monoubiquitination. Analyzing RAD18−/− cells, in addition to the function of TLS, we found a new function of RAD18 in DNA strand break repair.
We have observed that RAD18−/− cells, defective in the repair of X-ray-induced chromosomal aberrations, were significantly hypersensitive to X-ray irradiation and also to the topoisomerase I inhibitor camptothecin (CPT) capable of inducing SSBs but were not so sensitive to the topoisomerase II inhibitor etoposide capable of inducing DSBs as compared with parental HCT116 cells. Furthermore, PCNA was not monoubiquitinated after X-ray irradiation or treatment with topoisomerase I or II inhibitor even in the parental HCT116 cells. Our experiments showed that RAD18−/− cells were defective in SSB repair (SSBR), but functional for DSB repair (DSBR). Further, it turned out that, for SSBR, RAD18−/− cells were only defective in S phase but not in non-S phase or in non-replicating state, implying that RAD18 could have some unknown intriguing function particularly in S phase, exerting such tolerance to X-ray-induced or drug-induced SSBs, without PCNA monoubiquitination.
MATERIALS AND METHODS
Cell lines and culture conditions
The human colon carcinoma cell line HCT116, which was normal in p53 status but defective in mismatch repair gene hMLH1, and its RAD18−/− derivative were cultured in DMEM supplemented with 10% fetal bovine serum, 50 U/ml penicillin G and 50 μg/ml streptomycin in a humidified 5% CO2 incubator at 37°C.
Inactivation of RAD18 locus in HCT116
A promoterless neomycin resistance gene or poly(A)-signalless puromycin resistance gene was inserted into exon2 of RAD18 in frame. A 4.1 kb 5′-targeting element was amplified from the isogenic DNA of HCT116 cells using the primer set 5′-GGTGTCGACAGGTTGGCAGTCTTATGGCACCAGCT-3′ and 5′-AATTCGCGGCCGCAGCAAATCATCTATTGTCTGAAATGC-3′. A 3.3 kb 3′-targeting element was amplified using the primer set 5′-ACATTGCGGCCGCAATACCTCAGTGTTCACATAACTG-3′ and 5′-CAAAAGGCGCGCCTGATGGCTTACAGCAACTCTGGAGA-3′. Amplified PCR products were cloned into a modified pBC vector (Stratagene). Both neomycin and puromycin resistance cassettes were prepared from pIRESneo2 and pIRESpuro2 (Clontech), respectively. Targeting vectors linearized with AscI were introduced into cells as described previously (20). For the selection of targeted cells, G418 and puromycin were added at 350 and 0.35 μg/ml, respectively. After 14 days, colonies were isolated and expanded. Genomic DNA from individual colonies was screened for gene targeting by PCR using the primer sets 5′-TGGCAGTCATGAAGGTGAGTGCTTCG-3′ and 5′-ATTTTCCACCATGATATTCGGCAAGCAGGC-3′ for neomycin resistance, and 5′-AGGCGCACCGTGGGCTTGTACTCGGTCATG-3′ and 5′-CTTGTCGACACGCGTCCAGCTGGTTCTTTCCGCCTC-3′ for puromycin resistance. To confirm the gene targeting, genomic DNA from candidate clones was digested with SpeI and KpnI and was subjected to Southern blot analysis. The blots were hybridized with an external probe, a 3′-region of the targeting constructs as described previously (21).
Western blotting
Cell extracts were electrophoresed in 10% SDS–PAGE gels. Following transfer on to PVDF filters, the blots were probed with a mouse anti-PCNA antibody (PC10; Santa Cruz Biotechnology), an anti-RAD18 antibody raised in rabbits (16) or an anti-FLAG antibody (M2; Sigma).
Synchronization
Cells were synchronized by the thymidine-hydroxyurea block method. Briefly, 5 × 104 cells were seeded to 3.5 cm dishes and grown for 2 days. Thymidine was then added to a final concentration of 1.5 mM and cells were further incubated for 23–24 h. The cells were released from thymidine block by washing three times with the medium. After a 10 h incubation in the fresh medium, HU was added to a final concentration of 1 mM and cells were incubated for 15–16 h. Under these conditions, the cells were accumulated at the G1/S boundary. HU was removed by washing three times with the fresh medium. The cells were incubated for 1 or 5 h in the fresh medium to obtain cells at early S phase or late S/G2 phase, respectively. The synchronization of the cell cycle was confirmed using FACSCalibur (Becton Dickinson).
Growth rates and cell survival assays
Cells were plated at a density of 2 × 104 cells per 35 mm dish and cultured at 37°C.
On the days indicated, cells were counted using a hematocytometer. All measurements were performed in triplicate. Cell survival assay was carried out as follows. Cells were plated at a density of 2 × 102 or 103 cells per 60 mm dish and irradiated with X-ray or UV light, or cultured in the medium containing indicated doses of cisplatin, MMS, etoposide (VP-16) or CPT. In some experiments, synchronized or randomly growing cells were treated with indicated concentrations of CPT for 2 h. For aphidicolin (APH) experiments, APH (1 μg/ml) was added 15 min before CPT was added. The exponentially growing cells were incubated with the two drugs together for 2 h. After treatment, cells were washed twice with the culture medium, followed by a 2 h incubation in the medium containing 1 μg/ml APH. Then, the cells were washed twice with PBS and plated onto 60 mm dish. The cells were grown for 9–10 days and colonies were counted.
Analysis of chromosomal aberrations
After treatment with 0.05 μg/ml colcemid for 1 h, chromosomes were prepared according to the air-drying method as described previously (22). Chromosomal aberrations were scored according to the international nomenclature of aberrations (23). Chromatid-type aberrations included gaps, breaks and exchanges. Chromosome-type aberrations were classified into breaks, dicentrics, rings double minutes and pulverization. Translocations were omitted from the score because of ambiguity in non-banded preparations. Chromatid-type gaps were most abundantly induced by X-ray irradiation, but were excluded from the calculation of aberrations per cell because of ambiguity between true DNA break and aberrant chromatin packaging. All types of aberrations were scored as one event.
Wortmannin treatment
Cells were preincubated with 20 μM Wortmannin 3 h before X-ray irradiation. Wortmannin treatment was maintained for 3 days after treatment.
Assessment of stable transformation
HCT116 or RAD18−/− cells were transfected with the circular or linear form of pTK-Hyg (Clontech) plasmid DNA by electroporation and selected in the medium containing hygromycin B (400 μg/ml) for 2 weeks. The frequency of stable transformation was assessed on the basis of the frequency of drug-resistant colonies.
Evaluation of gene targeting at HPRT locus
The poly(A)-signalless hygromycin resistance gene was inserted into exon 2 of HPRT in frame. A 3.1 kb 5′-targeting element was amplified from the isogenic DNA of HCT116 cells using the primer set 5′-GTTGTCGACCAGGTATTACGGGCCAACCTGACAATACATG-3′ and 5′-ATGCAAGCGGCCGCAAGGTCATAACCTGGTTCATCATCAC-3′. A 1.8 kb 3′-targeting element was amplified using the primer set 5′-TGGAAAGCGGCCGCATTCCTCATGGACTAATTATGGACAGG-3′ and 5′-TTTATAGGCGCGCCTGAGCACACAGAGGGCTACAATGTGATG-3′. Amplified PCR products were cloned into the modified pBC vector (Stratagene). A hygromycin resistance cassette was prepared from pIREShygro2 (Clontech). Targeting vectors linearized with AscI were introduced into cells by electroporation. For the selection of targeted cells, hygromycin B was added at 400 μg/ml in the growth medium. After 14 days, colonies were isolated and expanded. Genomic DNA from individual colonies was screened for gene targeting by PCR using the primer set 5′-ATCAATATTGACTTCTGCCTGCTGTATAGC-3′ and 5′-GCGGCCATTGTCCGTCAGGACATTGTTGGA-3′.
Comet assay (single cell micro gel electrophoresis)
The alkaline comet assay was performed using a comet assay kit (CometAssay; Trevigen) according to the manufacturer's protocol. DNA was stained with Cyber green. Mean tail moment was quantified for 100 cells per sample in each experiment using Metafer Comet Scan software.
Expression of RAD18 gene
Human RAD18 cDNA was amplified from cDNA derived from HCT116 cells using the primer set 5′-TCCGAATTCGCGACCATGGACTCCCTGGCC-3′ and 5′-AAGGTCGACATTCCTATTACGCTTGTTTC-3′. The cDNA was inserted into pBC (Stratagene). RAD18 cDNA tagged with FLAG at the C-terminus was inserted downstream of the CMV enhancer and promoter of pCI-neo (Promega). RAD18−/− cells were transfected by electroporation with the RAD18 cDNA expression vector and pTK-Hyg, and selected in the medium containing 400 μg/ml hygromycin B (Wako, Japan) for 2 weeks. Surviving colonies were isolated and propagated individually. The expression of the RAD18 gene was examined by western blot analysis using the anti-FLAG and anti-RAD18 antibodies.
RESULTS
Generation of human RAD18−/− cells
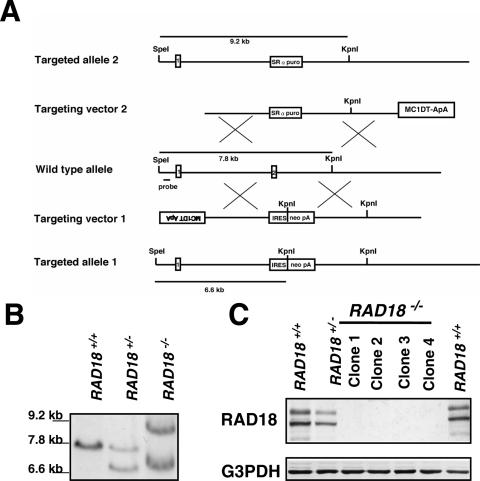
We have disrupted human RAD18 inserting a promoterless neomycin resistant gene (neo) or poly(A)-signalless puromycin-resistant (puro) gene into human colon carcinoma cell line HCT116. When the targeted cell DNAs were digested with both SpeI and KpnI, the correct insertion of the neo or puro gene into the locus was expected to yield a 6.6 kb or a 9.2 kb fragment, respectively, instead of a 7.8 kb wild-type fragment (Figure 1A). G418-resistant colonies generated with the RAD18-neo targeting vector were screened for targeted integration first by PCR then by Southern blot analysis. Six of fourty-eight G418-resistant clones showed the targeting bands (Figure 1B). The RAD18-puro targeting vector was introduced into one of these clones (RAD18+/−). Puromycin-resistant clones were screened for targeted integration. Eight homozygous knockout clones were obtained from 3000 puromycin-resistant clones. Southern blot analysis of one of the clones is shown in Figure 1B. Western blot analysis revealed that no RAD18 protein was expressed in the homozygous knockout clones and that the level of the RAD18 protein correlated with targeting events (Figure 1C). One homozygous knockout clone (clone 1) was named as RAD18−/− and was used for further studies.
Figure 1.
Generation of RAD18−/− HCT116 cells. (A) Schematic representation of RAD18 locus, disruption constructs and configuration of targeted alleles. Relevant restriction sites and the position of the probe used for Southern blot analysis are shown. MC1, MC1 promoter; DT-ApA, diphtheria toxin gene; puro, puromycin resistance gene; neo, neomycin resistance gene; SRα, SRα promoter; IRES, internal ribosome entry site; pA, poly (A) additional signal. Exons 1 and 2 are indicated by numbered boxes. (B) Southern blot analysis of KpnI- and SpeI-digested genomic DNA from cells with indicated genotypes of RAD18 gene. (C) Western blot analysis of whole-cell extracts from different HCT116 clones with indicated genotypes of RAD18 gene. G3PDH was used as an internal control.
Growth and sensitivity of RAD18−/− cells to various DNA-damaging agents
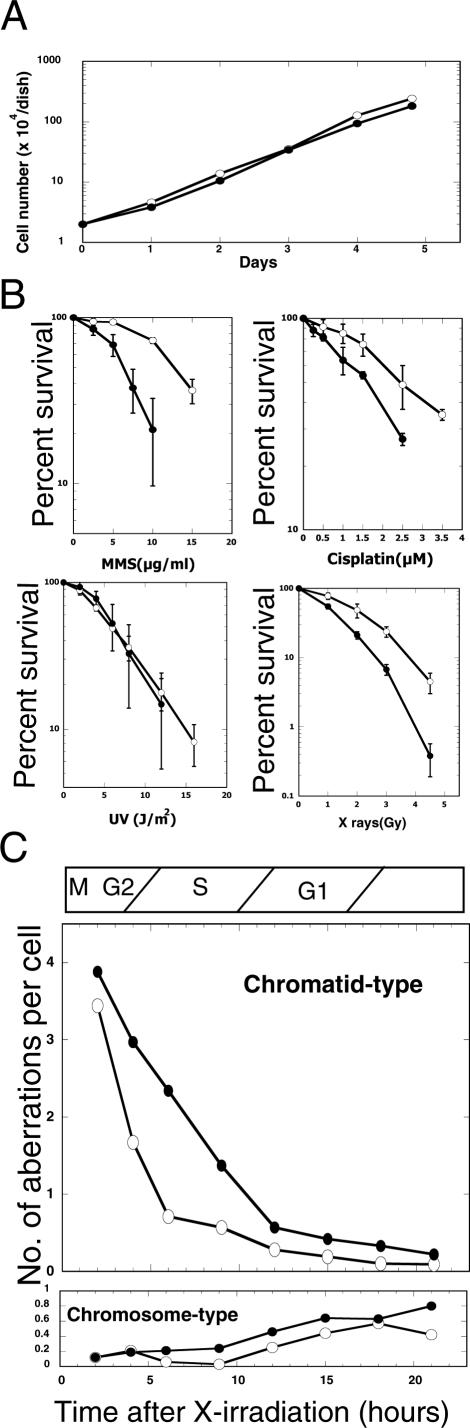
The growth rate of RAD18−/− cells was almost the same as that of wild-type cells (Figure 2A). The doubling time of the cells was ∼18 h. The sensitivity of RAD18−/− cells to various DNA-damaging agents was determined by colony formation assay. As shown in Figure 2B, RAD18−/− cells were significantly more sensitive than wild-type HCT116 to cisplatin, methyl methanesulfonate (MMS) or X-ray. In contrast, surprisingly, they showed no significant increase in sensitivity to UV light unlike the case with RAD18-knockout cells from mouse ES or chicken DT40 cells, suggesting that different mechanisms for tolerance to UV light may be in operation in human HCT116 cells.
Figure 2.
Growth curves, sensitivity to DNA-damaging agents and chromosomal aberrations induced by X-ray in RAD18−/− cells. (A) Growth curves of wild-type (closed circles) and RAD18−/− cells (open circles). The data show typical experimental results of three independent experiments. (B) Sensitivities of wild-type (closed circles) and RAD18−/− cells (open circles) to continuous treatment with cisplatin or MMS. Sensitivities to UV light or X-ray irradiation are also shown. The data shown are means of three independent experiments with standard deviations (error bars). (C) Chromosomal aberrations in HCT116 and RAD18−/− cells after X-ray irradiation. Cells were irradiated with 1.4 Gy of X-rays and cultured thereafter. Cells were treated with 0.05 μg/ml colcemid for 1 h prior to harvest and chromosomes were prepared as described in Materials and Methods. A total of 100 metaphase cells were examined in each sample. Chromatid-type (upper) and chromosome-type (lower) aberrations in HCT116 (open circles) and RAD18−/− (closed circles) cells were counted. Approximate cell cycle phases (determined by separate experiments) at the time of X-ray irradiation are shown at the top of the figure. The number of spontaneous chromatid-type aberrations in HCT116 and RAD18−/− cells were 0.06/cell and 0.06/cell, respectively. The number of spontaneous chromosome-type aberrations in HCT116 and RAD18−/− cells were 0.09/cell and 0.07/cell, respectively.
Chromosomal aberrations induced by X-ray irradiation
Exponentially growing wild-type HCT116 and RAD18−/− cells were irradiated, respectively, with 1.4 Gy of X-ray, and, at various intervals after irradiation, chromosomal aberrations were examined in metaphase cells accumulated by 1 h treatment with colcemid. As shown in Figure 2C, chromosomal aberrations induced by X-ray irradiation were mostly of the chromatid type until 10 h after irradiation. The number of chromatid-type aberrations in HCT116 cells accumulated in metaphase within 3–4 h after irradiation was slightly lower than or nearly the same as that in RAD18−/− cells. These cells accumulated at metaphase appeared to be in G2 phase at the time of irradiation judging from the cell cycle phases determined by separate experiments. The number of chromatid-type aberrations in HCT116 cells accumulated in metaphase from 4 to 11 h after irradiation decreased rapidly, whereas it decreased gradually in RAD18−/− cells at the same time intervals. These metaphase cells appeared to be in S phase at the time of irradiation. These results indicated that DNA lesions resulting in chromatid-type aberrations were repaired efficiently during the S phase in HCT116 but not in RAD18−/− cells, suggesting that the repair mechanism functioning in the S phase was defective in RAD18−/− cells, i.e. RAD18 was likely to have a particular repair function specific for S phase.
Targeted integration in RAD18−/− cells
Targeted integration (gene targeting) shares the homologous recombination (HR) repair pathway (24) with other different types of HR (such as between sisters, between homologues). To know whether RAD18 was involved in the common part of the HR pathway in human cells, we have examined targeted integration frequencies both in wild-type HCT116 and RAD18−/− cells.
To assess the frequency of targeted integration, the HPRT-targeting vector DNA was transfected into HCT116 or RAD18−/− cells. The frequency of targeted integration in RAD18−/− cells was ∼10 times higher than that in wild-type HCT116 cells (Table 1). These results indicated that the loss of RAD18 function caused the increase in frequency of targeted integration in human cells, suggesting that the common part of the HR pathway was functioning much more efficiently in RAD18−/− cells than in wild-type HCT116 cells. Thus, RAD18 appeared to be involved in protective function from homologous integration.
Table 1.
Frequency of targeted integrationa
| Expt no. | No. of colonies with targeted integration/total no. tested | |
|---|---|---|
| HCT116 (W.T.) | RAD18−/− | |
| 1 | 0/280 | 10/316 |
| 2 | 2/258 | 10/203 |
| 3 | 1/442 | 21/587 |
| Total (%) | 3/980 (0.31) | 41/1106 (3.71) |
aThe HPRT allele-targeting construct was transfected into cells. After selection with 400 μg/ml of hygromycin, targeted integration events were determined by PCR.
RAD18−/− cells appear normal in DSBR
In mammalian cells, DNA DSBs are mainly repaired via two pathways, HR and nonhomologous end joining (NHEJ) pathways. RAD18−/− cells were significantly hypersensitive to radiation (Figure 2B) and were defective in the repair of chromosomal aberrations (Figure 2C), suggesting that the NHEJ pathway was not properly functioning in RAD18−/− cells. In contrast, HR pathway in RAD18−/− cells appeared functioning efficiently judging from their efficient targeted integration activity (Table 1). Thus, next, we have attempted to clarify whether RAD18 was involved in the NHEJ pathway.
Since stable transformation processes through random integration of an exogenous gene into host cell genome supposedly need factors functioning in the NHEJ pathway (24), we have examined random integration frequencies both in wild-type HCT116 and RAD18−/− cells to know whether RAD18 was involved in the NHEJ pathway in human cells. For this experiment, HCT116 and RAD18−/− cells were both transfected with the circular or linear form of the plasmid pTK-Hyg containing the hygromycin B resistance gene. The frequency of stable transformation was then determined by counting the number of the hygromycin B-resistant colonies. The results obtained indicated that irrespective of plasmid DNA forms, the transformation frequencies of RAD18−/− cells were 6–12 times higher than those of wild-type HCT116 cells (Table 2), suggesting that factors needed for stable transformation appeared to be active in RAD18−/− cells.
Table 2.
Frequency of stable transformation
| Expt no. | Cells | Plasmid DNA (bC or cL) | No. of cells plated | Plating efficiency | No. of cells tested | No. of hygror clones | aS.T. frequency |
|---|---|---|---|---|---|---|---|
| 1 | HCT116 | pTK-hygro (C) | 2 × 106 | 0.63 | 1.27 × 106 | 8 | 6.3 × 10−6 |
| HCT116 | pTK-hygro (L) | 2 × 106 | 0.64 | 1.28 × 106 | 11 | 8.6 × 10−6 | |
| RAD18−/− | pTK-hygro (C) | 2 × 106 | 0.51 | 1.01 × 106 | 39 | 38.6 × 10−6 | |
| RAD18−/− | pTK-hygro (L) | 2 × 106 | 0.43 | 0.86 × 106 | 42 | 49.0 × 10−6 | |
| 2 | HCT116 | pTK-hygro (C) | 2 × 106 | 0.60 | 1.20 × 106 | 10 | 8.3 × 10−6 |
| HCT116 | pTK-hygro (L) | 2 × 106 | 0.74 | 1.47 × 106 | 10 | 6.8 × 10−6 | |
| RAD18−/− | pTK-hygro (C) | 2 × 106 | 0.41 | 0.82 × 106 | 74 | 90.6 × 10−6 | |
| RAD18−/− | pTK-hygro (L) | 2 × 106 | 0.43 | 0.85 × 106 | 70 | 82.4 × 10−6 |
aST, stable transformation; bC, circular; cL, linear.
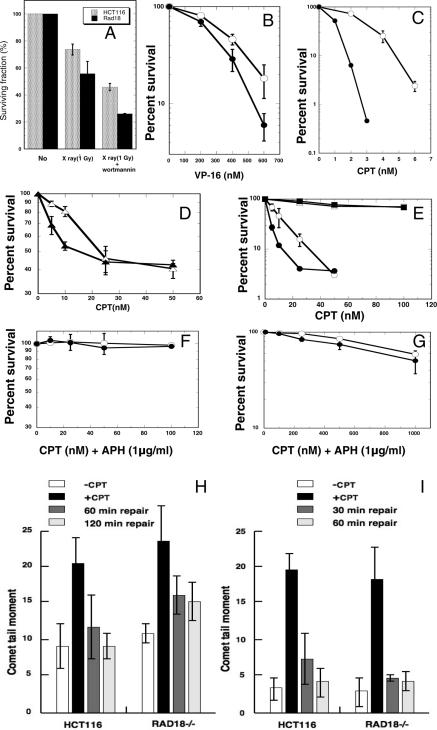
Wortmannin inhibits phosphatidylinositol-3 (PI-3) kinases including its family member DNA-dependent protein kinase (DNA-PK) involving the NHEJ pathway of DSBs. Treatment with Wortmannin increased the radiation sensitivity of eukaryotic cells, but showed no effects on their mutant cells defective in the NHEJ pathway such as DNA-PK catalytic subunit (DNA-PKcs)- or XRCC4-deficient cells (25). Thus, we have examined effects of Wortmannin on RAD18−/− cells. As shown in Figure 3A, when treated with Wortmannin, RAD18−/− cells were sensitized to radiation to the extents similar to those with HCT116 cells, indicating that the functioning Wortmannin-sensitive NHEJ pathway was present in RAD18−/− cells. The results suggested that RAD18 was not directly involved in the Wortmannin-sensitive NHEJ pathway.
Figure 3.
Survival after treatment with X-ray and Wortmannin or incubation with topoisomerase inhibitors, and comet assays. (A) Effects of Wortmannin treatment on the survival of HCT116 (gray bars) and RAD18−/− cells (closed bars). Cells were irradiated with 1 Gy of X-rays. Cells were preincubated with 20 μM Wortmannin for 3 h before irradiation and Wortmannin treatment was maintained after irradiation. (B and C) Survival curves for HCT116 (open circles) and RAD18−/− (closed circles) cells after continuous culture with indicated concentrations of VP-16 (B) or CPT (C). (D) Survival curves for exponentially growing HCT116 (open triangles) and RAD18−/− (closed triangles) cells treated for 2 h with indicated concentrations of CPT. (E) Survival curves for HCT116 (open circles and open squares) and RAD18−/− (closed circles and closed squares) cells synchronized at S phase (open circles and closed circles) or G2 phase (open squares and closed squares) and treated for 2 h with indicated doses of CPT. (F) Survival curves for HCT116 (open circles) and RAD18−/− (closed circles) cells treated for 2 h with CPT (up to 100 nM) and APH as described in Materials and Methods. (G) Broadening of low-dose range shown in (F) up to 1000 nM of CPT. (H) Strand breaks quantified in S phase cells untreated or treated with CPT (100 nM for 30 min) and then incubated for 0, 60 or 120 min in drug-free medium. S phase cells were prepared as described in Materials and Methods. (I) Strand breaks quantified in mock-treated or CPT-treated (2000 nM for 30 min) cells with APH (1 μg/ml) and incubated for 0, 30 or 60 min in CPT-free medium containing APH (1 μg/ml). Data shown are means of three independent experiments with standard deviations (error bars).
RAD18−/− cells become synthetic lethal when combined with heterozygous mutation at XRCC4 locus (XRCC4+/−)
To further investigate the functional relationship between the RAD18-dependent strand break repair and the NHEJ pathway, we first attempted to disrupt one of the XRCC4 (the gene involved in the NHEJ pathway) alleles in RAD18−/− cells inserting the promoterless hygromycin-resistant (hyg) gene (Supplementary Figure 1A). After transfection of RAD18−/− cells with the targeting vector DNA, we obtained 48 hygromycin-resistant clones and subsequently examined the targeting event by PCR analysis (Supplementary Figure 1B). This way, we have identified three candidate clones for the heterozygous disruptant of the XRCC4 locus. Because cells from all these three clones exhibited extremely decreased growth rates and eventually all ceased proliferation after forming very tiny colonies, we were unable to analyze the targeting event by Southern blotting. Alternatively, we have carried out nucleotide sequencing using PCR products amplified from the genomic DNA of these three clones. Sequencing analyses revealed that, in all the three cases, the sequence of XRCC4 gene was fused to the hygromycin resistance gene as expected (data not shown), indicating that these three clones were the heterozygous disruptants of the XRCC4 locus. These results strongly suggested that RAD18−/− cells became synthetic lethal due to the additional heterozygous disruption of the XRCC4 locus (XRCC4+/−). Therefore, RAD18 may function in a repair pathway different from the XRCC4-dependent NHEJ pathway.
RAD18−/− cells appears defective in SSBR
The major types of DNA damage resulted in chromatid-type aberrations are DSBs and SSBs. Topoisomerase inhibitors, CPT and etoposide (VP-16), interfere with the catalytic activity of topoisomerase I and II, and generate primarily SSBs and DSBs, respectively (26–28). To further identify which repair mechanism (SSBR or DSBR) was defective in RAD18−/− cells, we examined sensitivities of HCT116 and RAD18−/− cells to these drugs. As shown in Figure 3B and C, when compared with HCT116 cells, RAD18−/− cells were highly sensitive to the killing effect of CPT capable of inducing SSBs, but were slightly sensitive to VP-16 capable of inducing DSBs. Thus, these results strongly suggested that RAD18−/− cells were mainly defective in SSBR.
RAD18−/− cells are more sensitive to CPT than HCT116 cells in S phase but not in non-S phase or in non-replicating state
Exponentially growing HCT116 or RAD18−/− cells were pulse-treated for 2 h with CPT at various concentrations. As shown in Figure 3D, both HCT116 and RAD18−/− cells showed the persistent presence of drug-resistant cell populations. Thus, the cells tested could be divided into at least two populations: one was highly sensitive to CPT (∼50% of total cells); and the other highly resistant to CPT.
When the sensitive population of RAD18−/− cells was compared with that of HCT116, as shown in Figure 3D, the sensitive population of RAD18−/− cells was much more sensitive than that of HCT116 cells. On the other hand, the resistant population of RAD18−/− cells was as resistant as that of HCT116 cells (Figure 3D). These results suggested the possibilities that the sensitivity to CPT could be dependent on cell cycle phases, and that RAD18−/− cells were highly sensitive to CPT only in a certain cell cycle phase(s).
To identify the possible sensitive phase(s) in the cell cycle, we have synchronized the cells followed by pulse treatment with CPT, and examined the sensitivities. As shown in Figure 3E, both HCT116 and RAD18−/− cells in the G2 phase were similarly resistant to CPT. In contrast, both cell lines in the S phase were highly sensitive to CPT, though RAD18−/− cells were much more sensitive than HCT116 cells (Figure 3E), suggesting that RAD18 was involved in the SSBR mechanism in the S phase, i.e. the mechanism underlying the tolerance to the toxic effect of CPT.
To determine whether the sensitivity of RAD18−/− cells to CPT depends on DNA synthesis, we have examined CPT sensitivity in the presence of a DNA replication inhibitor APH. In the absence of DNA replication by treatment with APH, those cells in S phase showed perfect resistance to CPT (Figure 3F), implying that replicative DNA synthesis was necessary for the manifestation of CPT sensitivity. Further, as shown in Figure 3G, when both cell lines were treated with much higher doses of CPT in the presence of APH, >50% of both cells survived even after 1000 nM CPT treatment, although surviving fractions in both cell lines gradually decreased. These results implied that CPT-induced SSBs were removed at the same efficiency in both cell lines in the non-replicating state, even in the S phase.
SSBR in RAD18−/− cells is defective when examined by alkaline comet assay
To clarify the molecular basis of CPT hypersensitivity in RAD18−/− cells, the level of DNA strand breaks induced by CPT was determined by alkaline comet assay (alkaline single cell micro gel electrophoresis), and was compared between HCT116 and RAD18−/− cells. In both cell lines synchronized in the S phase, the same levels of strand breaks accumulated following the 30 min treatment with CPT. In HCT116 cells, CPT-induced strand breaks were repaired almost completely during 120 min repair period, whereas significant levels of induced breaks persisted in RAD18−/− cells even at 120 min period (Figure 3H), indicating that RAD18−/− cells were defective in strand break repair when cells were in the S phase. However, in non-replicating cells whose replication was inhibited by APH-treatment, strand breaks were repaired as efficiently in RAD18 cells as in HCT116 cells (Figure 3I), indicating that strand breaks were repaired normally in both cells in the non-replicating state. It should be pointed out that, although both SSBs and DSBs can be detected by alkaline comet assay, most of the CPT-induced strand breaks detected were SSBs (29). Thus, RAD18−/− cells were defective in SSBR in the S phase, particularly under the replication-ongoing state, implying that RAD18 has particular function in S phase for SSBR.
RAD18-dependent strand break repair does not require monoubiquitination of PCNA
At stalled replication forks caused by UV-induced lesions, PCNA has been shown to be monoubiquitinated in a RAD18-dependent manner in mammalian cells as well as in yeast cells, and this monoubiquitination subsequently caused a polymerase switch from replicative polymerase to translesion polymerase in TLS (12,13). PCNA has also been reported to be monoubiquitinated after the treatment of human cells with hydroxyurea (HU), which caused replication fork stalling, suggesting that such modification of PCNA could have been induced by the blockade of replication forks (12). Thus, we have examined the monoubiquitination of PCNA after treatment with UV or HU in RAD18−/− human cells.
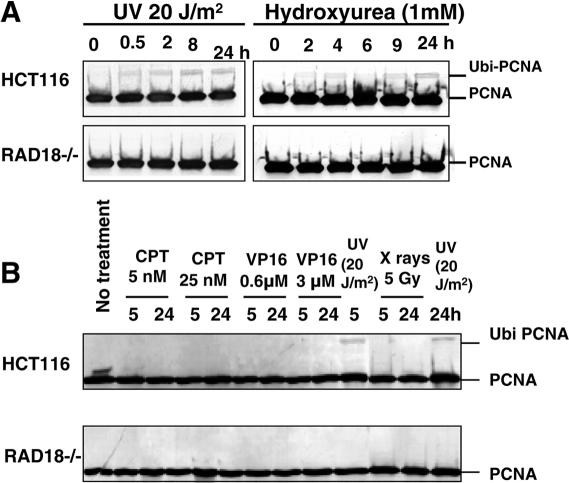
As shown in Figure 4A, the monoubiquitination of PCNA was observed when HCT116 cells were irradiated with UV light or cultured in the medium containing 1 mM HU. However, monoubiquitination was not observed in RAD18−/− cells, indicating that human RAD18 played a critical role in the monoubiquitination of PCNA following UV light irradiation or HU treatment, as reported previously (12,13).
Figure 4.
Monoubiquitination of PCNA. (A) UV light- or HU-induced monoubiquitination of PCNA as determined by western blot analysis. Wild-type HCT116 or RAD18−/− cells were irradiated with 20 J/m2 of UV light or cultured in the presence of 1 mM HU and harvested at indicated times. (B) Monoubiquitination of PCNA after treatment with UV light, X-ray, VP-16 or CPT. HCT116 or RAD18−/− cells were irradiated with UV light (20 J/m2) or X-rays (5 Gy) and harvested after 5 or 24 h incubation for western blot analysis. Cells were cultured in the medium containing CPT (5 or 25 nM) or VP-16 (0.6 or 3 μM) and harvested at indicated times.
As described above, RAD18−/− cells were moderately sensitive to X-ray irradiation or VP-16 treatment, and were highly sensitive to CPT treatment (Figures 2B, 3B and C). Thus, next, we have examined whether these treatments could also induce the monoubiquitination of PCNA. Treatment of RAD18−/− cells with CPT or VP-16, or X-ray irradiation did not induce the monoubiquitination of PCNA for up to 24 h (Figure 4B). Similar observation was made even in wild-type HCT116 cells (Figure 4B), although the monoubiquitination of PCNA was detected 5 h after UV light irradiation. These results strongly suggested that PCNA monoubiquitination was not required for the repair of strand breaks resulting from treatment with CPT or VP-16, or X-ray irradiation.
Ectopical overexpression of RAD18 cDNA complements defective monoubiquitination of PCNA and hypersensitivity to CPT in RAD18−/− cells
To confirm whether both the failure of PCNA monoubiquitination after UV light irradiation and the hypersensitivity to CPT in RAD18−/− cells were due to the defect of RAD18, we have established several RAD18−/− cell clones ectopically expressing wild-type human RAD18. In this experiment, RAD18−/− cells were co-transfected with both the plasmid pTK-Hyg and the RAD18 cDNA expression vector, in which RAD18 was tagged with FLAG to distinguish the introduced RAD18 from the endogenous RAD18 that might be produced by the reversion or alternative splicing of the target-disrupted RAD18 gene.
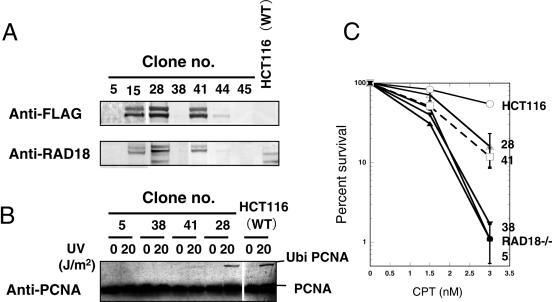
First, we selected hygromycin-resistant clones and then screened these drug-resistant clones by PCR analysis to identify clones stably integrating the full-length RAD18 cDNA. PCR-positive clones (clone nos 5, 15, 28, 38, 41, 44 and 45) were examined by western blot analysis using anti-FLAG and anti-RAD18 antibodies as probes. As shown in Figure 5A, clones 15, 28 and 41 expressed the RAD18 protein. In clone 28, PCNA was ubiquitinated at the same level as that in HCT116 cells after UV irradiation, but in clone 41, PCNA ubiquitination was very weak even after UV irradiation (Figure 5B). In non-expressing clones (clones 5 and 38), any ubiquitinated PCNA was not detected even after UV irradiation (Figure 5B).
Figure 5.
Restoration of CPT sensitivity and UV-induced PCNA monoubiquitination in RAD18−/− cells expressing FLAG-tagged human RAD18. (A) Detection of FLAG-tagged RAD18 using anti-FLAG antibody or anti-human RAD18 antibody. Seven hygromycin-resistant clones (clones 5, 15, 28, 38, 41, 44 and 45) were examined for the expression of the FLAG-tagged RAD18 by western blot analysis. (B) Restoration of UV light-induced monoubiquitination of PCNA in clones (clones 28 and 41) expressing FLAG-tagged RAD18, but not in non-expressing clones (clones 5 and 38). (C) Restoration of CPT sensitivity in clones expressing FLAG-tagged RAD18 (clones 28 and 48), but not in non-expressing clones (clones 5 and 38) as determined by colony-forming assay.
As shown in Figure 5C, either clone 5 or 38, into whose genome the full-length RAD18 cDNA had been integrated, respectively, did not express the RAD18 protein (Figure 5A), and was as sensitive to CPT as RAD18−/− cells. On the other hand, clones 28 and 41 both expressing the RAD18 protein were significantly more resistant to CPT than RAD18−/− cells, although still less resistant than HCT116 cells (Figure 5C). The intermediate resistance of clones 28 and 41 to CPT and the incomplete monoubiquitination of PCNA in clone 41 could be explained by the disruptive effect of FLAG on RAD18. These results described above indicated that high sensitivity to CPT and the defective monoubiquitination of PCNA after UV irradiation in RAD18−/− cells were caused by a defect of RAD18.
DISCUSSION
RAD18 is involved in the repair of X-ray-induced DNA damage independent of PCNA monoubiquitination
Recently, it was reported that PCNA was monoubiquitinated in a RAD18/RAD6-dependent manner at replication forks stalled by UV light-induced lesions in human and mouse cells as well as in yeast, and the monoubiquitination of PCNA caused a polymerase switch from a replicative to a translesion polymerase in TLS (12,13). RAD18 gene disruption in either yeast, chicken or mouse cells resulted in sensitivity to various DNA-damaging agents due probably to the lack of the polymerase switch. Thus, the mechanism underlying polymerase switching through the monoubiquitination of PCNA appears to be conserved in a variety of species.
As shown in Figure 2B, RAD18-knockout (RAD18−/−) cells generated from human HCT116 cells were significantly hypersensitive to X-ray irradiation (Figure 2B) and defective in the repair of X-ray-induced chromosomal aberrations (Figure 3C) when compared with wild-type HCT116 cells. However, X-irradiation, unlike UV irradiation, did not cause PCNA monoubiquitination in HCT116 cells (Figure 4B). These findings strongly suggested that, upon X-ray-induced DNA damage such as DNA strand breaks (DSBs and/or SSBs), RAD18 was involved in some other repair mechanisms independent of PCNA monoubiquitination.
RAD18 functions in repair pathway different from both HR and NHEJ pathways of DSBR
Forms of DNA damage that results in chromatid-type aberrations are DSBs and SSBs. As shown in Figure 3C, wild-type HCT116 cells have repaired these forms of damage (DSBs and/or SSBs) efficiently in the S phase but RAD18−/− cells have not, indicating that some strand break repair mechanism functioning in the S phase could have been defective in RAD18−/− cells. In mammalian cells, DSBs are mainly repaired through two major repair pathways, HR and NHEJ. RAD18−/− cells appeared to have functioning HR pathway because of their efficient homologous integration activity (Table 1) and also to have functioning NHEJ pathway since they showed efficient stable transformation activity (Table 2) and since a NHEJ inhibitor, Wortmannin, increased the radiation sensitivity of RAD18−/− cells to extents similar to that of HCT116 cells (Figure 3A), indicating that the Wortmannin-sensitive NHEJ pathway is working normally in RAD18−/− cells.
The synthetic lethality of RAD18- and RAD54-double knockout cells has been reported in chicken DT40 cells (19). Either of these single knockouts caused only a slight phenotypic change in terms of cell proliferation. Single knockout of most genes involved in HR caused a severe phenotype such as hypersensitivity to ionizing radiation, severe growth retardation or cellular lethality, whereas knockout of the RAD54 gene among genes involved in HR caused a relatively mild phenotype such as slight sensitivity to ionizing radiation and a normal cell growth rate in both murine embryonic stem cells and DT40 cells (30,31).
Furthermore, in our present study, we found the synthetic lethality of RAD18-knockout mutation in human HCT116 cells when the gene knockout was combined with the heterozygous disruption (+/−) of the NHEJ repair gene XRCC4. HCT116 cells with the heterozygous disruption of XRCC4 showed heteroinsufficiency, i.e. a slow cell growth rate and a slight sensitivity to ionizing radiation (M. Mori, unpublished data). Thus, when the minor defect in NHEJ caused by heterozygous disruption of XRCC4 was combined with RAD18-knockout, the resultant cells exhibited synthetic lethality as was observed in the cells with double knockouts of both RAD18 and RAD54 genes. As mentioned above, RAD54 is involved in HR repair whereas XRCC4 is in NHEJ repair. Thus, these synergistic phenotypes observed in double knockout cells indicate that a RAD18-dependent repair mechanism functions in a pathway different from both RAD54-dependent HR and XRCC4-dependent NHEJ pathways of DSBR.
RAD18 is involved in SSBR in S phase
A topoisomerase (Top 1) inhibitor CPT interferes with the catalytic cycle of Top 1 by stabilizing the covalent complex, called cleavage complex, formed between Top 1 and cleaved DNA (27). Cleavage complexes are repaired: first, tyrosyl phosphodiesterase1 (TDP1) cuts Top 1-DNA cross-links; and then resulting SSBs are repaired by XRCC1-dependent SSBR mechanisms (29,32). Because RAD18−/− cells were hypersensitive to CPT when compared with wild-type HCT116 cells (Figure 3C), RAD18 appeared to be involved in some step(s) of the repair mechanism for the cleavage complex.
The inherited disorder spinocerebellar ataxia with axonal neuropathy-1 (SCAN1) was caused by a mutation in TDP1 (33). SCAN 1 cells were reported to be defective in the repair of CPT-induced Top 1-DNA cross-links and SSBs, and highly sensitive to CPT when compared with wild-type cells (29,34). Furthermore, EM9 Chinese hamster ovary cells lacking XRCC1, a central factor for SSBR, were also hypersensitive to CPT, but not to VP-16 (35). On the other hand, Chinese hamster ovary mutant xrs1 (Ku80-deficient mutant) cells were defective in DSBR and highly sensitive to VP-16, but not to CPT when compared with wild-type cells (36). These findings imply that DNA damage induced by CPT is mainly repaired by SSBR.
As shown in Figure 3D–G, RAD18−/− cells were hypersensitive to CPT only in the S phase when replicative DNA synthesis was ongoing, whereas they were not sensitive in other phases of cell cycle or in the S phase when replicative DNA synthesis was inhibited. Thus, function of RAD18 appears to be tightly associated with SSBR on replication-ongoing sites. Indeed, in RAD18−/− cells, X-ray-induced SSBs appeared to be not repaired efficiently and resulted in chromatid-type aberrations via DNA replication (Figure 2C). Furthermore, the defective SSBR in the S phase was confirmed by the alkaline comet assay in RAD18−/− cells (Figure 3H and I). These findings strongly suggest that RAD18 plays an important role in the S-phase-specific SSBR process that still remains unresolved.
It has been reported that XRCC1-deficient Chinese hamster mutant EM9 cells showed higher sensitivity to CPT in the S phase than did wild-type AA8 cells. EM9 cells were also highly sensitivity to CPT in the non-S phase or non-replicating state when compared with AA8 cells (37). Furthermore, TDP1-deficient human SCAN1 cells have been reported to be deficient in their ability to repair SSBs caused by CPT both in the S phase and in the non-S phase (29,34). These findings indicate that if cells are deficient in factors functioning in the general repair pathways for SSBs, then the cells are sensitive to CPT throughout the cell cycle, which is different from our present finding that RAD18 involved the S phase-specific repair function.
Because SSBs are converted to more severe secondary damage, i.e. DSBs, via replication (38) when SSBs are generated in the S phase or when unrepaired SSBs are persisting in the S phase, cells need to remove SSBs as rapidly as possible before SSBs are converted into DSBs to avoid the toxic effect of such damage. To facilitate SSBR in the S phase, RAD18 may function in the S phase-specific SSBR pathway (RAD18-dependent SSBR).
Although SSBR functions efficiently in the S phase, SSBs may have already been converted into DSBs. In such a situation, the HR and NHEJ pathways could serve as backup repair pathways for removing generated DSBs; thereby, cells can escape from the toxic effect of such DNA damage. As mentioned above, double mutant cells defective in both RAD18 and a gene functioning in HR or NHEJ were synthetic lethal. In these double knockout cells, SSBR in the S phase alone may be insufficient, since unrepaired SSBs could be converted into DSBs more frequently than in normal cells. DSBs are fatal for mutant cells deficient in the DSBR pathway, even though the DSBR deficiency is marginal (as in XRCC4+/−). Thus, the double knockout cells are likely to show such a lethal phenotype.
How does RAD18 recognize SSBs and function in S phase-specific SSBR?
RAD18 has been shown to accumulate quite rapidly at the site of SSBs independent of DNA replication and also independent of accumulation of the general SSBR proteins PARP and XRCC1 (39). In the non-S phase, SSBs are repaired efficiently through the rapid XRCC1-dependent SSBR pathway irrespective of the existence of RAD18, because RAD18−/− cells in the non-S phase were as resistant to CPT as wild-type HCT116 cells (Figure 3D–G). On the other hand, in the S phase, RAD18-dependent SSBR could be the one working. During DNA replication, a replication fork encounters an unrepaired SSB to which RAD18 is already bound, and this may serve as a signal for the activation (or initiation) of RAD18-dependent SSBR.
RAD18 is known as a ubiquitin ligase. PCNA was monoubiquitinated in a RAD18- dependent manner in cooperation with RAD6 (a ubiquitin-conjugating enzyme) at replication forks stalled by DNA lesions, and the monoubiquitination of PCNA caused a polymerase switch from a replicative to a translesion polymerase in TLS (12,13). In S phase-specific SSBR, RAD18 could ubiquitinate some protein other than PCNA in cooperation with RAD6 at SSBs, thereby stimulating the activity of SSBR. Further biochemical analysis is required to determine the role of RAD18 in the S phase-specific SSBR pathway in human cells. Our RAD18−/− cells could serve as a useful tool for such studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank K. Noshiro, A. Nakamura and T. Tanaka for technical assistance. We also thank M. Goto-Nakawatari for comet assay and A. Tanaka for critical reading of the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding to pay the Open Access publication charges for this article was provided by National Institute of Radiological Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T., Wood R.D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C., Feaver W.J., Gerlach V.L. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc. Natl Acad. Sci. USA. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann A.R. Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 5.Bailly V., Lamb J., Sung P., Prakash S., Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 6.Bailly V., Prakash S., Prakash L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enxzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailly V., Lauder S., Prakash S., Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 8.Haracska L., Kondratic C.M., Unk I., Prakash S., Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoege C., Pfender B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 10.McDonald J.P., Levine A.S., Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Eschrichia coli dinB and umuC is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 12.Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. Rad18 guides pol eta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes R.H., Kunz B.A. DNA repair and mutagenesis. In: Strathern J.N., Jones E.W., Broach J.R., editors. The Molecular Biology of the Yeast Saccharomyces. Plainview, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 371–414. [Google Scholar]
- 15.Broomfield S., Hryciw T., Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 16.Tateishi S., Sakuraba Y., Masuyama S., Inoue H., Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl Acad. Sci. USA. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 2003;23:474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xin H., Lin W., Sumanasekera W., Zhang Y., Wu X., Wang Z. The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res. 2000;28:2847–2854. doi: 10.1093/nar/28.14.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita Y.M., Okada T., Matsusaka T., Sonoda E., Zhao G.Y., Araki K., Tateishi S., Yamaizumi M., Takeda S. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada Y.-N., Shiomi N., Koike M., Ikawa M., Okabe M., Hirota S., Kitamura Y., Kitagawa M., Matsunaga T., Nikaido O., et al. Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 1999;19:2366–2372. doi: 10.1128/mcb.19.3.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiomi N., Kito S., Oyama M., Matsunaga T., Haradam Y.-N., Ikawa M., Okabe M., Shiomi T. Identification of the XPG region that causes the onset of Cockayne syndrome by using xpg mutant mice generated by the cDNA-mediated knock-in method. Mol. Cell. Biol. 2004;24:3712–3719. doi: 10.1128/MCB.24.9.3712-3719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji H., Takahashi E.-I., Tsuji S., Tobari I., Shiomi T., Sato K. Chromosomal instability in mutagen-sensitive mutants isolated from mouse lymphoma L5178Y cells. Mutat. Res. 1987;178:107–116. doi: 10.1016/0027-5107(87)90092-3. [DOI] [PubMed] [Google Scholar]
- 23.Harnden D.G, Klinger H.P., Standing Committee on Human Cytogenetic Nomenclature. An International System for Human Cytogenetic Nomenclature (ISCN) Report of the standing committee on human cytogenetic nomenclature. Karger in collaboration with Cytogenetics and Cell Genetics and the March of Dimes Birth Defects Foundation (Basel) Cytogenet. Cell. Genet. 1985 [Google Scholar]
- 24.Vasquez K.M., Marburger K., Intody Z., Wilson J.H. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delacôte F., Han M., Stamato T.D., Jasin M., Lopez B.S. An xrcc4 defect or wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Froelich-Ammon S.J., Oshroff N. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 1995;270:21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 27.Pommier Y., Pourquier P., Fan Y., Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochem. Biophys. Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 28.Strumberg D., Nitiss J.L., Dong J., Kohn K.W., Pommier Y. Molecular analysis of yeast and human type II topoisomerase. Enzyme–DNA and drug interactions. J. Biol. Chem. 1999;274:28246–28255. doi: 10.1074/jbc.274.40.28246. [DOI] [PubMed] [Google Scholar]
- 29.El-Khamisy S.F., Saifi G.M., Weinfeld M., Johansson F., Helleday T., Lupski J.R., Caldecott K.W. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 30.Bezzubova O., Silbergleit A., Yamaguchi-Iwai Y., Takeda S., Buerstedde J.M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 31.Essers J., Hendriks R.W., Swagemakers S.M., Troelstra C., de Wit J., Bootsma D., Hoeijmakers J.H., Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 32.Caldecott K.W. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 33.Takashima H., Boekoel C.F., John J., Saifi G.M., Salih M.A., Armstrong D., Mao Y., Quiocho F.A., Rao B.B., Nakagawa M., et al. Nature Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 34.Interthal H., Chen H.J., Kehl-Fie T.E., Zotzmann J., Leppard J.B., Champoux J.J. SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caldecott K., Jeggo P. Cross-sensitivity of γ-ray-sensitive hamster mutants to cross-linking agents. Mutat. Res. 1991;255:111–121. doi: 10.1016/0921-8777(91)90046-r. [DOI] [PubMed] [Google Scholar]
- 36.Jeggo P.A., Caldecott K., Pidsley S., Banks G.R. Sensitivity of Chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res. 1989;49:7057–7063. [PubMed] [Google Scholar]
- 37.Barrows L.R., Holden J.A., Anderson M., D'Apra P. The CHO XRCC1 mutant, EM9, deficient in DNA ligase III activity, exhibits hypersensitivity to camptothecin independent of DNA replication. Mutat. Res. 1998;408:103–110. doi: 10.1016/s0921-8777(98)00022-6. [DOI] [PubMed] [Google Scholar]
- 38.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA. 2001;98:8241–8246. doi: 10.1073/pnas.131009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakajima S., Lan L., Kannno S., Usami N., Kobayashi K., Mori M., Shiomi T., Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]