Abstract
The small GTPase RAB4 regulates endocytic recycling, a process that contributes to Major Histocompatibility Complex (MHC)-mediated antigen presentation by specialized antigen presenting cells (APC) of the immune system. The gene encoding the RAB4B isoform of RAB4 was singled out by two complementary genome-wide screens. One of these consisted of a computer scan to identify genes containing characteristic MHC class II-related regulatory sequences. The second was the use of chromatin immunoprecipitation coupled to microarrays (ChIP-on-chip) to identify novel targets of a transcriptional co-activator called the MHC class II transactivator (CIITA). We show that the RAB4B gene is regulated by a typical MHC class II-like enhancer that is controlled directly by both CIITA and the multiprotein transcription factor complex known as the MHC class II enhanceosome. RAB4B expression is thus activated by the same regulatory machinery that is known to be essential for the expression of MHC class II genes. This molecular link between the transcriptional activation of RAB4B and MHC class II genes implies that APC boost their antigen presentation capacity by increasing RAB4-mediated endocytic recycling.
INTRODUCTION
The partitioning of eukaryotic cells into distinct compartments by lipid membranes necessitates the existence of an elaborate transport system capable of transferring membrane components and cargos between different membrane-bounded organelles and between these organelles and the plasma membrane (PM). The various intracellular trafficking routes are served by specialized lipid vesicles that travel to their destination along cytoskeleton filaments and fuse with their target membranes by tightly regulated mechanisms. Proteins belonging to the Rab family of small Ras-related GTPases play pivotal roles in the regulation of intracellular vesicular traffic (1–3). Rab proteins associate with specific vesicles and regulate multiple steps in their trafficking, including their formation and transport, cargo selection, as well as docking to and fusion with their target membranes. To fulfill these functions, Rab proteins are tightly regulated at the level of their localization, membrane association, activation and expression. The importance of the latter is underscored by the fact that deregulated Rab expression is tightly correlated with several pathologies, including vascular, lung and thyroid disorders as well as a number of cancers (4,5).
Vesicular traffic is of central importance for the presentation of peptides by Major Histocompatibility Complex (MHC) molecules to the antigen receptors (TCRs) of T lymphocytes. MHC-mediated peptide presentation is essential for the adaptive immune system because it governs the development and activation of T cells. The engagement of MHC-peptide complexes by the TCR of thymocytes during their development in the thymus directs the positive and negative selection processes that shape the mature T cell repertoire. In the periphery, the recognition of peptides presented by MHC molecules controls the initiation and development of T cell-mediated immune responses directed against pathogens and tumors. MHC-mediated peptide presentation is also critical for the maintenance of tolerance to self-antigens and is implicated in the breakdown of this self-tolerance during autoimmune diseases.
There are two classes of MHC molecules—MHC class I (MHC-I) and MHC class II (MHC-II)—which differ with respect to their pattern of expression, the nature of the peptides they present, the T cell subsets that recognize them and their functions in the immune system. MHC-I molecules are expressed by all nucleated cells, and are specialized for the presentation of peptides derived from the degradation of intracellular proteins to the TCR of CD8+ T cells (6,7). Peptides presented by MHC-I molecules are generated in the cytoplasm by the proteasome and transported by a peptide transporter (TAP) into the endoplasmic reticulum (ER), where they are loaded onto newly synthesized MHC-I molecules. The MHC-I-peptide complexes are then transported via the golgi apparatus to the PM. MHC-II molecules are expressed mainly by thymic epithelial cells and specialized antigen presenting cells (APC; dendritic cells (DC), macrophages, B cells) and present peptides derived from extracellular proteins to the TCR of CD4+ T cells (7,8). These peptides are generated in endocytic compartments by the proteolysis of internalized proteins and are loaded onto MHC-II molecules that have been transported into the endocytic compartments thanks to their association with an accessory protein called the invariant chain (Ii). Peptide-loaded MHC-II complexes are then transported to the cell surface. The classical MHC-I and MHC-II antigen presentation pathways depend on vesicular transport routes that are regulated by well known members of the Rab family.
In addition to the classical antigen presentation pathways, there is growing evidence that endocytic recycling processes play important roles in antigen presentation (9). The presentation of certain antigens requires the loading of peptides onto MHC-II molecules that have been internalized by endocytosis, followed by recycling of these peptide-MHC-II complexes back to the cell surface (10–12). Such a recycling pathway has notably been documented to be required for certain antigens taken up by receptor-mediated endocytosis in B cells (13). Recycling of internalized cell surface MHC-I molecules is also one of the mechanisms that allows peptides derived from endocytosed proteins to be presented by MHC-I molecules. This cross-presentation process is most efficient in DC (7,14,15). Finally, recent results have demonstrated that a recycling pathway is involved in the presentation of intact proteins by DC to the antigen receptors of B cells (16).
RAB4 is a key player in endocytic recycling. It is associated with early endosomes (EE) and recycling endosomes (RE), and regulates the recycling of membranes and proteins from these compartments back to the PM (1,17). There are two highly homologous RAB4 isoforms—RAB4A and RAB4B—encoded by two separate genes (RAB4A and RAB4B). RAB4A and RAB4B are localized in the same cellular compartments and are believed to have similar or identical functions in recycling (18,19). Importantly, RAB4 has been implicated directly in MHC-II-restricted antigen presentation; a dominant negative mutant of RAB4 was shown to block the presentation of antigens internalized by receptor-mediated uptake in B cells (13).
We show here that transcription of the RAB4B gene is controlled by the same regulatory machinery that is critical for the expression of MHC-II genes and enhances the expression of MHC-I genes. The promoter of RAB4B contains a strongly conserved enhancer known as the S-Y module, which was until now believed to be highly specific for MHC-II and related genes. This RAB4B S-Y module is regulated by the same transcription factors that are dedicated for the activation of MHC-II expression. These notably include the transcription factor called Regulatory Factor X (RFX) and a transcriptional co-activator known as the MHC class II transactivator (CIITA). This molecular link between the transcriptional activation of RAB4B and genes involved in antigen presentation implies that APC boost their antigen presentation capacity by increasing the efficiency of endocytic recycling.
MATERIALS AND METHODS
Identification of the RAB4B S-Y module
The generation of profiles used to search for S-Y sequences has been described (20). The profile used here is provided in Supplementary Figure 1A. Scans of the human Genomic Reference Sequence, build 29 (International Human Genome Sequencing Consortium), were performed with the program pfscan from the pftools package (available at ftp://ftp.isrec.isb-sib.ch/pub/software/unix/pftools/) which is a software implementation of the generalized profile method (21).
Cells
RJ2.2.5 B cells (22,23), SJO B cells (24), SJO cells complemented with RFX5 (25), RJ2.2.5 cells complemented as described (25,26) with lentiviral vectors encoding CIITA isoforms I, III or IV and Me67.8 melanoma cells (27) were grown in RPMI+Glutamax medium (Invitrogen) supplemented with 10% fetal calf serum and antibiotics. Me67.8 cells were induced with 200 U/ml of IFN-γ (Invitrogen). HUVEC were prepared as described (28) and induced with 1000 U/ml of IFN-γ (Invitrogen). Human monocyte-derived DC were prepared, matured with LPS and analyzed by FACS as described (29).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described using rabbit RFX, CREB and CIITA antisera (20,30). Results were quantified by real-time PCR as described (20) using the primers listed in Supplementary Table 1A. All results are presented as the mean ± SD of at least two independent ChIP experiments and three independent amplifications.
ChIP-on-chip experiments
The quality of CIITA-ChIP samples was verified by assessing the enrichment of HLA–DRA sequences. Under our conditions, the CIITA antibody precipitates ∼2% of the input HLA–DRA promoter fragments but <0.01% of non-specific DNA. DNA extracted from CIITA-ChIP samples were blunted for 30 min at 72°C with 3 U of Pfu polymerase (Promega) and phosphorylated with T4 Polynucleotide kinase (New England Biolabs). 120 pmoles of adaptors consisting of annealed oligonucleotides A (5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and B (5′-GAATTCAGATC-3′) were ligated to the DNA by overnight incubation at 16°C with 2000 U of T4 DNA ligase (New England Biolabs). Two rounds of PCR amplification with oligonucleotide A were performed using 1.25 U of Taq polymerase (New England Biolabs) and 0.025 U of Pfu Turbo polymerase (Stratagene). The cycle used was: 1 × (2′ at 55°C, 5′ at 72°C, 2′ at 95°C), 28 × (1′ at 95°C, 1′ at 60°C, 2′ at 72°C), 5′ at 72°C. Four micrograms of each DNA were purified and sent to NimbleGen Inc for probe preparation and hybridization. The arrays were either the NimbleGen 5 kb human promoter array set or a custom array of our own design carrying selected regions of human genomic DNA. Genomic sequences on the custom array are covered at high density with overlapping TM-matched oligonucleotides (∼50 bp long) spaced such that their 5′ ends are situated ∼10 bp apart. Each comparison between CIITA-ChIP and control probes was repeated at least twice.
Plasmids and reporter gene assays
The complete and minimal HLA–DRA luciferase constructs have been described (30). Plasmids containing the RAB4B S-Y and RAB4B Y-S sequences were created by replacing the MluI-BglII fragment spanning the S-Y region of HLA–DRA with the PCR fragments containing the RAB4B S-Y motif. Primers used to generate these constructs are listed in Supplementary Table 1B. Mutations were introduced into the RAB4B S-Y module by site-directed mutagenesis. Luciferase reporter gene assays were done as described (20). The results are presented as the mean ± SD of three assays.
mRNA quantification
RNA extraction, cDNA synthesis and real-time PCR were done as described (30). TBP mRNA was used for normalization. The results are presented as the mean ± SD of three independent quantifications. Primer sequences are listed in Supplementary Table 1C.
RESULTS
A typical MHC-II-like S-Y module is found upstream of the RAB4B gene
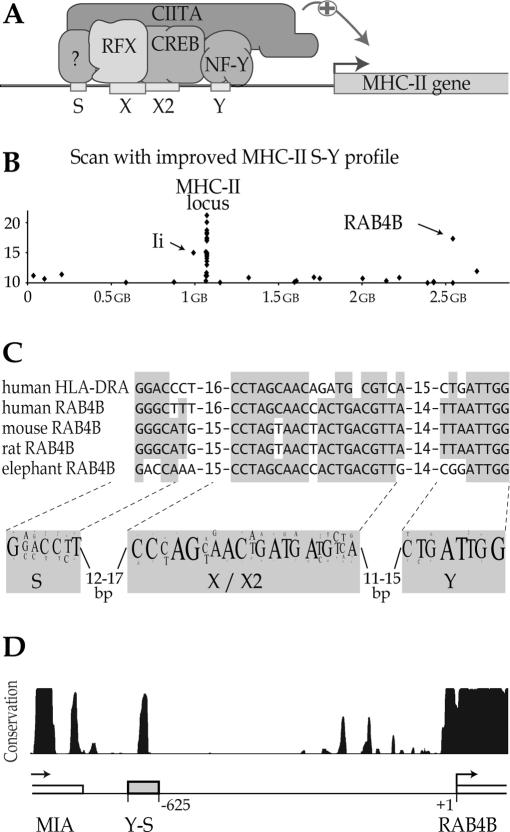
In man there are three classical MHC-II isotypes—HLA–DR, HLA–DP and HLA–DQ—each of which is a heterodimer consisting of an α chain and a β chain. There are also two non-classical MHC-II heterodimers—HLA–DO and HLA–DM—playing intracellular roles in peptide loading of the classical MHC-II molecules. The genes encoding the α and β chains of all classical and non-classical MHC-II isotypes, as well as the gene encoding the MHC-II-associated invariant chain (Ii), are tightly co-regulated by a highly conserved regulatory element called the S-X-X2-Y (S-Y) module, whichis located near to the transcription initiation site of each gene (Figure 1A) (31,32). Similar S-Y sequences also contribute, albeit to a lesser degree, to the expression of MHC-I genes (33).
Figure 1.
Identification of an S-Y module in the upstream region of the RAB4B gene. (A) Schematic drawing of the regulatory machinery controlling the transcription of MHC-II genes. The promoters of all MHC-II genes contain a conserved regulatory module (S-X-X2-Y) that is recognized by an enhanceosome complex consisting of RFX, CREB and NF-Y. Transcription is activated by recruitment of the co-activator CIITA to this enhanceosome. (B) The 3 kb upstream regions of all human genes were scanned with the MHC-II S-Y profile. Sequences bearing similarity to S-Y modules are plotted with respect to their position in the genome and the score attributed by the search algorithm. S-Y sequences in the RAB4B, invariant chain (Ii) and MHC-II genes are indicated. (C) Alignment between the human HLA–DRA S-Y module and the S-Y sequences found in the RAB4B genes of the indicated organisms. The most conserved residues are highlighted. A sequence logo representing the consensus MHC-II S-Y motif is shown below; the font size reflects the frequency at which a nucleotide is found at a given position. (D) Sequence conservation in the intergenic region between the RAB4B and MIA genes is represented by a graph derived from the Conservation track available at the UCSC Genome Browser at genome.ucsc.edu. The track shows a measure of the evolutionary conservation between 17 vertebrates. A peak corresponding to the most strongly conserved sequence in the intergenic region overlaps with the X–X2 region of the S-Y module.
The X, X2, Y and S sequences in the S-Y module are bound, respectively, by RFX, cAMP response element binding protein (CREB), nuclear factor Y (NF-Y) and an as yet unidentified S-binding factor (Figure 1A) (24,34–39). These transcription factors bind cooperatively to the S-Y module to form a multiprotein complex called the MHC-II enhanceosome, which constitutes a platform to which a transcriptional co-activator called the class II transactivator (CIITA) is recruited (Figure 1A) (40,41). CIITA coordinates most of the processes that are required to activate the transcription of MHC-II genes, including chromatin remodeling, recruitment of the general transcription machinery (GTM), promoter clearance by RNA polymerase II and transcription elongation (42,43).
The S-Y module is strictly conserved with respect to the sequence, orientation, position and spacing of its four subelements (31,32). This unique architecture allowed us to generate a stringent sequence profile that can be used to predict the presence of novel S-Y like sequences with a high degree of confidence. We previously validated this approach by identifying novel S-Y enhancers in the Ii gene and within the MHC-II locus (20). The sequence information derived from these new S-Y modules was incorporated into a new profile that improved the correlation between the score assigned by the search algorithm and the in vivo binding efficiency of CIITA and RFX (20).
We have used the improved profile to search for new S-Y modules in the 3 kb upstream regions of all human genes (Figure 1B). Outside of the MHC locus and Ii gene, the scan detected only 21 matches with scores greater than a value of 10. This threshold was chosen on the basis of the scores obtained for known bona fide target sequences of RFX and CIITA (20). Most of the 21 candidates were assigned relatively low scores (10 to 12). However, a putative S-Y module having a high score (17.3) was identified 625 bp upstream of the predicted transcription initiation site of the human RAB4B gene (Figure 1B).
The RAB4B S-Y motif exhibits strong resemblance to the S-Y sequences of MHC-II genes, as demonstrated by its alignment with the S-Y module of the prototypical HLA–DRA gene (Figure 1C). As observed when S-Y modules from different MHC-II genes are aligned, similarity between the RAB4B and HLA–DRA S-Y motifs is greatest in the core X and Y sequences. The RAB4B S-Y motif is highly conserved in the orthologous genes of other vertebrates (Figure 1C). In fact, sequence conservation between the upstream regions of vertebrate RAB4B genes is strongest at a position that coincides with the X-X2 sequence of the S-Y module (Figure 1D). This conservation is as strong as that of the core-promoter situated near the transcription start site.
The RAB4B S-Y module is a direct target of CIITA
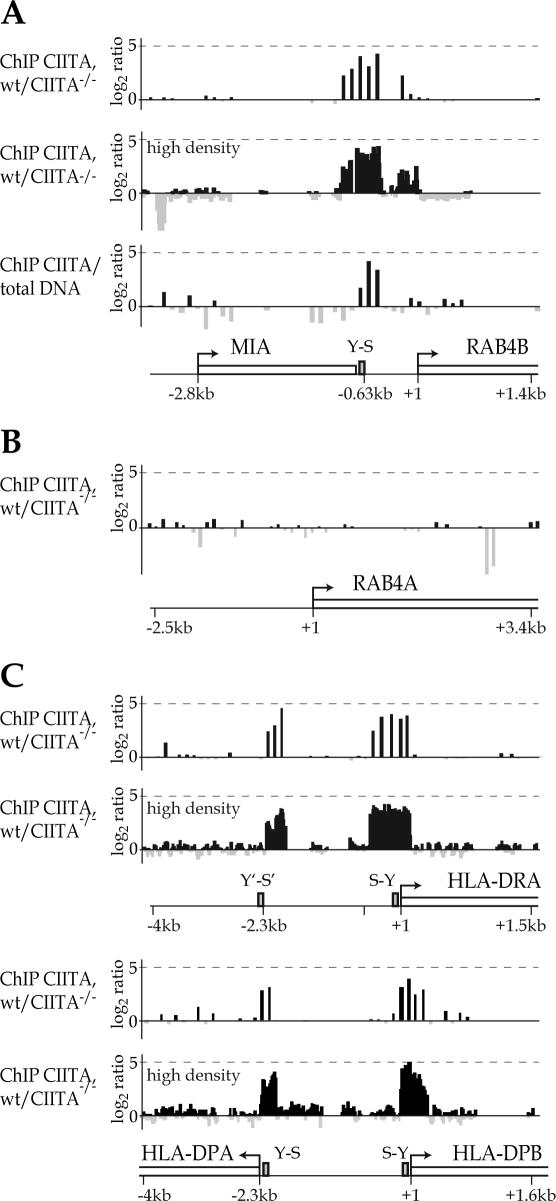
As a second approach for identifying new target genes regulated by the MHC-II regulatory machinery we used a ChIP-on-chip method relying on the use of microarrays (chip) to analyze the results of ChIP experiments (44). DNAs extracted from ChIP samples obtained with a CIITA-specific antiserum were used to prepare probes that were hybridized to mircoarrays carrying genomic sequences. Two types of arrays were used. The first was the human 5 kb promoter array set from NimbleGen Inc. which carries 5 kb promoter regions (−4 kb to +1 kb relative to transcription start site) of 27 434 human genes. The second was a high density array of our own design carrying selected genomic regions relevant to the regulation of MHC-II gene expression.
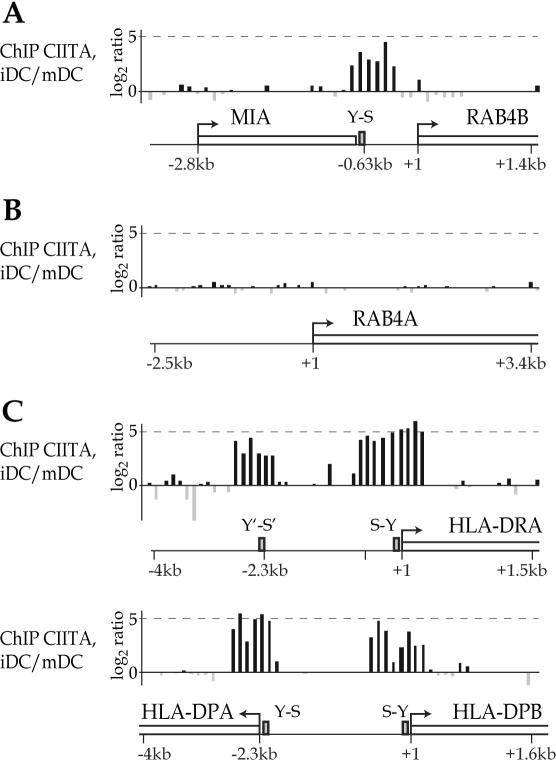
CIITA-ChIP samples were prepared from two different CIITA-positive cell types, a wild-type human B cell line (Raji) and human monocyte-derived DC. Hybridization signals obtained with these CIITA-ChIP probes were compared with those obtained with control probes. As controls for the B cell experiments we used genomic DNA or CIITA-ChIP samples prepared from a CIITA-deficient mutant of Raji (RJ2.2.5). As controls for the DC experiments we used CIITA-ChIP samples prepared from DC in which maturation had been induced by treatment with LPS for 24 h. Maturation of DC induces rapid silencing of the CIITA gene (45).
The ChIP-on-chip experiments revealed that CIITA binds efficiently to the upstream region of the RAB4B gene in B cells (Figure 2A) and immature DC (Figure 3A). The signals obtained for binding of CIITA coincide with the RAB4B S-Y module identified by the computer search. Specificity is demonstrated by the absence of signals in the upstream region of the closely related RAB4A gene, which does not contain an S-Y module (Figures 2B and 3B). The signals obtained for the RAB4B S-Y module are similar in strength to those observed for the typical S-Y modules found upstream of the HLA–DRA, HLA–DPA and HLA–DPB genes (Figures 2Cand 3C). The strength of the signals observed at RAB4B are well within the range of those obtained for the S-Y modules of other classical MHC-II genes, the non-classical HLA–DM and HLA–DO genes, and the Ii gene (Table 1).
Figure 2.
Binding of CIITA to the RAB4B promoter region in B cells. Binding of CIITA to the RAB4B (A), RAB4A (B) and HLA–DRA, HLA–DPA and HLA–DPB (C) promoter regions was assessed by ChIP-on-chip experiments. The results are represented as log2 ratios between the hybridization signals obtained for CIITA-ChIP DNAs derived from CIITA-positive B cells (wt) and those obtained with control DNAs. Controls were CIITA-ChIP DNAs from CIITA-/- B cells or input genomic DNA. Arrays were either the standard NimbleGen promoter arrays or a high density custom array. Each bar corresponds to a single oligonucleotide on the array. Grey bars correspond to log2 ratios lower than zero and reflect stronger signals for the control samples. The latter probably result from sporadic random amplification of short DNA fragments in the control samples. Schematic maps with positions of the genes (open boxes), transcription start sites (arrows) and S-Y modules (grey boxes) are shown.
Figure 3.
(A–C) Binding of CIITA to the RAB4B promoter region in DC. Binding of CIITA in immature DC (iDC) was assessed by ChIP-on-chip experiments as in Figure 2. The control hybridization was done with CIITA-ChIP DNA from mature DC (mDC).
Table 1.
Scores assigned by the profile search and binding of RFX and CIITA to S-Y modules
| S-Y modulea | Score | Binding of RFX (ChIP)b | Binding of CIITA (ChIP)b | Binding of CIITA in B cells (ChIP-on-chip)c | Binding of CIITA in DC (ChIP-on-chip)c | Binding of CIITA in B cells (ChIP-on-chip HD)c,d |
|---|---|---|---|---|---|---|
| DRB5 | 20.79 | —e | — | 59 | 40 | 57 |
| DRB1 | 18.73 | — | — | 81 | 45 | 57 |
| DRA | 18.15 | 100 | 100 | 100 | 100 | 100 |
| DRAe | 17.91 | 50 | 30 | 47 | 61 | 38 |
| DQB | 17.44 | — | — | 34 | 11 | 41 |
| RAB4B | 17.33 | 47 | 22 | 48 | 52 | 69 |
| DPA | 17.27 | 103 | 79 | 29 | 62 | 19 |
| #4 | 17.02 | 59 | 49 | 28 | 31 | 33 |
| Ii | 15.02 | 41 | 33 | 32 | 33 | 54 |
| DOB | 14.65 | — | 4 | 44 | 27 | 79 |
| DMA | 14.63 | — | — | 61 | 49 | 62 |
| DPB | 14.07 | 41 | 13 | 28 | 80 | 40 |
| DOA | 13.57 | — | 15 | 30 | 19 | 29 |
| #8 | 13.24 | 103 | 108 | — | — | 27 |
| DMB | 13.02 | 14 | 7 | 43 | 40 | 46 |
| #2 | 12.59 | 93 | 92 | — | — | 47 |
| Ii S′-Y′ | 12.45 | 80 | 19 | — | — | 28 |
| DQA | 11.18 | — | 3 | 19 | 18 | 1 |
| Ii Y′′-S′′ | 10.42 | 19 | 6 | — | — | 33 |
| #6 | 9.33 | 52 | 16 | — | — | 42 |
aS-Y modules found in the promoter-proximal regions of the indicated MHC-II genes, or in distal enhancers (DRAe, Ii S′-Y′, Ii Y′′-S′′, #2, #4, #6 and #8) described previously (20,30).
bin B cells, relative to DRA (set at 100%).
cvalues were calculated as peak surfaces (peak length multiplied by the average intensity obtained for separate probes) and represented relative to DRA (set at 100%).
dHD, high density array.
e—, not done (ChIP) or not on array (ChIP-on-chip).
The signals obtained for binding of CIITA in ChIP-on-chip and classical ChIP experiments vary substantially between the different promoter-proximal and -distal S-Y modules (Table 1). This variability is likely to reflect a combination of differences in multiple parameters, including enhanceosome stability, efficiency of CIITA recruitment, cell-type specific expression patterns, accessibility of the epitopes recognized by the antibodies and crosslinking efficiency.
In addition to RAB4B, the ChIP-on-chip experiments identified a relatively small number (>40) of other loci containing potential CIITA binding sites. None of these correspond to genes proposed previously to be targets of CIITA. As done here for RAB4B, these potential new target genes will have to be validated by classical ChIP experiments and expression studies comparing wild-type and CIITA-deficient cells.
RAB4B expression is regulated by CIITA
CIITA is called the master regulator of MHC-II genes because its pattern of expression dictates the cell-type specificity and level of MHC-II expression (32,42,46). APC such as B cells are MHC-II positive because they express CIITA constitutively. Most cells of non-hematopoietic origin do not express CIITA and are consequently MHC-II negative. However, the latter can generally be induced to express MHC-II genes by stimulation with interferon γ (IFN-γ), which activates the expression of CIITA.
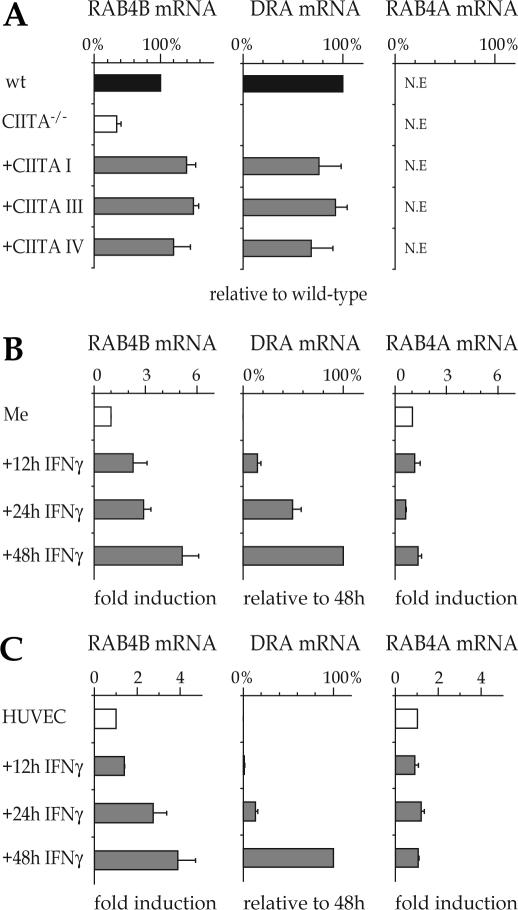
To determine whether RAB4B is regulated by CIITA in B cells we quantified RAB4B mRNA levels by real-time RT–PCR in the Raji B cell line, its isogenic CIITA-deficient mutant RJ2.2.5 and RJ2.2.5 cells complemented by transfection with CIITA expression vectors. Expression of RAB4B mRNA was reduced 3-4 fold in RJ.2.2.5 cells but was restored to levels exceeding those observed in wild-type Raji cells by complementation of the RJ2.2.5 cells with CIITA expression vectors (Figure 4A). All three isoforms of CIITA (isoforms I, III and IV) restored RAB4B expression. The dependence of RAB4B expression on CIITA is very similar to that observed for the HLA–DRA gene (Figure 4A). The only notable difference is that a basal level of RAB4B mRNA is retained in RJ2.2.5 whereas HLA–DRA expression is completely lost in these cells. This is reminiscent of human MHC-I genes, which also show only partially reduced expression in CIITA-deficient cells.
Figure 4.
Expression of RAB4B is controlled by CIITA and is induced by IFN-γ. mRNA levels for the RAB4B, HLA-DRA and RAB4A genes were determined for (A) wt B cells (Raji), CIITA-/- B cells (RJ2.2.5) and CIITA-/- cells complemented with expression vectors encoding the three isoforms of CIITA (CIITA I, III and IV), (B) Me67.8 melanoma cells (Me) induced with IFN-γ for 0, 12, 24 and 48 h and (C) HUVEC cells induced with IFN-γ for 0, 12, 24 and 48 h. Results were normalized using TBP mRNA. Values for RAB4B and RAB4A are expressed as % of the levels found in wt B cells (A) or relative to uninduced cells (B and C). Values for HLA–DRA mRNA are expressed as % of the levels found in wt B cells (A) or cells induced with IFN-γ for 48 h (B and C).
To determine whether RAB4B expression is induced by IFN-γ, we performed real-time RT–PCR experiments with a human melanoma cell line (Figure 4B) and primary human umbilical vein endothelial cells (HUVEC, Figure 4C). In both cell types, RAB4B mRNA expression is increased 4–5 fold by IFN-γ according to a time course that is very similar to the one observed for the robust induction of HLA–DRA mRNA. The major difference between the two induction patterns is that there is a basal level of RAB4B but not HLA–DRA mRNA in non-induced cells. This is again similar to MHC-I genes, which are expressed at a basal level in most cell types in the absence of IFN-γ induced CIITA expression.
No significant amount of RAB4A mRNA was detected in either Raji or RJ2.2.5 cells (Figure 4A). Furthermore, no increase in RAB4A mRNA abundance was observed in IFN-γ induced cells (Figure 4B and C). This is consistent with the fact that the RAB4A gene does not contain an S-Y module and is not bound by CIITA (Figures 2B and 3B). Regulation by CIITA is thus specific for the RAB4B gene.
We also analyzed expression of the MIA gene, which is located close to RAB4B (see Figures 1–3). MIA mRNA levels were negligibly low (>1000-fold less than RAB4B) in B cells (data not shown). Moreover, no significant basal or IFN-γ induced MIA expression was observed in HUVEC (data not shown). The MIA gene is thus not active in cells that express CIITA and is therefore not likely to be a target of the MHC-II regulatory machinery.
Enhanceosome assembly and CIITA recruitment at the RAB4B S-Y module
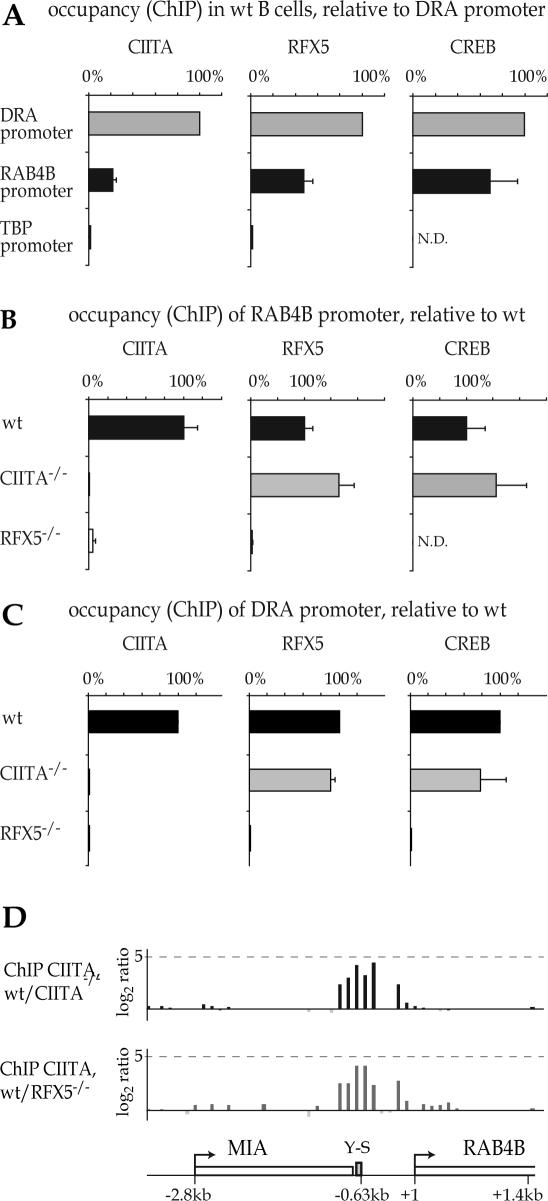
CIITA is recruited to the promoters of MHC-II genes by protein–protein interactions with the enhanceosome complex that assembles at their S-Y modules. To determine whether the MHC-II enhanceosome also assembles at the RAB4B S-Y module we performed ChIP experiments using antibodies directed against CIITA, RFX and CREB. All three factors associate with the RAB4B promoter in Raji B cells (Figure 5A). Occupation of the RAB4B promoter relative to the HLA–DRA promoter ranged from ∼20% for CIITA to ∼70% for CREB. Although lower, this occupation remains well within the range observed for known MHC-II genes, which is (as discussed above for CIITA) quite variable (Table 1). Specificity is demonstrated by the absence of detectable binding of the three factors to the promoter of the control TATA-binding protein (TBP) gene.
Figure 5.
Occupation of the RAB4B promoter by the MHC-II specific regulatory machinery. (A) The association of CIITA, RFX5 and CREB with the HLA–DRA, RAB4B and TBP promoters was assessed by ChIP in wt B cells. Results are expressed as % of occupation observed at the HLA-DRA promoter. (B) Occupancy of the RAB4B promoter by CIITA, RFX5 and CREB was analyzed in wt B cells and in mutant B cells lacking CIITA or RFX5. Results are expressed as % of the occupation observed in the wt cells. (C) Occupancy of the HLA–DRA promoter was analyzed as in (B). (D) Binding of CIITA to the RAB4B promoter was analyzed by ChIP-on-chip. Results are represented as ratios between the signals obtained with CIITA-ChIP DNAs from wt and CIITA-/- B cells (top) or wt and RFX5-/- B cells (bottom). N.D., not done.
RFX is a hetero-trimeric factor consisting of RFX5, RFX associated protein (RFXAP) and RFX associated protein containing ankyrin repeats (RFXANK) (24,34–36). Mutations in any one of these three subunits abolish binding of the RFX complex, and thus eliminate both enhanceosome assembly and CIITA recruitment at the S-Y modules of MHC-II genes (Figure 5C) (30,41). Conversely, mutations in CIITA do not abrogate binding of RFX and enhanceosome assembly (Figure 5C) (30). A similar pattern is observed at the RAB4B S-Y module. Binding of CIITA and RFX5 are completely lost in RFX5-deficient SJO cells. This is revealed by classical ChIP experiments using antibodies against CIITA and RFX5 (Figure 5B) and ChIP-on-chip experiments comparing binding of CIITA in wild-type and SJO cells (Figure 5D). On the other hand, RFX5 and CREB bind normally to the RAB4B S-Y module in CIITA-deficient B cells, indicating that enhanceosome assembly is independent of CIITA (Figure 5B). These results indicate that assembly of the enhanceosome and recruitment of CIITA at the RAB4B promoter follows the same rules as those previously established for MHC-II genes.
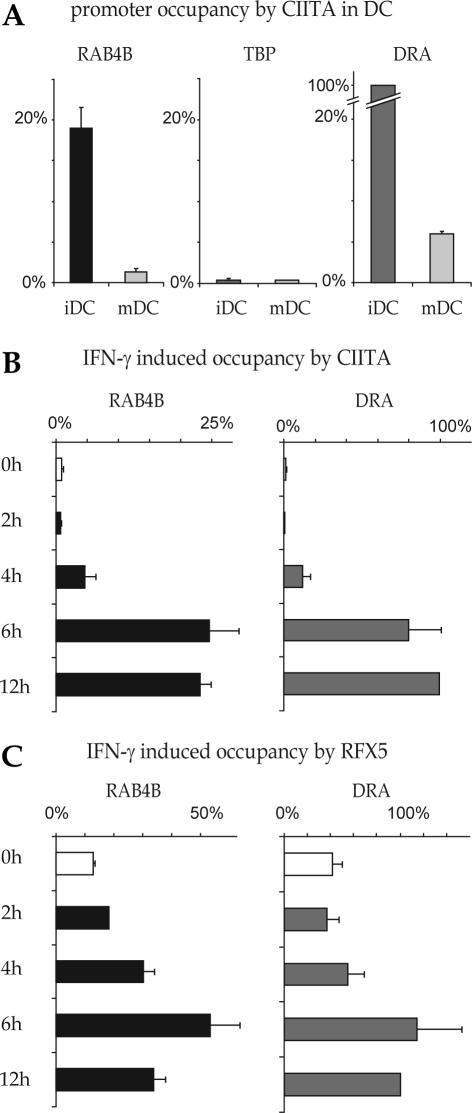
RAB4B promoter occupancy by RFX and CIITA in DC and IFN-γ induced cells
The maturation of DC is accompanied by rapid silencing of the CIITA gene. Binding of CIITA to the HLA–DRA promoter in ChIP experiments is consequently strongly reduced in DC that have been induced to mature by exposure to LPS (Figure 6A). This reduction is also evident at the RAB4B promoter, where the occupancy by CIITA is reduced more than 10-fold during DC maturation (Figure 6A).
Figure 6.
Occupation of the RAB4B promoter by the MHC-II regulatory machinery in DC and IFN-γ induced cells. (A) Binding of CIITA to the RAB4B, HLA-DRA and TBP promoters was analyzed by ChIP in DC before (iDC) and after (mDC) maturation with LPS. Results are represented as % of the occupation observed at the HLA-DRA promoter in iDC. The TBP promoter was used as a negative control. (B and C) ChIP was used to measure occupancy of the RAB4B promoter by CIITA (B) and RFX5 (C) in Me67.8 melanoma cells induced with IFN-γ for 0, 2, 4, 6 and 12 h. Results are represented as % of the occupation observed at the HLA–DRA promoter after 12 h of induction.
Binding of CIITA to the RAB4B promoter was also assessed in IFN-γ induced melanoma cells (Figure 6B and C).IFN-γ induced occupancy of the RAB4B promoter by CIITA follows a time course that is identical to that observed for the HLA–DRA promoter. Binding is not detected prior to induction, first becomes evident after 4 h and reaches a plateau by 6 h (Figure 6B). At the HLA–DRA promoter, stability of the enhanceosome is enhanced by CIITA in IFN-γ induced cells. This manifests itself in ChIP experiments by a 2–3 fold increase in the signals obtained for binding of RFX5 (Figure 6C). A similar increase in RFX occupancy is detected at the RAB4B promoter (Figure 6C). The time course of the IFN-γ induced increase in RFX binding at the RAB4B promoter closely parallels that observed at the HLA–DRA promoter.
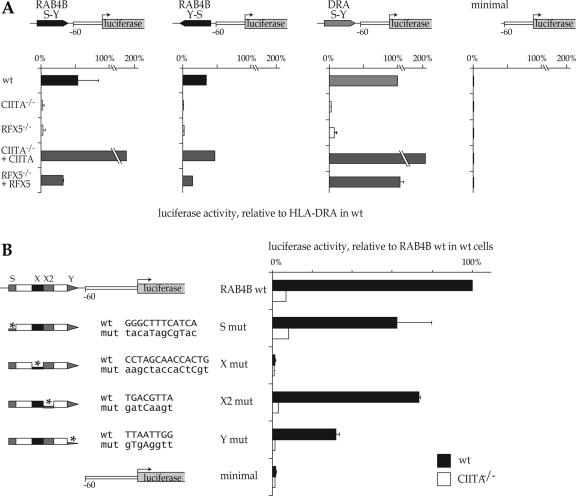
The RAB4B S-Y module functions as a CIITA and RFX-dependent enhancer
To confirm that the RAB4B S-Y module functions as an RFX- and CIITA-dependent enhancer we performed luciferase reporter gene assays using a strategy that was first devised to validate the novel S-Y enhancers discovered in the MHC-II locus and Ii gene (20,30). We generated reporter gene constructs in which the S-Y module of the HLA–DRA promoter was replaced with the RAB4B S-Y module in the forward (S-Y) or reverse (Y-S) orientation (Figure 7A). These two RAB4B constructs, the HLA–DRA construct and a control construct driven by a minimal DRA promoter lacking an S-Y module were transfected into five cell types; wild-type Raji B cells, CIITA-deficient RJ2.2.5 cells, RFX5-deficient SJO cells, RJ2.2.5 cells corrected by complementation with CIITA and SJO cells corrected by complementation with RFX5 (Figure 7A). In wild- type Raji cells, the RAB4B S-Y and Y-S constructs attained activities exceeding 50 and 30%, respectively, of that observed for the HLA–DRA construct. Although lower than for HLA–DRA, these levels were nevertheless 30-fold greater than the basal activity of the minimal promoter construct. Both RAB4B constructs exhibited a pattern of activity in the different cell types that was very similar to that observed for the HLA–DRA construct. The strong expression observed in wild-type B cells was reduced to negligible levels in the mutant B cells lacking CIITA or RFX. This loss in expression was restored by re-introducing CIITA or RFX in the corresponding mutant cells. Taken together, these results indicate that the RAB4B S-Y module functions as transcriptional enhancer regulated by RFX and CIITA.
Figure 7.
The RAB4B S-Y module functions as a CIITA and RFX-dependent transcriptional enhancer. (A) Transient transfections were performed with luciferase reporter gene constructs containing the HLA–DRA regulatory region or hybrid promoters in which the S-Y module from HLA–DRA was replaced with the RAB4B S-Y sequence in the forward or inverse orientations. A construct containing the basal HLA–DRA promoter lacking the S-Y module (minimal) was used as a negative control. Constructs were transfected into wt, CIITA-/- and RFX5-/- B cells, and into CIITA-/- and RFX-/- cells complemented with CIITA or RFX5 expression vectors, respectively. Results are expressed as % of the activity of the HLA–DRA S-Y construct in wt cells. (B) Mutations were introduced into the S, X, X2 and Y sequences of the luciferase construct containing the RAB4B S-Y module. The wt and mutated S, X, X2 and Y sequences are provided. Activities of the resulting constructs were tested after transfection into wt and CIITA-/- cells. The minimal construct was used as a negative control. Results are expressed as % of the activity of intact RAB4B construct in wt cells.
To confirm that the RAB4B S-Y module functions as a typical S-Y enhancer we performed reporter gene assays with constructs carrying mutations in its S, X, X2 or Y sequences (Figure 7B). The mutations were chosen on the basis of their previously documented ability to affect activity of the HLA–DRA S-Y module and to interfere with assembly of the enhanceosome complex. All four mutations decreased activity of the RAB4B construct in Raji cells (Figure 7B). The X mutation abrogated activity completely, underscoring the pivotal importance of the RFX target sequence. The Y, S and X2 box mutations decreased activity by approximately 70%, 40% and 30%, respectively. The remaining activities of the mutated Y, S and X2 constructs were eliminated in CIITA-deficient cells and must thus result from partial CIITA recruitment. These results are in full agreement with previous studies on MHC-II promoters, which have demonstrated that destruction of the X box is always extremely detrimental whereas mutations in the S, X2 and Y motifs generally inhibit promoter function only partially (39,47).
DISCUSSION
We have used two strategies to identify genes that are co-regulated with MHC-II genes. First, we used a computer scan to search for genes that contain MHC-II-like S-Y enhancers in their upstream regions. Second, we performed ChIP-on-chip screens to identify genes that are direct targets of CIITA, the master regulator of MHC-II genes. These two approaches have converged on RAB4B, a gene encoding a protein implicated in endosome recycling. A combination of ChIP, ChIP-on-chip, expression analysis and functional studies in wild-type and mutant cells lacking CIITA or RFX have confirmed that RAB4B expression is indeed driven by a typical S-Y enhancer that is regulated by both CIITA and the MHC-II enhanceosome complex composed of RFX, CREB and NF-Y.
Searching for potential binding sites in genomic DNA sequences is generally of limited usefulness for identifying target genes regulated by specific transcription factors. A major problem with such approaches is that transcription factor binding sites are often short and degenerate with respect to their consensus sequence, such that computer searches for potential binding sites tend to yield overwhelmingly large numbers of irrelevant hits. Our searches with the MHC-II S-Y profile constitute an exception to this rule because they have proved to be remarkably reliable—in this report and in a previous study (20)—for the identification of novel enhancers regulated by the MHC-II enhanceosome complex and CIITA. The success of our approach implies that similar strategies could be valuable for identifying genes regulated by higher order transcription factor complexes, particularly in systems where gene expression is controlled by well-defined composite regulatory modules.
The use of genome-scale ChIP-on-chip-based binding studies to identify target genes of specific transcription factors in eukaryotic organisms has become feasible only recently thanks to the availability of microarrays covering entire genomes, large genomic regions or the promoter regions of comprehensive sets of genes. To date, ChIP-on-chip studies have investigated the target gene specificity of DNA-binding transcription factors (48–52). We demonstrate here that such ChIP-on-chip experiments can also be very powerful for studying the target genes of non-DNA-binding transcriptional co-activators such as CIITA.
Past efforts to identify new genes regulated by CIITA have led to a certain amount of controversy concerning the target gene specificity of CIITA (53). CIITA was initially believed to be dedicated for the expression of classical MHC-II genes and related genes implicated in antigen presentation, including the Ii, HLA–DO, HLA–DM and MHC-I genes. Subsequent reports suggested that CIITA might be more pleiotropic in its function. Several genes were suggested to be repressed by CIITA in various cell types, including those encoding IL4 and FasL in T cells, cathepsin E and IL-10 in B cells and DC, and collagen type I α2, thymidine kinase and cyclin D1 in IFN-γ induced cells (54–57). A microarray experiment suggested that at least 16 other genes of diverse functions are repressed by CIITA in a human B cell line (58). Finally, microarray experiments have suggested that CIITA enhances the expression of Plexin-A1 in mouse DC and numerous other genes, including RAB4B, in a human B cell line and in IFN-γ induced cells (58,59). However, none of these candidate genes have as yet been demonstrated to be controlled by S-Y enhancers or been validated as direct targets of the MHC-II regulatory machinery by ChIP experiments. In fact, for several candidates an indirect mechanism involving sequestration of the general co-activator CBP by CIITA has been proposed (54–57). Moreover, conflicting reports concerning their dependence on CIITA have been published for several candidates (60). Therefore, with the exception of the Ii gene, the RAB4B gene identified here is the first example of a non-MHC gene that contains a typical S-Y enhancer and is regulated by the same regulatory machinery as MHC-II genes.
RAB4 has a well established function in recycling proteins and lipids from EE and RE back to the PM. The RAB4A and RAB4B isoforms co-localize to the same compartments and are highly homologous, suggesting that they have largely redundant functions in recycling. Enhancing RAB4B expression by placing it under the control of the MHC-II regulatory machinery may thus simply be a way of increasing the recycling capacity of APC by raising the total level of RAB4 protein. However, an alternative possibility is suggested by an intriguing difference between the two RAB4 isoforms at amino acid 199. In RAB4A there is a serine (S) at this position. Phosphorylation of S199 by the mitotic cdc2 kinase causes the dissociation of RAB4A from membranes and its accumulation in the cytoplasm (61,62). This was proposed to be responsible for the inactivation of RAB4A during mitosis. Interestingly, RAB4B has a glutamine (Q) at position 199, a substitution that was shown to prevent the phosphorylation and cytoplasmic accumulation of RAB4A. RAB4B is thus likely to remain active during mitosis. This difference could underlie a specific function of RAB4B. Regulating RAB4B expression by the MHC-II regulatory machinery may thus be a way of ensuring this specific function in APC.
Endocytic recycling contributes to antigen presentation by MHC-II molecules. Cell surface MHC-II molecules can be internalized by endocytosis, loaded with new peptides in early endosomes and recycled back to the PM instead of being targeted for lysosomal degradation (9). Certain MHC-II restricted peptides do not require extensive processing in late endosomal compartments and are generated and loaded onto MHC-II molecules in early endosomes, from which the MHC-II-peptide complexes are recycled to the cell surface (10–12). This recycling-dependent antigen presentation pathway is presumably under the control of RAB4. Direct evidence for a role of RAB4 in antigen presentation has been provided by studies using a dominant negative mutant of RAB4A. Transfection of mouse A20 B cells with a dominant negative RAB4A mutant was found to inhibit the processing and presentation of certain antigens internalized by B-cell-receptor (BCR) or Fc-receptor-mediated uptake (13). Interestingly, we have found that A20 B cells actually express only RAB4B mRNA (data not shown). The dominant negative RAB4A mutant must therefore have interfered with the activity of RAB4B, thereby providing a functional link between the latter and antigen presentation.
In addition to MHC-II restricted antigen presentation, recycling is known to play key roles in other antigen presentation processes. Several studies have demonstrated that recycling is one of the mechanisms that accounts for the ability of DC to cross-present peptides derived from exogenous antigens in the context of MHC-I molecules (7,14,15). DC have recently also been shown to internalize antigens and the recycle them back to PM for direct presentation to B cells (16). Enhancing the expression of RAB4B in APC by the MHC-II regulatory machinery is thus likely to contribute to the efficiency of several different antigen presentation processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are very grateful to M. Strubin for helpful discussions and to P. Bradfield and S. Jemelin for providing us with HUVEC cells. Work in the laboratory of Walter Reith was supported by grants from the Swiss National Science foundation. Funding to pay the Open Access publication charges for this article was provided by grant 3100A0-105895.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zerial M., McBride H. Rab proteins as membrane organizers. Nature Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 2.Pfeffer S.R. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 3.Jordens I., Marsman M., Kuijl C., Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–1077. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 4.Cheng K.W., Lahad J.P., Gray J.W., Mills G.B. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005;65:2516–2519. doi: 10.1158/0008-5472.CAN-05-0573. [DOI] [PubMed] [Google Scholar]
- 5.Stein M.P., Dong J., Wandinger-Ness A. Rab proteins and endocytic trafficking: potential targets for therapeutic intervention. Adv. Drug Deliv. Rev. 2003;55:1421–1437. doi: 10.1016/j.addr.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Lehner P.J., Cresswell P. Processing and delivery of peptides presented by MHC class I molecules. Curr. Opin. Immunol. 1996;8:59–67. doi: 10.1016/s0952-7915(96)80106-3. [DOI] [PubMed] [Google Scholar]
- 7.Trombetta E.S., Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 8.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 9.Robinson J.H., Delvig A.A. Diversity in MHC class II antigen presentation. Immunology. 2002;105:252–262. doi: 10.1046/j.0019-2805.2001.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinet V., Vergelli M., Martin R., Bakke O., Long E.O. Antigen presentation mediated by recycling of surface HLA–DR molecules. Nature. 1995;375:603–606. doi: 10.1038/375603a0. [DOI] [PubMed] [Google Scholar]
- 11.Lindner R., Unanue E.R. Distinct antigen MHC class II complexes generated by separate processing pathways. EMBO J. 1996;15:6910–6920. [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak S.S., Blum J.S. Endocytic recycling is required for the presentation of an exogenous peptide via MHC class II molecules. Traffic. 2000;1:561–569. doi: 10.1034/j.1600-0854.2000.010706.x. [DOI] [PubMed] [Google Scholar]
- 13.Lazzarino D.A., Blier P., Mellman I. The monomeric guanosine triphosphatase rab4 controls an essential step on the pathway of receptor-mediated antigen processing in B cells. J. Exp. Med. 1998;188:1769–1774. doi: 10.1084/jem.188.10.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groothuis T.A., Neefjes J. The many roads to cross-presentation. J. Exp. Med. 2005;202:1313–1318. doi: 10.1084/jem.20051379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ackerman A.L., Cresswell P. Cellular mechanisms governing cross-presentation of exogenous antigens. Nature Immunol. 2004;5:678–684. doi: 10.1038/ni1082. [DOI] [PubMed] [Google Scholar]
- 16.Bergtold A., Desai D.D., Gavhane A., Clynes R. Cell surface recycling of internalized antigen permits dendritic cell priming of B cells. Immunity. 2005;23:503–514. doi: 10.1016/j.immuni.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 17.van der Sluijs P., Hull M., Webster P., Male P., Goud B., Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 18.Novick P., Zerial M. The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 19.Rzomp K.A., Scholtes L.D., Briggs B.J., Whittaker G.R., Scidmore M.A. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczyk M., Peyraud N., Rybtsova N., Masternak K., Bucher P., Barras E., Reith W. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J. Immunol. 2004;173:6200–6210. doi: 10.4049/jimmunol.173.10.6200. [DOI] [PubMed] [Google Scholar]
- 21.Bucher P., Karplus K., Moeri N., Hofmann K. A flexible motif search technique based on generalized profiles. Comput. Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 22.Accolla R.S. Human B cell variants immunoselected against a single Ia antigen subset have lost expression in several Ia antigen subsets. J. Exp. Med. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steimle V., Otten L.A., Zufferey M., Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 24.Steimle V., Durand B., Barras E., Zufferey M., Hadam M.R., Mach B., Reith W. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (Bare Lymphocyte Syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 25.Villard J., Peretti M., Masternak K., Barras E., Caretti G., Mantovani R., Reith W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell Biol. 2000;20:3364–3376. doi: 10.1128/mcb.20.10.3364-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krawczyk M., Masternak K., Zufferey M., Barras E., Reith W. New functions of the major histocompatibility complex class II-specific transcription factor RFXANK revealed by a high-resolution mutagenesis study. Mol. Cell Biol. 2005;25:8607–8618. doi: 10.1128/MCB.25.19.8607-8618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlethaler-Mottet A., Otten L.A., Steimle V., Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson-Leger C.A., Aurrand-Lions M., Beltraminelli N., Fasel N., Imhof B.A. Junctional adhesion molecule-2 (JAM-2) promotes lymphocyte transendothelial migration. Blood. 2002;100:2479–2486. doi: 10.1182/blood-2001-11-0098. [DOI] [PubMed] [Google Scholar]
- 29.LeibundGut-Landmann S., Waldburger J.M., Reis e Sousa C., Acha-Orbea H., Reith W. MHC class II expression is differentially regulated in plasmacytoid and conventional dendritic cells. Nature Immunol. 2004;5:899–908. doi: 10.1038/ni1109. [DOI] [PubMed] [Google Scholar]
- 30.Masternak K., Peyraud N., Krawczyk M., Barras E., Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nature Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 31.Reith W., Mach B. The bare lymphocyte syndrome and the regulation of mhc expression. Annu. Rev. Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 32.Ting J.P., Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl.):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 33.van den Elsen P.J., Holling T.M., Kuipers H.F., van der Stoep N. Transcriptional regulation of antigen presentation. Curr. Opin. Immunol. 2004;16:67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Durand B., Sperisen P., Emery P., Barras E., Zufferey M., Mach B., Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masternak K., Barras E., Zufferey M., Conrad B., Corthals G., Aebersold R., Sanchez J.C., Hochstrasser D.F., Mach B., Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nature Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 36.Nagarajan U.M., Louis-Plence P., DeSandro A., Nilsen R., Bushey A., Boss J.M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 37.Moreno C.S., Beresford G.W., Louis-Plence P., Morris A.C., Boss J.M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 38.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 39.Muhlethaler-Mottet A., Krawczyk M., Masternak K., Spilianakis C., Kretsovali A., Papamatheakis J., Reith W. The S box of major histocompatibility complex class II promoters is a key determinant for recruitment of the transcriptional co-activator CIITA. J. Biol. Chem. 2004;279:40529–40535. doi: 10.1074/jbc.M406585200. [DOI] [PubMed] [Google Scholar]
- 40.Zhu X.S., Linhoff M.W., Li G., Chin K.C., Maity S.N., Ting J.P. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB To cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masternak K., Muhlethaler-Mottet A., Villard J., Zufferey M., Steimle V., Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 42.Reith W., LeibundGut-Landmann S., Waldburger J.M. Regulation of MHC class II gene expression by the class II transactivator. Nature Rev. Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 43.Zika E., Ting J.P. Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. Curr. Opin. Immunol. 2005;17:58–64. doi: 10.1016/j.coi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Ren B., Dynlacht B.D. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Meth. Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 45.Landmann S., Muhlethaler-Mottet A., Bernasconi L., Suter T., Waldburger J.M., Masternak K., Arrighi J.F., Hauser C., Fontana A., Reith W. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class ii transactivator (ciita) expression. J. Exp. Med. 2001;194:379–392. doi: 10.1084/jem.194.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boss J.M., Jensen P.E. Transcriptional regulation of the MHC class II antigen presentation pathway. Curr. Opin. Immunol. 2003;15:105–111. doi: 10.1016/s0952-7915(02)00015-8. [DOI] [PubMed] [Google Scholar]
- 47.Tsang S.Y., Nakanishi M., Peterlin B.M. Mutational analysis of the DRA promoter: cis-acting sequences and trans-acting factors. Mol. Cell Biol. 1990;10:711–719. doi: 10.1128/mcb.10.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beima K.M., Miazgowicz M.M., Lewis M.D., Yan P.S., Huang T.H., Weinmann A.S. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J. Biol. Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber J., Jenner R.G., Murray H.L., Gerber G.K., Gifford D.K., Young R.A. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl Acad. Sci. USA. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 53.LeibundGut-Landmann S., Waldburger J.M., Krawczyk M., Otten L.A., Suter T., Fontana A., Acha-Orbea H., Reith W. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 54.Zhu X.S., Ting J.P. A 36-amino-acid region of CIITA is an effective inhibitor of CBP: novel mechanism of gamma interferon-mediated suppression of collagen alpha(2)(i) and other promoters. Mol. Cell Biol. 2001;21:7078–7088. doi: 10.1128/MCB.21.20.7078-7088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yee C.S., Yao Y., Li P., Klemsz M.J., Blum J.S., Chang C.H. Cathepsin E: a novel target for regulation by class II transactivator. J. Immunol. 2004;172:5528–5534. doi: 10.4049/jimmunol.172.9.5528. [DOI] [PubMed] [Google Scholar]
- 56.Sisk T.J., Gourley T., Roys S., Chang C.H. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF–AT to bind the co-activator CREB binding protein (CBP)/p300. J. Immunol. 2000;165:2511–2517. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- 57.Gourley T.S., Patel D.R., Nickerson K., Hong S.C., Chang C.H. Aberrant expression of Fas ligand in mice deficient for the MHC class II trans-activator. J. Immunol. 2002;168:4414–4419. doi: 10.4049/jimmunol.168.9.4414. [DOI] [PubMed] [Google Scholar]
- 58.Nagarajan U.M., Bushey A., Boss J.M. Modulation of gene expression by the MHC class II trans-activator. J. Immunol. 2002;169:5078–5088. doi: 10.4049/jimmunol.169.9.5078. [DOI] [PubMed] [Google Scholar]
- 59.Wong A.W., Brickey W.J., Taxman D.J., van Deventer H.W., Reed W., Gao J.X., Zheng P., Liu Y., Li P., Blum J.S., et al. CIITA-regulated plexin-A1 affects T-cell–dendritic cell interactions. Nature Immunol. 2003;4:891–898. doi: 10.1038/ni960. [DOI] [PubMed] [Google Scholar]
- 60.Otten L.A., Leibundgut-Landmann S., Huarte J., Kos-Braun I.C., Lavanchy C., Barras E., Borisch B., Steimle V., Acha-Orbea H., Reith W. Revisiting the specificity of the MHC class II trans-activator CIITA in vivo. Eur. J. Immunol. 2006;36:1548–1558. doi: 10.1002/eji.200535687. [DOI] [PubMed] [Google Scholar]
- 61.van der Sluijs P., Hull M., Huber L.A., Male P., Goud B., Mellman I. Reversible phosphorylation–dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J. 1992;11:4379–4389. doi: 10.1002/j.1460-2075.1992.tb05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gerez L., Mohrmann K., van Raak M., Jongeneelen M., Zhou X.Z., Lu K.P., van Der Sluijs P. Accumulation of rab4GTP in the cytoplasm and association with the peptidyl–prolyl isomerase pin1 during mitosis. Mol. Biol. Cell. 2000;11:2201–2211. doi: 10.1091/mbc.11.7.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]