Abstract
Signaling-protein mRNAs tend to have long untranslated regions (UTRs) containing binding sites for RNA-binding proteins regulating gene expression. Here we show that a PUF-family RNA-binding protein, Mpt5, represses the yeast MAP-kinase pathway controlling differentiation to the filamentous form. Mpt5 represses the protein levels of two pathway components, the Ste7 MAP-kinase kinase and the Tec1 transcriptional activator, and negatively regulates the kinase activity of the Kss1 MAP kinase. Moreover, Mpt5 specifically inhibits the output of the pathway in the absence of stimuli, and thereby prevents inappropriate cell differentiation. The results provide an example of what may be a genome-scale level of regulation at the interface of signaling networks and protein-RNA binding networks.
Introduction
Mitogen-activated protein-kinase (MAPK) signal-transduction pathways function in biological responses ranging from yeast cell differentiation to mammalian immunity [1]. In diploid cells of the budding yeast, Saccharomyces cerevisiae, growth on low-ammonium solid media stimulates a signal-transduction network [2] mediating a switch from the ovoid single-cell yeast form of growth to a pathogen-like filamentous form characterized by cell elongation, unipolar distal budding to form cell chains, and substrate invasion [3]. A central module of this network is the filamentation MAP-kinase (fMAPK) pathway [4]. The fMAPK cascade includes the kinases Ste11, Ste7, and the Kss1 MAP kinase. Activated Kss1 derepresses the Ste12 Tec1 transcription-factor heterodimer [5], [6], which activates filamentation genes [7], [8] including TEC1 itself.
The yeast MPT5 gene, also known as HTR1 [9], UTH4 [10], and PUF5 [11], encodes a Pumilio and fem-3 binding (PUF) family RNA-binding protein required for multiple cellular processes. This is apparent in the pleiotropic phenotype of mpt5 mutants. MPT5 is required for longevity [10], [12], cell-wall integrity [13], [14], recovery from mating-pheromone arrest [15], [16], stress tolerance [9], [17], and post-transcriptional regulation of the HO gene [18]. The Mpt5 protein could affect these diverse processes through its ability to bind various mRNAs [19], or through its interactions with other proteins such as MAP kinases [15].
In the fruit fly, Drosophila melanogaster, the pumilio gene, a namesake of the PUF gene family, encodes a repressor of the translation of germ-line cell-differentiation proteins [20]. In yeast, links between MPT5 and filamentous-form cell differentiation are suggested by network analyses [2], [4] integrating multiple data sources. In addition, we noticed in a genome-wide RNA-binding data set [19] that several of the 224 mRNAs bound by Mpt5 encode key filamentation signaling proteins. Based on these observations, we investigated a possible functional connection between Mpt5 and yeast filamentation. Here, we have established a major regulatory role for the Mpt5 RNA-binding protein in yeast cell differentiation. Specifically, we found that Mpt5 prevents inappropriate cell differentiation through the inhibition of fMAPK pathway activity.
Results
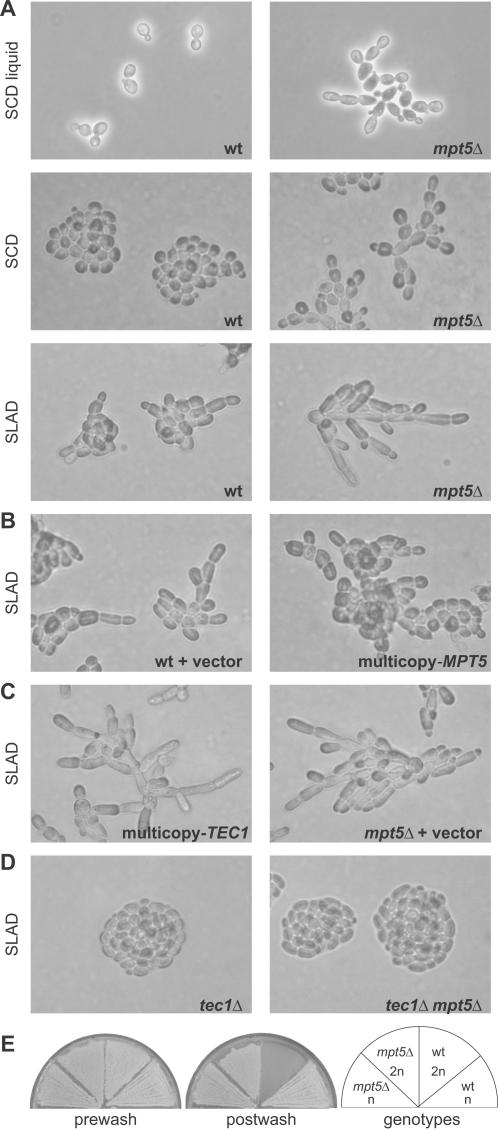
We found that the MPT5 gene encodes a repressor of yeast cell differentiation to the filamentous form. Deletion of MPT5 from filamentation-competent diploid yeast results in a constitutively filamentous phenotype. Mutant mpt5Δ yeast filament in the absence of filamentation stimuli, and are hyper-filamentous under filamentous-form growth conditions (Fig. 1A). Overexpression of the MPT5+ gene on a multicopy plasmid (Materials and Methods) antagonizes filamentation under filamentous-form growth conditions (Fig. 1B). The striking phenotypes of these mutants suggest that MPT5 may exert its effects through regulation of the fMAPK pathway. Supporting this suggestion, the mpt5Δ phenotype resembles the multicopy TEC1+ phenotype (Fig. 1C) and requires an intact TEC1 gene (Fig. 1D).
Figure 1. Repression of yeast filamentous-form phenotypes by MPT5.
(A–D) Diploid yeast were grown under yeast-form conditions (SCD liquid and SCD) or filamentous-form conditions (SLAD) and microscopically imaged. (E) Patches of yeast were grown on rich medium agar and subjected to a washing-off assay of adhesion.
MPT5 is also a repressor of adhesion, a filamentation-associated process. Haploid yeast cells adhere to and invade an agar surface when grown for an extended time in patches on rich medium [21]. The fMAPK pathway activates this adhesion [21], which is repressed in diploids by the mating-type genes and by increased ploidy [22]. In an assay of resistance to washing off a rich-medium agar surface (Materials and Methods), wild-type diploids wash off readily whereas mpt5Δ diploids adhere avidly (Fig. 1E); haploids serve as positive controls. Thus, deletion of MPT5 derepresses two fMAPK-related phenotypes, filamentous-form growth and adhesion.
Mpt5, a PUF protein, binds to the mRNAs of several filamentation signaling genes, including PHD1, RAS2, and STE7 [19]. The epistasis of tec1Δ to mpt5Δ (Fig. 1D), and the strong resemblance of the mpt5Δ and multicopy TEC1+ phenotypes (Figs. 1A and C), led us to investigate the possibility of a link of MPT5 with TEC1. We found that the TEC1 mRNA immunoprecipitates with Mpt5 protein in vivo (Fig. 2). We appended a 13-myc epitope tag to the endogenous MPT5 gene. Tagged Mpt5 protein was immunoprecipitated from diploid yeast cells (Text S1). To detect mRNAs, the immunoprecipitate was subjected to reverse transcription and polymerase chain reaction. PHD1 served as a positive control. Negative-control experiments lacking either reverse transcriptase or the 13-myc tag assured that the detected sequences were neither DNA nor unbound co-purifying mRNA. No other fMAPK pathway components were tested; it remains possible that the mRNAs of other components are bound by Mpt5.
Figure 2. In vivo TEC1 mRNA binding by Mpt5.
An immunoprecipitate of epitope-tagged Mpt5 protein was subjected to reverse transcription and gene-specific polymerase chain reaction to detect the presence of bound mRNAs. Control experiments lacked either the epitope tag or reverse transcriptase.
The results suggest that the repression of yeast cell differentiation by the Mpt5 protein is due to effects on the fMAPK pathway. Nonetheless, the repression of filamentation by MPT5 might involve the binding of the Mpt5 protein to the mRNAs of major regulators of filamentation that are outside the fMAPK pathway [19], notably Phd1, a transcription factor whose overexpression induces filamentous growth [23], and Ras2, a GTPase whose activation stimulates filamentation by activating the fMAPK and cyclic-AMP/protein-kinase-A pathways [3]. However, in contrast with the requirement for an intact TEC1 gene (Fig. 1D), the mpt5Δ mutant phenotype requires neither PHD1 nor RAS2 (Fig. S1). Thus, the fMAPK pathway is a major mediator of the control of yeast cell differentiation by MPT5.
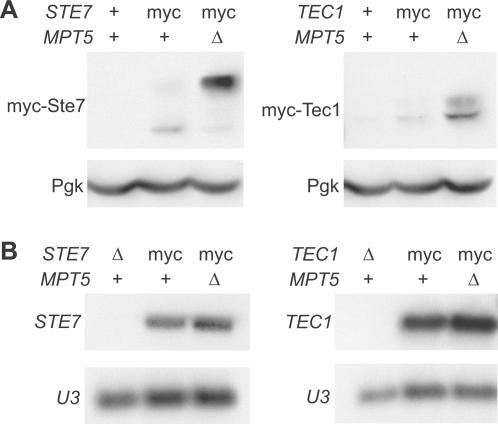
The interaction of the Mpt5 protein with the STE7 and TEC1 mRNAs, combined with the molecular activity of PUF proteins as translational repressors [24], [25] and mRNA de-adenylation factors [26], raises the possibilities that Mpt5 represses Ste7 and Tec1 protein expression or that Mpt5 destabilizes the mRNAs of these proteins. To test these possibilities, we constructed (Text S1) diploid strains with triple-myc epitope tags on the 5′ ends of the endogenous STE7 and TEC1 coding sequences. The modified genes are under the control of their native promoters, terminators, and UTRs. MPT5+ and mpt5Δ strain pairs were constructed. Protein and total-RNA extracts were prepared from cultures grown under yeast-form conditions, and were subjected to western-blot (Fig. 3A) and northern-blot (Fig. 3B) analyses. MPT5 represses Ste7 and Tec1 protein levels (Fig. 3A), and has a minor (Fig. 3B) but reproducible (data not shown) negative effect on STE7 and TEC1 mRNA levels. These results suggest that the Mpt5 protein represses Ste7 and Tec1 protein levels primarily at the level of protein translation from their respective mRNAs.
Figure 3. Repression of Ste7 and Tec1 protein levels by MPT5.
(A) Yeast strains were grown under yeast-form conditions. Protein extracts were analyzed by western blot, with Pgk serving as a loading control. (B) RNA extracts were analyzed by northern blot, with U3 serving as a loading control.
Note also that loss of MPT5 activity results in an increase in low-mobility forms of the Ste7 and Tec1 proteins (Fig. 3A). For Ste7, nearly all of the protein is in the low-mobility form. These low-mobility forms are phosphorylated protein. Treatment with phosphatase converts them to high-mobility forms (Fig. S2; Text S1).
Mpt5 and other PUF proteins are known to bind to sequence motifs in the 3′ untranslated regions (3′ UTR) of mRNAs [18], [19], [27]. Gerber et al. [19] have identified an 11-base sequence motif in 3′ UTRs of 33% of the mRNAs bound by the Mpt5 protein. We mapped the 3′ ends of the STE7 and TEC1 mRNAs (Text S1). The STE7 and TEC1 3′ UTRs extend 133 and 107 bases past their respective stop codons (data not shown), and thus are above the median 3′UTR length of 91 nucleotides [28]. Within the STE7 3′ UTR, there is a single site (AUGUAACAAUA, starting at the 119th base after the stop codon) matching the Mpt5 RNA-binding motif. We found that precise deletion [29] of this site results in derepression of Ste7 protein levels in an MPT5+ strain (Fig. 4A). Also, deletion of this site results in a partial increase in Ste7 phosphorylation (Fig. 4A). The combination of the binding-site-sequence deletion and deletion of MPT5 results in both derepression of Ste7 protein levels and maximum Ste7 phosphorylation (Fig. 4A).
Figure 4. UTR sequences and repression by MPT5.
Yeast strains were grown under yeast-form conditions. Protein and RNA extracts were prepared and analyzed by (A) western blot and (B) northern blot. Pgk protein and U3 RNA served as loading controls.
Like 67% of the mRNAs bound by Mpt5 [19], the TEC1 3′ UTR (and the 5′ UTR) contains no match to the Mpt5 binding-site motif. Mpt5 presumably binds either directly or indirectly to some other sequence in these mRNAs. Alternatively, the binding-site motif may require refinement. We tested the hypothesis that repression of Tec1 protein levels involves its UTR sequences. We replaced myc-Tec1 protein-coding sequences with myc-Ura3 protein-coding sequences (Text S1). In the mRNA expressed from this hybrid TEC1::myc-URA3 gene, TEC1 UTR sequences flank myc-Ura3 protein-coding sequences. As observed for the TEC1 gene encoding myc-Tec1 protein (Fig. 3A), MPT5 represses the levels of myc-Ura3 protein translated from an mRNA with TEC1 UTRs (Fig. 4A), and exerts a minor negative effect on the levels of the Ura3-encoding mRNA (Fig. 4B). Thus, the effect of MPT5 on Tec1 protein levels is independent of Tec1-protein sequences and TEC1-protein-coding nucleic-acid sequences. As a control, we tested the effect of MPT5 deletion on the levels of myc-Ura3 protein expressed from the URA3 gene and mRNA (with URA3 UTRs) (Fig. 4). The results show that MPT5-dependent repression of myc-Ura3 expression is imparted by TEC1 UTR sequences but not URA3 UTR sequences. We conclude that direct or indirect interaction of Mpt5 protein with UTR sequences mediates repression of Tec1 protein levels.
Deletion of the Mpt5 binding sequence in the STE7 message results not only in derepression of Ste7 protein levels, but also in an increase of Ste7 phosphorylation. However, only when MPT5 is deleted is Ste7 maximally phosphorylated (Fig. 4A). These observations suggest MPT5 has an effect on Ste7 phosphorylation through a mechanism that is separate from its effect on Ste7 protein levels. The effect on Ste7 phosphorylation depends on neither RAS2 nor PHD1 (Fig. 5A). However, we found that maximal phosphorylation of Ste7 depends entirely on Kss1. We constructed strains that harbored the kinase-dead allele kss1K42R [5] and were either MPT5+ or mpt5Δ. Extracts were prepared from cultures grown under yeast-form conditions, and were subjected to western-blot analysis detecting myc-Ste7. Whereas Ste7 protein levels are higher in mpt5Δ strains regardless of the KSS1 allele, accumulation of maximally phosphorylated forms of Ste7 in mpt5Δ strains requires the kinase activity of Kss1 (Fig 5B). These results suggest that Mpt5 represses Ste7 and Tec1 protein levels, and in addition exerts a negative effect on Kss1 kinase activity.
Figure 5. Inhibition of Kss1-dependent phosophorylation of Ste7 by MPT5.
Yeast strains were grown under yeast-form conditions. Protein extracts were analyzed by western blot, with Pgk serving as a loading control. The effect of MPT5 deletion on Ste7 phosphorylation is independent of RAS2 and PHD1 (A) but depends on KSS1 (B).
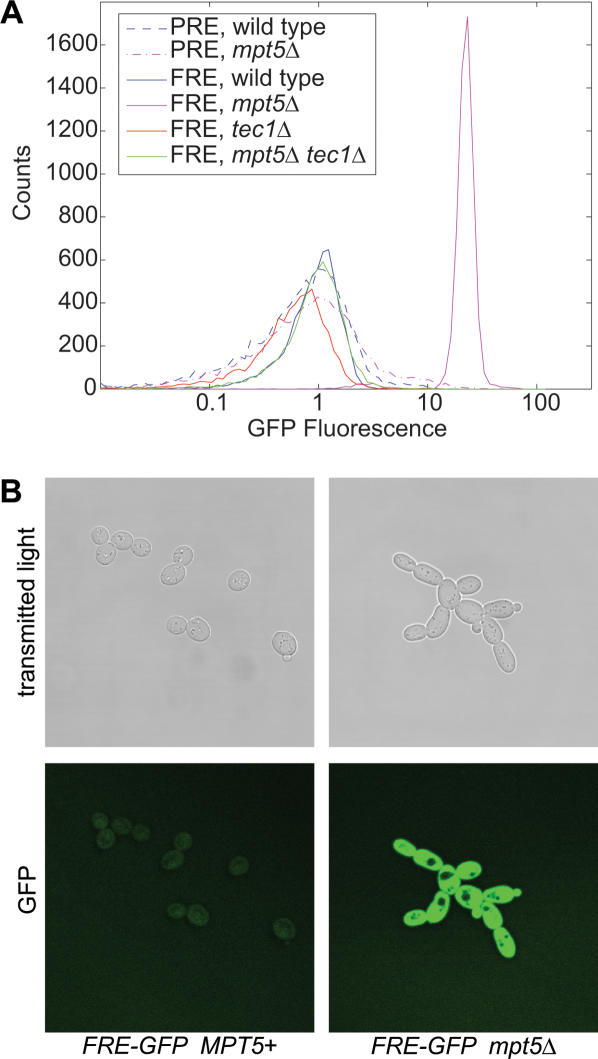
The genetics and molecular biology of the MPT5 gene suggest that it encodes an inhibitor of fMAPK pathway signaling. We tested this suggestion directly. fMAPK signaling derepresses the Ste12-Tec1 heterodimer, which binds to the Filamentation Response Element (FRE) in filamentation-gene promoters [7]. We fused a minimal FRE-dependent promoter [7] to Green Fluorescent Protein (GFP) coding sequences and integrated this fMAPK-pathway output reporter in the genome (Text S1). This diploid strain, plus mpt5Δ, tec1Δ, and mpt5Δ tec1Δ mutant derivatives, were grown under yeast-form conditions and analyzed by flow cytofluorometry (Materials and Methods). Deletion of MPT5 derepresses the fMAPK output reporter by 16-fold in the absence of pathway stimuli (Fig. 6). This effect requires TEC1 (Fig. 6A) and is accompanied by filamentous cell morphology (Fig. 6B). We conclude that Mpt5 is an inhibitor of signaling in the fMAPK pathway.
Figure 6. Specific inhibition of fMAPK pathway output by MPT5.
(A) Diploid strains with either a minimal filamentation-MAPK-pathway output reporter (FRE-GFP) or a mating-MAPK reporter (PRE-GFP) and the indicated genotypes were grown under yeast-form conditions and subjected to flow cytofluorometry. (B) The morphology and fluorescence of MPT5+ and mpt5Δ diploid cells with FRE-GFP were imaged by transmitted light and confocal fluorescence microscopy.
The fMAPK pathway shares multiple components, including Ste7 and Ste12, with the mating MAP-kinase (mMAPK) pathway, which is inactive in diploids. Because deletion of MPT5 activates the fMAPK pathway in the absence of stimuli, it was possible that it also results in inappropriate activation of mMAPK pathway output in diploids. We tested this possibility. In the GFP-based pathway reporter, FRE sequences were replaced with the triple Pheromone Response Element (PRE) sequences of the PRM1 promoter [30]. Though this reporter is induced sharply by pheromone in haploids (Fig. S3), it shows essentially no response to deletion of MPT5 in diploids (Figure 6A), and haploids (Fig. S3). Thus, Mpt5 inhibits signaling in the fMAPK pathway specifically.
Discussion
The combination of the amplification effect of MAPK cascades [31] and random noise, a substantial feature of signaling pathways [32], creates the risk of pathway off-state instability. This observation suggests the existence of mechanisms to prevent pathway output in the absence of a pathway stimulus. Our work suggests that Mpt5 serves this purpose in the fMAPK pathway and thereby prevents inappropriate filamentous-form differentiation.
We have shown that Mpt5 represses the protein levels of the MAP-kinase kinase Ste7 and the downstream transcription factor Tec1, and that this regulation is mediated by UTR sequences in their respective mRNAs. These effects may limit the signal capacity of the fMAPK pathway and thereby stabilize the pathway ‘off’ state. Furthermore, Mpt5 negatively regulates Kss1-dependent phosphorylation of Ste7. Kss1 phosphorylates Ste7 in vitro [33]. Thus, the simplest model to explain the effect on Ste7 phosphorylation is that Mpt5 negatively regulates the kinase activity of Kss1. At present, it is unclear whether the effect on Kss1 is direct or indirect. Because the Mpt5 and Kss1 proteins interact in a two-hybrid assay [15], a direct effect is possible. Also requiring further study is the question of how the activity of Mpt5 is regulated. Mpt5 protein abundance, as determined by western-blot analysis, does not differ in yeast-form cells and constitutively filamentous STE11-4 cells (S. Prinz, unpublished). Changes in the subcellular localization of Mpt5 or its target mRNAs could provide a regulatory mechanism. Also, the physical interaction of Mpt5 with the Kss1, Fus3, and Cdc28 kinases [15] raises the possibility that Mpt5 is a kinase substrate regulated by phosphorylation.
Our work may indicate a positive feedback operating in the fMAPK pathway. This is suggested by the observation that the increase of Ste7 protein due to the deletion of the Mpt5 binding site results also in increased Kss1-dependent phosphorylation of Ste7 protein. Though the functional consequences of Ste7 phosphorylation by Kss1 are not clear, Maleri et al. [34] have shown that it does not result in attenuation, as had been assumed previously [35]. Previous studies have shown maximal phosphorylation of Ste7 to be dependent on pheromone stimulation and Kss1 and Fus3 activity [36], [37]. Similarly, we found the phosphorylation state of Ste7 to be indicative of the activity of the fMAPK pathway. In mutants that are constitutively filamentous such as mpt5Δ (Fig. 3A) or STE11-4 [36] (data not shown), Ste7 is maximally phosphorylated, whereas in yeast-form cells (Fig. 3) or kinase-dead kss1K42R mutants (Fig. 4B) Ste7 is mainly unphosphorylated. Thus, we speculate that Kss1 acts on Ste7 in a positive feedback promoting the switch to the pathway ‘on’ state. Negative regulation by Mpt5 could stifle this effect and stabilize the ‘off’ state. Further molecular genetics, in conjunction with pathway modeling studies, could test these possibilities.
In conclusion, the combination of the ubiquity of RNA-binding proteins, the long binding-site-containing UTRs of mRNAs encoding regulatory proteins [19], [28], [38], and the emerging role of RNA-binding proteins in signaling [39], [40], suggests that there is a genome-scale level of regulation to be studied. Post-transcriptional control of gene activity by RNA-binding proteins may be critical for tight regulation of signaling and, consequently, the control of cell differentiation. Our results establish an example.
Materials and Methods
Growth conditions
Standard media and conditions were used for yeast-form [41] and filamentous-form [3] growth. Tests of filamentation and adhesion were done as described previously [2].
Immunoprecipitation and RT-PCR
Immunoprecipitation of Mpt5-13myc and bound RNAs from yeast-form cells with ensuing RT-PCR was done as described [40], except for the following: protease inhibitors were added as in Ren et al. [42]; the antibody was mouse anti-myc monoclonal 9E10 (Covance); beads were ImmunoPure Immobilized Protein G beads (Pierce); incubation with beads was for 2 hours; 1st strand cDNA synthesis was primed with a polyT oligonucleotide. Gene-specific PCR amplification used the following primers: TEC1_RACE (GAAAGTAATCCTGAGTT CAGTTCCA); TEC1_3UTR_R (GTTCGAGAACTGGTAATGTTTGACT); PHD1_RACE (AAGTTGTTGAATGTCACGAAGATG); and PHD1_3UTR_R (ATTG TACGAATCCTATCAGCCTTTC).
Protein extracts and western-blot analyses
Protein-extract preparations and western-blot analyses were done as described previously [2], except that extracts included Phosphatase Inhibitor Cocktails 1 and 2 (Sigma). The myc epitope was detected using mouse monoclonal antibody 9E10 (Covance). High-Range Rainbow (GE Healthcare) molecular weight markers were used. Triple-myc epitope-tagged Ste7 and Tec1 proteins showed gel mobilities corresponding to estimated molecular weights of 72 kD and 73 kD, respectively. The mobilities of phosphorylated Ste7 and Tec1 correspond to apparent molecular weights of approximately 84 kD and 81 kD.
RNA preparation and northern-blot analyses
RNA-extract preparations and northern-blot analyses were done as described previously [2]. TEC1 and STE7 probes were made by PCR using primers: TEC1_C_F (AAGTGCGTTCCGTCAAAGAG), TEC1_C_R (ATTGGTTGGATGGCGTAAAG ), Ste7_probe_F (TCTTAACCTGCAC CCAGATG), Ste7_probe_R (ATTTGAAGTTCCCGACAACG). Primers for the URA3 probe are described in Text S1. The U3 (SNR17A) probe was made using primers described elsewhere [43].
GFP fluorescence measurement and imaging
Exponential-phase cells grown in SCD medium were inspected and imaged with transmitted light and fluorescence using a Leica confocal microscope. For flow cytofluorometry, cells were briefly sonicated to disrupt cell aggregates. GFP fluorescence was measured using a Becton Dickinson FACSCalibur flow cytometer. Wild-type and mpt5Δ strains lacking GFP were included as autofluorescence controls. The scatter plots of fluorescence intensity (FLI) and forward light scatter (FLS) for the autofluorescence control strains were used to fit a function of FLI versus FLS using least squares. The control strains' FLS data were used to calculate FLS window sizes using the overall standard deviation of FLS. To ensure a homogeneous population of events and remove outliers (multi-cell events, etc.), this window was applied to the flow cytometry data for each strain, selecting only those events within the FLS window centred at each strain's mean FLS. The cell-size-dependent bias in FLI data was corrected by subtracting the FLI-versus-FLS function from the fluorescence of each event based on the event's FLS value. An additive correction was applied to ensure that the mean background-subtracted FLI for events within the window is the same as the raw (uncorrected) FLI for events within the window. For each event within the window, the background-subtracted FLI and the FLS were normalized relative to the overall mean FLI and FLS of the control strain, respectively. The average FLS ratios for the four strains were: 1.05 (wild type), 1.02 (tec1), 1.45 (mpt5), and 1.22 (mpt5 tec1). Background subtraction did not change the median FLI, but reduced cell-to-cell variation in FLI by approximately 5%.
Supporting Information
Supporting Materials and Methods
(0.14 MB PDF)
mpt5Δ phenotype requires neither PHD1 nor RAS2. Diploid yeast of the indicated genotypes were grown under filamentous-form conditions (SLAD agar) and microscopically imaged to show their filamentation phenotypes.
(0.93 MB PDF)
Phosphorylation of Tec1 and Ste7 in mpt5Δ strains. N-terminally tagged Tec1 or Ste7 protein, or a tagless control, was immunoprecipitated from an mpt5Δ strain with subsequent phosphatase treatment or mock treatment and analyzed by western blot.
(0.92 MB PDF)
fMAPK and mMAPK pathway output in haploids. Haploid MATa strains with either a minimal filamentation-MAPK-pathway output reporter (FRE-GFP) or a mating-MAPK reporter (PRE-GFP) and the indicated genotypes were grown under yeast-form conditions in the absence of alpha factor and subjected to flow cytofluorometry. As a control, PRE-GFP output of an alpha-factor stimulated strain is shown.
(0.52 MB PDF)
Acknowledgments
The authors thank A. Aderem, A. Amon, and G. Fink for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by grant P50 GM076547 from NIH. S.A. Ramsey was supported by grant U54 AI054523R from NIH. R.J. Taylor was supported by a junior graduate studentship from the Michael Smith Foundation for Health Research. T. Galitski is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
References
- 1.Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 2.Prinz S, Avila-Campillo I, Aldridge C, Srinivasan A, Dimitrov K, et al. Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 2004;14:380–390. doi: 10.1101/gr.2020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 4.Rives AW, Galitski T. Modular organization of cellular networks. Proc Natl Acad Sci U S A. 2003;100:1128–1133. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 6.Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, et al. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikuchi Y, Oka Y, Kobayashi M, Uesono Y, Toh-e A, et al. A new yeast gene, HTR1, required for growth at high temperature, is needed for recovery from mating pheromone-induced G1 arrest. Mol Gen Genet. 1994;245:107–116. doi: 10.1007/BF00279756. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy BK, Austriaco NR, Jr., Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 11.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. Embo J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, et al. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart MS, Krause SA, McGhie J, Gray JV. Eukaryot Cell. 2006. Mpt5p, a Stress Tolerance- and Lifespan-Promoting PUF Protein in Yeast, Acts Upstream of the Cell Wall Integrity Pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Kurjan J. Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol Cell Biol. 1997;17:3429–3439. doi: 10.1128/mcb.17.6.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu BE, Skowronek KR, Kurjan J. The N terminus of Saccharomyces cerevisiae Sst2p plays an RGS-domain-independent, Mpt5p-dependent role in recovery from pheromone arrest. Genetics. 2001;159:1559–1571. doi: 10.1093/genetics/159.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohkuni K, Kikuchi Y, Hara K, Taneda T, Hayashi N, et al. Suppressor analysis of the mpt5/htr1/uth4/puf5 deletion in Saccharomyces cerevisiae. Mol Genet Genomics. 2006;275:81–88. doi: 10.1007/s00438-005-0064-x. [DOI] [PubMed] [Google Scholar]
- 18.Tadauchi T, Matsumoto K, Herskowitz I, Irie K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. Embo J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 21.Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 22.Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 23.Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 27.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 30.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang CY, Ferrell JE., Jr. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAdams HH, Arkin A. It's a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 1999;15:65–69. doi: 10.1016/s0168-9525(98)01659-x. [DOI] [PubMed] [Google Scholar]
- 33.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maleri S, Ge Q, Hackett EA, Wang Y, Dohlman HG, et al. Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol Cell Biol. 2004;24:9221–9238. doi: 10.1128/MCB.24.20.9221-9238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Errede B, Ge QY. Feedback regulation of map kinase signal pathways. Philos Trans R Soc Lond B Biol Sci. 1996;351:143–149. doi: 10.1098/rstb.1996.0010. [DOI] [PubMed] [Google Scholar]
- 36.Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 37.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 38.Hurowitz EH, Brown PO. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasko P. Gene regulation at the RNA layer: RNA binding proteins in intercellular signaling networks. Sci STKE. 2003;2003:RE6. doi: 10.1126/stke.2003.179.re6. [DOI] [PubMed] [Google Scholar]
- 40.Sugiura R, Kita A, Shimizu Y, Shuntoh H, Sio SO, et al. Feedback regulation of MAPK signalling by an RNA-binding protein. Nature. 2003;424:961–965. doi: 10.1038/nature01907. [DOI] [PubMed] [Google Scholar]
- 41.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [Google Scholar]
- 42.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 43.Heikkinen HL, Llewellyn SA, Barnes CA. Initiation-mediated mRNA decay in yeast affects heat-shock mRNAs, and works through decapping and 5′-to-3′ hydrolysis. Nucleic Acids Res. 2003;31:4006–4016. doi: 10.1093/nar/gkg474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Materials and Methods
(0.14 MB PDF)
mpt5Δ phenotype requires neither PHD1 nor RAS2. Diploid yeast of the indicated genotypes were grown under filamentous-form conditions (SLAD agar) and microscopically imaged to show their filamentation phenotypes.
(0.93 MB PDF)
Phosphorylation of Tec1 and Ste7 in mpt5Δ strains. N-terminally tagged Tec1 or Ste7 protein, or a tagless control, was immunoprecipitated from an mpt5Δ strain with subsequent phosphatase treatment or mock treatment and analyzed by western blot.
(0.92 MB PDF)
fMAPK and mMAPK pathway output in haploids. Haploid MATa strains with either a minimal filamentation-MAPK-pathway output reporter (FRE-GFP) or a mating-MAPK reporter (PRE-GFP) and the indicated genotypes were grown under yeast-form conditions in the absence of alpha factor and subjected to flow cytofluorometry. As a control, PRE-GFP output of an alpha-factor stimulated strain is shown.
(0.52 MB PDF)