Abstract
The antifolate drugs sulfadoxine and pyrimethamine are commonly used to treat Plasmodium falciparum malaria. However, they can also affect the Plasmodium vivax parasite if it coexists with P. falciparum, as both species have common drug targets. Resistance to the antifolate drugs arises due to point mutations in the target enzymes of the respective parasite. To assess the cross-species impact of antifolate drug treatment, we describe here the dihydrofolate reductase (DHFR) mutations among field isolates of P. vivax and P. falciparum. The overall DHFR mutation rate for P. vivax was lower than that for P. falciparum. However, both species of Plasmodium followed similar trends of DHFR mutations. Similar to P. falciparum, the DHFR mutation rate of P. vivax also varied from region to region. It was lower in P. vivax-dominant regions but higher in the P. falciparum-dominated areas and highest where antifolates are used as the first line of antimalarial treatment. In conclusion, the antifolate treatment of falciparum malaria is proportionately affecting the DHFR mutations of P. vivax, suggesting that the drug should be used with caution to minimize the development of cross-species resistance in the field.
Sulfadoxine and pyrimethamine (SP), commonly used for Plasmodium falciparum malaria treatment, interfere with the enzymes involved in the folate biosynthesis pathway of the parasite (8). Point mutations in Plasmodium falciparum dihydrofolate reductase (PFDHFR) are known to give rise to pyrimethamine resistance. The first mutation occurs at the S108N codon, followed by C59R, N51I, and I164L mutations (8). One could observe single, double, triple, or quadruple PFDHFR mutations depending on the level of drug resistance. P. falciparum isolates with increased numbers of PFDHFR mutations were found to show a higher degree of pyrimethamine resistance (8). Indeed, the falciparum malaria patients containing quadruple PFDHFR mutations showed a treatment failure against this drug (8, 23).
This antifolate drug also affects the Plasmodium vivax parasite since both contain the same target enzyme (7). Pyrimethamine resistance in P. vivax is associated with two key DHFR mutations, S58R and S117N, which are equivalent to the C59R and S108N mutations of the PFDHFR (6). In addition, other mutations (I13L, P33L, F57L/I, T61M, S117T, I172V, and I173L) in P. vivax DHFR (PVDHFR) have also been reported (4-7, 9-12, 14, 16, 18, 22). Similar PVDHFR mutations at these codons except for I13L, P33L, I172V, and I173L have also been reported in India by various workers (11, 14, 22).
The in vitro and in vivo data on PVDHFR mutations have now clearly established the association between pyrimethamine resistance and these mutations (9-11, 15, 20, 21). For example, the wild-type PVDHFR enzyme expressed in Escherichia coli showed a higher catalytic activity than the mutant type enzyme (15, 20). Similarly, wild-type and different mutant alleles of PVDHFR were expressed in Saccharomyces cerevisiae and tested for their susceptibilities to pyrimethamine (9, 10). The yeast cells harboring the mutant PVDHFR constructs showed their survival at a higher concentration of pyrimethamine than those cells that were harboring the wild-type PVDHFR construct (9). The degree of pyrimethamine resistance in P. vivax also increases with the sequential addition of each mutation in the dhfr gene, and extremely resistant parasites carry an S117T mutation (9, 10, 12). Therefore, the PVDHFR allele with quadruple mutations (F57L plus S58R plus T61M plus S117T) exhibited the highest level of pyrimethamine resistance in the Saccharomyces cerevisiae system (9). The P. vivax malaria patients harboring this quadruple mutation were also unable to clear the parasite with pyrimethamine (21). The DHFR mutations of both Plasmodium species have now been used as markers to assess antifolate drug pressure in the field (1-3, 5, 8, 10, 11, 14, 16, 18, 21-23).
Although DHFR mutations have been described separately for P. vivax and P. falciparum from various countries, there is no report showing DHFR mutations of both species together from the same region at a given time point, which can assess the cross-species effect of the antifolate treatment. Therefore, to asses the cross-species effect of antifolate usage, we decided to investigate the DHFR mutations among P. vivax and P. falciparum isolates from different parts of India, including those areas where we have recently reported the PFDHFR mutations (2).
MATERIALS AND METHODS
Parasite collection.
Patients with fever were attending malaria clinics at Car Nicobar Island (Andaman and Nicobar Islands [A&N]), Kamrup (Assam), Aligarh (Uttar Pradesh [UP]), Panjim (Goa), Panna (Madhya Pradesh [MP]), Cuttack (Orissa), and Chennai (Tamil Nadu [TN]) during 2003 and 2004 (Fig. 1). Thick and thin blood smears of these patients were screened by light microscopy after Giemsa staining. Patients were treated with antimalarial drugs according to the National Drug Policy as described previously (2). Briefly, the vivax malaria patients from Aligarh, Panna, Panjim, and Chennai were treated with a single dose of chloroquine (10 mg per kg body weight) followed by primaquine (0.25 mg per kg body weight) per day for 5 days. However, the P. vivax patients from Kamrup, Cuttack, and Car Nicobar were treated with a total dose of 1,500 mg chloroquine over a 3-day period (600 mg, 600 mg, and 300 mg for the first, second, and third days, respectively) along with the same dose of primaquine over a 5-day period. The P. falciparum malaria cases from these areas except Kamrup were treated with similar doses of chloroquine but a single dose of primaquine (0.75 mg per kg body weight). The P. falciparum malaria patients from Kamrup were treated with a single dose of sulfadoxine (25 mg per kg body weight) and pyrimethamine (1.25 mg per kg body weight). One hundred to 200 μl of heparinized blood was collected from the malaria parasite-positive patients. Informed consent was obtained from the patients prior to blood collection according to institutional ethical guidelines. Previously, we used the P. falciparum-infected blood samples from UP, Assam, and A&N for the PFDHFR mutation studies (2). The remaining P. falciparum isolates from MP, Goa, and Orissa and the P. vivax-infected blood samples from all seven areas were used here for the DHFR mutation analysis.
FIG. 1.
Map of India showing study sites.
Extraction of parasite DNA and PCR amplification of the dhfr gene.
Parasite DNA was extracted from the blood of P. vivax and P. falciparum patients using an AccuPrep Genomic DNA extraction kit (Bioneer Corporation, Korea) according to the manufacturer's instructions. The DNA was eluted in Tris-EDTA buffer (pH 8.0), and an aliquot was used for the PCR amplification. A 1,014-bp fragment of the P. vivax dhfr-ts gene was amplified using primers PvDA (5′-ACCGCACCAGTTGATTCCTAC-3′) (forward) and PvDB (5′-TGTTAAAGCTGAAGTACACGAG-3′) (reverse) with the following cycling parameters: a 10-min initial denaturation step at 94°C was followed by 35 cycles with a 30-s denaturation step at 94°C, a 1-min annealing step at 58°C, a 1-min extension step at 72°C, and a final 10-min extension step at 72°C. This primary PCR product was diluted 10 times and used as a DNA template for nested PCR to amplify the 784-bp region, which covers the entire DHFR domain. Primers PvDF (5′-ATGGAGGACCTTTCAGATGT-3′) (forward) and PvDR (5′-AACGCATTGCAGTTCTCCGA-3′) (reverse) were used for nested PCR with the following cycling parameters: a 10-min initial denaturation step at 94°C followed by 35 cycles with a 30-s denaturation step at 94°C, a 30-s annealing step at 54°C, a 1-min extension step at 72°C, and a final 10-min extension step at 72°C. Primers PvDA (covering −75 to −55 bp upstream to the pvdhfr-ts start codon), PvDB, PvDF, and PvDR were designed from the known pvdhfr-ts sequence of pyrimethamine-sensitive P. vivax isolate ARI/Pakistan (X98123). Amplification of the dhfr gene from P. falciparum isolates was carried out under the same conditions as those described previously (2).
Nucleotide sequencing.
An individual band of each PCR product was excised from the agarose gel and purified using an AccuPrep gel purification kit (Bioneer Corporation, Korea) according to the manufacturer's instructions. The entire pvdhfr gene was sequenced using primers PvDM (5′-GTTAGCGTCTTGGAAAGCAC-3′) and PvDR, while primers for the pfdhfr gene sequencing were same as those described previously (2). Sequencing parameters and other downstream protocols were the same as those described previously (13). The nucleotide sequences were translated into amino acids using the Edit sequence program (Lasergene, version 5.1, July 1999; DNASTAR Inc., Madison, WI). Amino acid sequences were aligned using the GeneDoc Multiple Sequence Alignment Editor and Shading Utility (version 2.6.002).
Statistical analysis.
The chi-square test was applied to find significant differences between two groups. Furthermore, the chi-square test was also applied to assess the trends of DHFR mutations across the P. falciparum and P. vivax groups. A P value of <0.05 was considered significant.
RESULTS
Mutation analysis of the pvdhfr gene.
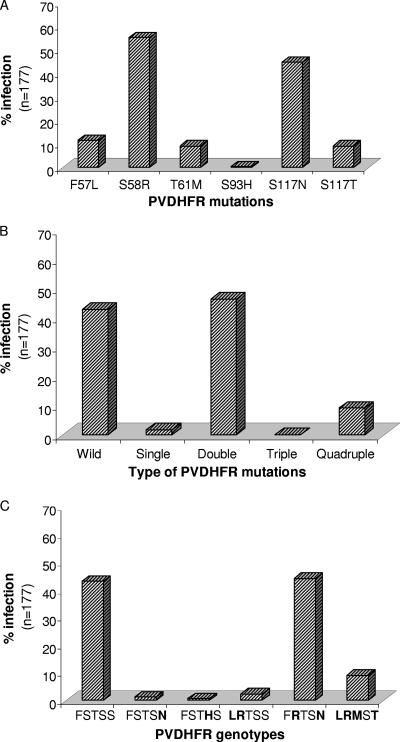
We have sequenced the entire DHFR domain of the pvdhfr-ts gene from 177 P. vivax clinical isolates. Among these isolates, mutations were observed at F57L, S58R, T61M, S93H, S117N, and S117T codons (Fig. 2A). The mutation frequency at S58R and S117N was higher among the isolates. Seventy-six (42.94%; n = 177) isolates contained the wild-type amino acids at these positions. Among the remaining isolates, the frequency of double DHFR mutations was highest, while no isolate was found to contain triple DHFR mutations (Fig. 2B). The frequency of quadruple DHFR mutations was higher than the frequency of single and triple DHFR mutations. There were six different genotypes among these 177 isolates, where genotype F57R58T61S93N117 was most prevalent, followed by L57R58M61S93T117 (mutated amino acids are in boldface type) (Fig. 2C).
FIG. 2.
Mutation rates in the P. vivax DHFR enzyme. (A) Mutation rate in individual PVDHFR codons. (B) Prevalence rate of the total number of mutations in PVDHFR. (C) Prevalence rates of the PVDHFR genotypes. The amino acid sequence (mutated amino acids are in boldface type) shown in the lower panel corresponds to PVDHFR codons 57, 58, 61, 93, and 117.
Besides the nonsynonymous mutations, there was a synonymous mutation at codon 69 (TAT→TAC) coding for tyrosine. TAT at codon 69 was associated with all six different PVDHFR genotypes and was present in the majority of the isolates (85.88%; n = 177). On the contrary, 24 of 25 isolates containing TAC at this codon were associated with the wild-type PVDHFR genotype.
There were variable numbers of GGDN repeats at amino acid position 88 of PVDHFR among the P. vivax isolates. The majority of them (91.53%; n = 177) were found to contain three GGDN repeats, whereas the rest of them contained two (6.78%; n = 177) or four (1.69%; n = 177) repeats. While isolates with the wild-type PVDHFR genotype had a variable number of GGDN repeats, the majority of the isolates with mutated genotypes (98.02%; n = 101) contained only three GGDN repeats.
Regional distribution of PVDHFR alleles.
The majority of isolates from MP and UP contained the wild-type amino acids at the above-mentioned five codons of PVDHFR (Table 1). On the contrary, the majority of isolates from Goa, Orissa, and TN were found to contain the double DHFR mutation genotype F57R58T61S93N117. This genotype was also predominant in Assam but not in A&N. The latter had a predominance of the quadruple DHFR mutation genotype L57R58M61S93T117. Genotypes L57R58M61S93T117 and L57R58T61S93S117 were present only among Assam and AN isolates, albeit with variable frequencies, whereas F57S58T61S93N117 and F57S58T61H93S117 were present only in UP (Table 1).
TABLE 1.
Geographical distributions of DHFR genotypes among Indian P. vivax isolates
| Genotypea | No. of mutations | No. (%) of isolates
|
||||||
|---|---|---|---|---|---|---|---|---|
| UP (n = 38) | MP (n = 24) | Goa (n = 38) | Orissa (n = 13) | Assam (n = 24) | TN (n = 10) | A&N (n = 30) | ||
| F57S58T61S93S117 | 0 | 22 (57.89) | 22 (91.66) | 8 (21.05) | 3 (23.08) | 7 (29.16) | 3 (30.00) | 11 (36.66) |
| F57S58T61S93N117 | 1 | 2 (5.26) | ||||||
| F57S58T61H93S117 | 1 | 1 (2.63) | ||||||
| L57R58T61S93S117 | 2 | 1 (4.16) | 3 (10.00) | |||||
| F57R58T61S93N117 | 2 | 13 (34.21) | 2 (8.33) | 30 (78.94) | 10 (76.92) | 12 (50.00) | 7 (70.00) | 4 (13.33) |
| L57R58M61S93T117 | 4 | 4 (16.66) | 12 (40.00) | |||||
Mutated amino acids are shown in boldface type.
Thirteen different PVDHFR alleles were observed among the 177 P. vivax isolates when variations at three different loci were considered, i.e., nonsynonymous mutations together with a synonymous mutation (TAT→TAC) at codon 69 and GGDN repeats (Table 2). The most common alleles had a TAT codon at 69 and three GGDN repeats associated with the F57R58T61S93N117 (43.50%; n = 177) or F57S58T61S93S117 (22.60%; n = 177) genotype. These two alleles (A1 and A11) were present in all the seven regions of the country. Certain alleles were rare and region specific, while others were present in more than one region (Table 2).
TABLE 2.
Allelic variation in the pvdhfr gene among Indian P. vivax isolates
| Allele | No. (%) of isolates (n = 177) | Genotypea | Synonymous mutation at codon 69 | Type of GGDN repeatb | Region(s) |
|---|---|---|---|---|---|
| A1 | 40 (22.60) | F57S58T61S93S117 | TAT | 1 | All regions studied |
| A2 | 23 (12.99) | F57S58T61S93S117 | TAC | 1 | Goa, UP, TN, MP, A&N, and Assam |
| A3 | 7 (3.95) | F57S58T61S93S117 | TAT | 2 | UP, TN, MP, Orissa, and A&N |
| A4 | 4 (2.26) | F57S58T61S93S117 | TAT | 3 | UP, Orissa, and MP |
| A5 | 1 (0.56) | F57S58T61S93S117 | TAT | 4 | A&N |
| A6 | 1 (0.56) | F57S58T61S93S117 | TAC | 4 | A&N |
| A7 | 1 (0.56) | F57S58T61S93N117 | TAT | 1 | UP |
| A8 | 1 (0.56) | F57S58T61S93N117 | TAC | 1 | UP |
| A9 | 1 (0.56) | F57S58T61H93S117 | TAT | 5 | UP |
| A10 | 4 (2.26) | L57R58T61S93S117 | TAT | 1 | A&N and Assam |
| A11 | 77 (43.50) | F57R58T61S93N117 | TAT | 1 | All regions studied |
| A12 | 1 (0.56) | F57R58T61S93N117 | TAT | 3 | A&N |
| A13 | 16 (9.04) | L57R58M61S93T117 | TAT | 1 | A&N and Assam |
Mutated amino acids are shown in bold face.
Types of GGDN repeats are defined as follows: type 1, GGDNTSGGDNTHGGDN; type 2, GGDNTHGGDN; type 3, GGDNTSGGDN; type 4, GGDNTSGGDNTHGGDNTHGGDN; type 5, GGDNTHGGDNTHGGDNTHGGDN.
Analysis of PFDHFR mutations.
Previously, we described the PFDHFR mutations among 117 P. falciparum isolates from three regions of India (UP, Assam, and A&N) collected during 2003 and 2004 (2). Here, we describe the PFDHFR mutations among 73 additional isolates collected from three other areas of the country (MP, Goa, and Orissa) during the same period (Table 3). Isolates from all three additional areas showed a predominance of the double PFDHFR mutations (Table 3), similar to the isolates from UP and Assam (2). Quadruple PFDHFR mutations from A&N reported previously (2) were not observed in any of the isolates, while triple PFDHFR mutations were present in Orissa (Table 3).
TABLE 3.
Geographical distributions of PFDHFR genotypes among Indian P. falciparum isolatesa
| Genotypeb | No. of mutations | No. (%) of isolates
|
||
|---|---|---|---|---|
| MP (n = 24) | Goa (n = 22) | Orissa (n = 27) | ||
| A16N51C59S108I164 | 0 | 1 (4.16) | ||
| A16N51C59N108I164 | 1 | 6 (25.00) | 4 (18.81) | 4 (14.81) |
| A16N51R59N108I164 | 2 | 17 (70.83) | 18 (81.81) | 22 (81.48) |
| A16N51R59N108L164 | 3 | 1 (3.70) | ||
The PFDHFR mutations among P. falciparum isolates from UP, Assam, and A&N have been described in our previous studies (2). The PFDHFR mutation data from three other regions (MP, Goa, and Orissa) are from this study.
Mutated amino acids are shown in boldface type.
Relationship between PVDHFR and PFDHFR mutations.
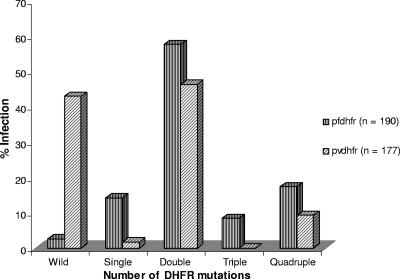
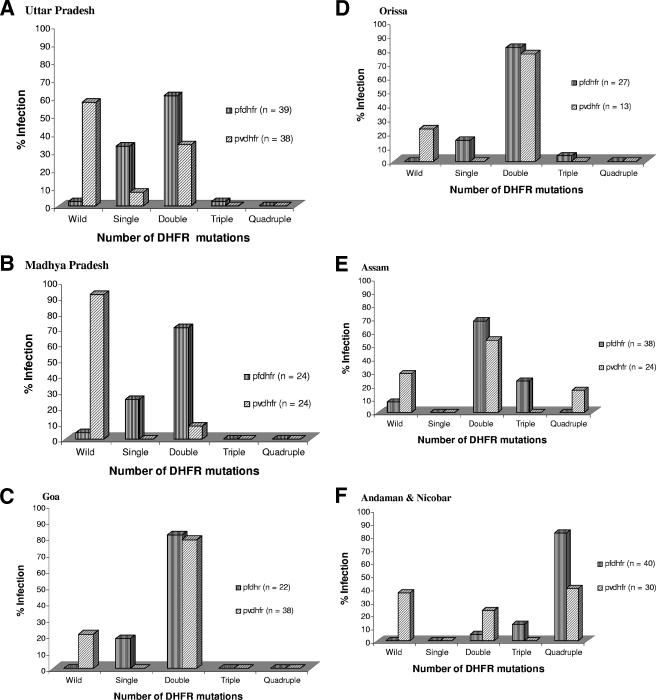
P. falciparum and P. vivax infections occur in all the seven study areas where we have analyzed the PVDHFR mutations (Table 1). We have already described PFDHFR mutations among 117 P. falciparum isolates from UP, Assam, and A&N (2), while the remaining 73 isolates from MP (n = 24), Goa (n = 22), and Orissa (n = 27) were sequenced here (Table 3). Therefore, we were able to analyze the DHFR mutations of P. falciparum (190 isolates) and P. vivax (177 isolates) combined together from these areas, except TN. As shown in Fig. 3 and as confirmed by the chi-square test, there was a specific trend of DHFR mutations across the P. falciparum and P. vivax groups (P = 0.0001). Nevertheless, the frequency of the wild-type PVDHFR genotype was significantly higher than that of the PFDHFR genotype (P = 0.0001) among all the isolates combined together (Fig. 3) as well as in each region (Fig. 4). The numbers of isolates containing double DHFR mutations for each species were higher than the numbers of isolates with single, triple, or quadruple DHFR mutations among all the isolates combined together (Fig. 3) as well as isolates of each region separately, except A&N, where quadruple DHFR mutations were predominant (Fig. 4). Furthermore, the numbers of P. vivax isolates with double DHFR mutations were greater than those of P. falciparum (P = 0.023) in A&N. Elsewhere, the frequencies of double DHFR mutations for both the species were almost same (P > 0.05), except for MP (P = 0.001). Interestingly, the quadruple DHFR mutations were observed in P. vivax but not in P. falciparum isolates from Assam (Fig. 4).
FIG. 3.
Mutation patterns of P. vivax and P. falciparum DHFR enzyme sequences among total Indian isolates. The chi-square test was applied to assess the DHFR mutation trends across the species.
FIG. 4.
Mutation patterns of P. vivax and P. falciparum DHFR enzyme sequences among the regional isolates. The PFDHFR data for UP, Assam, and A&N are derived from our previous studies (2). The chi-square test was applied to assess DHFR mutation rates among the groups.
DISCUSSION
Antifolate drugs are commonly used for the treatment of falciparum malaria. According to the National Drug Policy of India, chloroquine-resistant uncomplicated falciparum malaria cases should be treated with SP. In those regions of India where chloroquine resistance has reached a very high level, SP is the first line of antimalarial drug, except for a few primary health centers, where artesunate-based combined therapy has recently been introduced as an alternative treatment. If P. falciparum and P. vivax species coexist in the region, both of them are at risk of being exposed to the antifolate drugs, as they have common target enzymes (7, 8). We found here that the antifolate drug pyrimethamine was also affecting the P. vivax parasite, as PVDHFR mutations were observed among isolates of all seven regions (Table 1). This may be due to the fact that both of these species coexist together in these regions, and chloroquine-resistant falciparum malaria is treated with SP (17). Exposure of P. vivax to SP can occur in these areas for several reasons. First, there could be a situation where the clinician prescribes the antimalarial drug based on clinical grounds but lacks the opportunity of immediate diagnosis, and the inaccurate diagnosis of the parasite species also leads to a similar situation. Second, economic issues can also lead to the treatment of P. vivax malaria with SP because it is cost-effective, easily available, and simple to administer due to its single-dose treatment regimen. Third, mixed P. falciparum and P. vivax infections may also be a reason for such a situation. Mixed infections are quite common in India despite the fact that there are separate transmission peaks for each species (19). This may be due to the fact that there is a window period that overlaps between the peaks and provides a congenial environmental condition for the transmission of both the species. Indeed, 26% of our malaria-infected blood samples from A&N showed mixed P. falciparum and P. vivax infections when detected by species-specific PCR for 18S rRNA gene sequences (data not shown).
There was a regional variation in the PVDHFR mutation rates (Table 1). A similar regional variation in the PVDHFR mutations was also described previously by Kaur et al. (14). Rates of PVDHFR mutations reported from Chennai and Goa were similar to that of our data. However, the same group simultaneously reported a higher rate of PVDHFR mutations from Chennai (22) using a different methodology, i.e., PCR-restriction fragment length polymorphism, which could not be as accurate as sequencing. Our data from Cuttack and Car Nicobar (Table 1) could not be compared with the data reported previously by Kaur et al. (14) due to differences in sample sizes; for example, they analyzed only one isolate from Car Nicobar. Furthermore, we describe here PVDHFR mutations from additional regions of India, i.e., Aligarh (UP), Kamrup (Assam), and Panna (MP), and we also report here the prevalence of quadruple PVDHFR mutations in A&N and Assam that could be highly resistant to SP (9-12). Imwong et al. (11) also analyzed a few Indian isolates (n = 16) and found double PVDHFR mutations in 19% of them, but the origin of these isolates is not known. Similar to the data reported previously by Kaur et al. (14), we also did not find any mutation at 13, 33, and 173 codons of the pvdhfr gene among Indian isolates. However, we did not find any isolate with PVDHFR mutations at codons 37, 109, 131, 159, 171, and 188, which were considered novel by Kaur et al. (14). This discrepancy could arise due to different approaches used for sequencing, as Kaur et al. cloned the PCR products and then sequenced them, whereas we employed direct sequencing of the PCR products. These novel mutations have also not been reported from any other country by other workers in the field. Kaur et al. (14) reported PVDHFR mutation rates (29.75%; n = 121) for their isolates that were lower than those of the present study (57.06%; n = 177). The reason for this discrepancy may be the sample distribution, since the majority of their isolates, i.e., 96 of 121 (79.34%), were obtained from Northern India, where wild-type PVDHFR predominates.
It seems that there is a selective pressure (positive natural selection) on the pvdhfr gene leading to the increased frequency of some of the alleles (Table 1, Table2). However, the selective sweep of any particular allele cannot be suggested at this time point, since the flanking neutral markers for each of the mutant PFDHFR alleles are not yet fixed in the population. This is because we have observed an enormous heterogeneity in these markers for each PVDHFR mutant allele among the isolates (M. T. Alam et al., unpublished data).
Regional variations in the PVDHFR mutation rate observed here (Table 1) were similar to those for PFDHFR reported previously (2) and in Fig. 4. Since PFDHFR mutation data were available from these regions (except TN), we analyzed them along with PVDHFR mutations to assess the cross-species impact of the drug. Interestingly, the PVDHFR mutation pattern was similar to that of the PFDHFR mutation pattern among the isolates (Fig. 3). There was a specific trend in DHFR mutations across the species. Isolates from each region also followed similar PVDHFR and PFDHFR mutation patterns (Fig. 4). Similar DHFR mutation trends across species indicate that SP treatment of P. falciparum patients also affected the P. vivax parasite population.
Although we have observed similar DHFR mutation patterns for both the species, there was a significant difference in the rate of DHFR mutations between P. vivax and P. falciparum. The overall DHFR mutation rate for P. vivax was significantly lower than that for P. falciparum (Fig. 3). Considering the fact that SP is used for P. falciparum malaria treatment and not for P. vivax, the latter parasite expectedly showed the lower rate of DHFR mutations among all these regions of the country. However, the frequency of PVDHFR mutations was higher in the P. falciparum-dominated areas and lower in the P. vivax-dominated areas except Goa and TN (Table 1). SP resistance also varies in these regions, and it is higher in the P. falciparum-dominated areas due to its expectedly higher usage (1-3, 17). Among the mainland isolates, the quadruple PVDHFR mutations were observed only in Assam, where the SP pressure was higher since it is the first-line drug to treat falciparum malaria in this area (17). Therefore, it seems that the rate of PVDHFR mutations increased in P. falciparum-dominated areas, which had higher usage of SP, and vice versa. Surprisingly, so far, no P. falciparum isolate from Assam has been found to contain the quadruple PFDHFR mutations as seen in A&N (2). Indeed, we have not detected any quadruple PFDHFR mutations among the mainland isolates so far (1, 2). These results indicate that the P. falciparum strains from A&N are probably different from those from mainland India and therefore contain quadruple DHFR mutations similar to those of Southeast Asian strains, or quadruple mutations in PVDHFR arise faster than those in PFDHFR. This requires further investigations.
It seems from our results that the antifolates used for P. falciparum treatment are also affecting P. vivax mutations. This warrants the careful usage of antifolates in order to check the development of cross-species resistance.
Acknowledgments
Financial support for the work came from the Department of Biotechnology (Government of India) and the Indian Council of Medical Research. M.T.A. and P.K.B. acknowledge the Council of Scientific and Industrial Research and University Grants Commission for Senior Research Fellowships, respectively. H.B. received a Junior Research Fellowship from the Indian Council of Medical Research.
We are grateful to S. N. Dwivedi for statistical analysis of the data and M. R. Ranjit and Sumiti Vinayak for help and discussions. We also acknowledge facilities of the Biotechnology Information System (BTIS).
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Ahmed, A., D. Bararia, S. Vinayak, M. Yameen, S. Biswas, V. Dev, A. Kumar, M. A. Ansari, and Y. D. Sharma. 2004. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine pyrimethamine resistance. Antimicrob. Agents Chemother. 48:879-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, A., M. K. Das, V. Dev, M. A. Saifi, Wajihullah, and Y. D. Sharma. 2006. Quadruple mutations in dihydrofolate reductase of Plasmodium falciparum isolates from Car Nicobar Island, India. Antimicrob. Agents Chemother. 50:1546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, A., V. Lumb, M. K. Das, V. Dev, Wajihullah, and Y. D. Sharma. 2006. Prevalence of mutations associated with higher levels of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Car Nicobar and Assam, India. Antimicrob. Agents Chemother. 50:3934-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brega, S., F. de Monbrison, C. Severini, R. Udomsangpetch, I. Sutanto, P. Ruckert, F. Peyron, and S. Picot. 2004. Real-time PCR for dihydrofolate reductase gene single-nucleotide polymorphisms in Plasmodium vivax isolates. Antimicrob. Agents Chemother. 48:2581-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Pecoulas, P. E., R. Tahar, P. Yi, K. H. Thai, and L. K. Basco. 2004. Genetic variation of the dihydrofolate reductase gene in Plasmodium vivax in Snoul, northeastern Cambodia. Acta Trop. 92:1-6. [DOI] [PubMed] [Google Scholar]
- 6.de Pecoulas, P. E., L. K. Basco, R. Tahar, T. Ouatas, and A. Mazabraund. 1998. Analysis of the Plasmodium vivax dihydrofolate reductase thymidylate synthase gene sequence. Gene 211:177-185. [DOI] [PubMed] [Google Scholar]
- 7.de Pecoulas, P. E., R. Tahar, T. Ouatas, A. Mazabraud, and L. K. Basco. 1998. Sequence variations in the Plasmodium vivax dihydrofolate reductase thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol. Biochem. Parasitol. 92:265-273. [DOI] [PubMed] [Google Scholar]
- 8.Gregson, A., and C. V. Plowe. 2005. Mechanism of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 9.Hastings, M. D., J. D. Maguire, M. J. Bangs, P. A. Zimmerman, J. C. Reeder, J. K. Baird, and C. H. Sibley. 2005. Novel Plasmodium vivax dhfr alleles from the Indonesian Archipelago and Papua New Guinea: association with pyrimethamine resistance determined by a Saccharomyces cerevisiae expression system. Antimicrob. Agents Chemother. 49:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings, M. D., K. M. Porter, J. D. Maguire, I. Susanti, W. Kania, M. J. Bangs, C. H. Sibley, and J. K. Baird. 2004. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J. Infect. Dis. 189:744-750. [DOI] [PubMed] [Google Scholar]
- 11.Imwong, M., S. Pukrittakayamee, S. Looareesuwan, G. Pasvol, J. Poirriez, N. J. White, and G. Snounou. 2001. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 45:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imwong, M., S. Pukrittayakamee, L. Renia, F. Letourneur, J. P. Charlieu, U. Leartsakulpanich, S. Looareesuwan, N. J. White, and G. Snounou. 2003. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: Evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 47:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jalah, R., R. Sarin, N. Sud, M. T. Alam, N. Parikh, T. K. Dash, and Y. D. Sharma. 2005. Identification, expression, localization and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol. Biochem. Parasitol. 142:158-169. [DOI] [PubMed] [Google Scholar]
- 14.Kaur, S., S. K. Prajapati, K. Kalyanaraman, A. Mohmmed, H. Joshi, and V. S. Chauhan. 2006. Plasmodium vivax dihydrofolate reductase point mutations from the Indian subcontinent. Acta Trop. 97:174-180. [DOI] [PubMed] [Google Scholar]
- 15.Leartsakulpanich, U., M. Imwong, S. Pukrittayakamee, N. J. White, G. Snounou, W. Sirawaraporn, and Y. Yuthavong. 2002. Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol. Biochem. Parasitol. 119:63-73. [DOI] [PubMed] [Google Scholar]
- 16.Na, B. K., H. W. Lee, S. U. Moon, T. S. In, K. Lin, M. Maung, G. T. Chung, J. K. Lee, T. S. Kim, and Y. Kong. 2005. Genetic variations of the dihydrofolate reductase gene of Plasmodium vivax in Mandalay Division, Myanmar. Parasitol. Res. 96:321-325. [DOI] [PubMed] [Google Scholar]
- 17.Sharma, V. P. 1999. Current scenario of malaria in India. Parasitologia 41:349-353. [PubMed] [Google Scholar]
- 18.Schunk, M., W. P. Kumma, I. B. Miranda, M. E. Osman, S. Roewer, A. Alano, T. Loscher, U. Bienzle, and F. P. Mockenhaupt. 2006. High prevalence of drug-resistance mutations in Plasmodium falciparum and Plasmodium vivax in Southern Ethiopia. Malar. J. 5:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh, N., S. S. Mishra, M. P. Singh, and V. P. Sharma. 2000. Seasonality of Plasmodium vivax and P. falciparum in tribal villages in central India (1987-1995). Ann. Trop. Med. Parasitol. 44:101-112. [DOI] [PubMed] [Google Scholar]
- 20.Tahar, R., P. E. de Pecoulas, L. K. Basco, M. Chiadmi, and A. Mazabraud. 2001. Kinetic properties of dihydrofolate reductase from wild-type and mutant Plasmodium vivax expressed in Escherichia coli. Mol. Biochem. Parasitol. 113:241-249. [DOI] [PubMed] [Google Scholar]
- 21.Tjitra, E., J. Baker, S. Suprianto, Q. Cheng, and N. M. Anstey. 2002. Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob. Agents Chemother. 46:3947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valecha, N., H. Joshi, A. Eapen, J. Ravinderan, A. Kumar, S. K. Prajapati, and P. Ringwald. 2006. Therapeutic efficacy of chloroquine in Plasmodium vivax from areas with different epidemiological patterns in India and their Pvdhfr gene mutation pattern. Trans. R. Soc. Trop. Med. Hyg. 100:831-837. [DOI] [PubMed] [Google Scholar]
- 23.Wang, P., C. S. Lee, R. Bayoumi, A. Djimde, O. Doumbo, G. Swedberg, L. D. Das, H. Mshinda, M. Tanner, W. M. Watkins, P. F. G. Sims, and J. E. Hyde. 1997. Resistance to antifolate in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a larger number of field samples of diverse origin. Mol. Biochem. Parasitol. 89:161-177. [DOI] [PubMed] [Google Scholar]