Abstract
The artemisinin-based combination therapies artemether-lumefantrine (AL) and amodiaquine (AQ) plus artesunate have been adopted for treatment of Plasmodium falciparum malaria in many African countries. Molecular markers of parasite resistance suitable for surveillance have not been established for any of the component drugs in either of these combinations. We assessed P. falciparum mdr1 (Pfmdr1) alleles present in 300 Tanzanian children presenting with uncomplicated falciparum malaria, who were enrolled in a clinical trial of antimalarial therapy. Pfmdr1 genotype analysis was also performed with isolates from 182 children who failed AQ monotherapy and 54 children who failed AL treatment. Pfmdr1 alleles 86Y, 184Y, and 1246Y were more common among treatment failures in the AQ group than among pretreatment infections. The converse was found in the AL-treated group. Children presenting with the 86Y/184Y/1246Y Pfmdr1 haplotype and treated with AQ were significantly more likely to retain this haplotype if they were parasite positive during posttreatment follow-up than were children treated with AL (odds ratio, 33.25; 95% confidence interval, 4.17 to 1441; P, <0.001). We conclude that AL and AQ exert opposite within-host selective effects on the Pfmdr1 gene of P. falciparum.
The emergence and spread of Plasmodium falciparum resistance to antimalarial drugs is now one of the greatest challenges facing the global effort to control malaria. In Africa, until recently, there has been a reliance on the cheap drugs chloroquine and sulfadoxine-pyrimethamine. To protect drugs from resistance, there is now clear evidence that combining them can improve their efficacy without increasing their toxicity (13), and with the development of highly effective artemisinin derivatives, there is renewed hope for the treatment of malaria in the form of artemisinin-based combination therapy (ACT). In 2001, the World Health Organization recommended ACTs as the first-line treatment for uncomplicated malaria (31).
ACTs protect the individual drugs from resistance by relying on the principle of combining two drugs with different mechanisms of action (30). The fast-acting artemisinin derivative rapidly clears the main parasite load within the few hours that it remains at therapeutic levels and thus reduces subsequent gametocyte carriage (28), while the partner drug, which is generally longer lasting, remains to clear the rest of the parasites. The combination of artesunate and mefloquine has been used with success in Southeast Asia (17). In sub-Saharan Africa, most countries have now adopted either artemether-lumefantrine (AL), which is coformulated as Coartem, or artesunate (AS)-amodiaquine (AQ) as their first-line ACT, although the transition to widespread deployment is only just beginning. Treatment success with an ACT will depend largely on the parasite's existing level of tolerance to the partner drug, and therefore, molecular markers of drug resistance can be useful tools in the population surveillance of drug efficacy. There is significant interest in describing genetic mutations that are associated with resistance to ACT partner drugs and to the artemisinins themselves.
AQ is a 4-aminoquinoline related to chloroquine (CQ), and the mode of action of the two drugs appears to be similar; both are known to be concentrated in the parasite's lysosome (5). Although CQ resistance is now widespread across the African continent, the efficacy of AQ is variable in treatment trials (12). A mutation from lysine (K) to threonine (T) at codon 76 of the P. falciparum chloroquine resistance transporter (Pfcrt) gene is associated with resistance to both CQ and AQ (1, 8, 11). However, in vitro and in vivo evidence suggests that cross-resistance between the two drugs is incomplete (12, 23). Therefore, AQ resistance is likely to be determined by additional genotypic changes found in some, but not all, CQ-resistant parasites. The identification of such changes would facilitate the rational deployment of AQ-containing combinations in settings where resistance-associated alleles were rare.
Numerous in vitro studies of polymorphisms in the P. falciparum multidrug-resistant 1 (Pfmdr1) gene have concluded that its protein product, Pgh1, plays a role in modulating levels of resistance to several structurally unrelated drugs (4, 21, 24). Five distinct single-nucleotide substitutions (SNPs), leading to amino acid changes at codons 86, 184, 1034, 1042, and 1246, have been described. The N86Y change of the Pfmdr1 gene product is favorable for resistance to both CQ and AQ and is concomitantly associated with increased sensitivity to mefloquine and halofantrine (4). Sidhu et al. (24) have shown, using reverse genetics, that the 1042D and 1034C/1042D/1246Y alleles are associated with increased resistance to quinine and increased sensitivity to mefloquine and artemisinin drugs. This finding supported the earlier work of Reed et al. (21) using allelic replacement experiments.
Studies from Southeast Asia have demonstrated an association between the increased copy number of the Pfmdr1 gene and treatment failure with mefloquine (15, 19), supporting in vitro studies showing that laboratory lines with amplified Pfmdr1 loci have reduced susceptibility to mefloquine and related drugs such as halofantrine (18). Recent evidence from Thailand suggests that amplification of Pfmdr1 can also modulate susceptibility to the artemether-lumefantrine ACT, leading to poor treatment response in some patients (20).
Two African clinical trial reports provide evidence for the selection of particular Pfmdr1 alleles after AL treatment. In Zanzibar, the posttreatment prevalence of 86N was significantly higher than pretreatment levels in parasites that reinfected patients within the 42-day follow-up period (25). This finding is supported by evidence from Uganda (2), where AL treatment for uncomplicated malaria selected newly infecting parasites carrying the 86N, 184F, and 1246D alleles. Dokomajilar et al. found no evidence for an increased Pfmdr1 copy number in either pre- or posttreatment infections, suggesting that amplification of the Pfmdr1 locus may be less important in African parasite populations (2).
To further investigate the effect of antimalarial treatment on different alleles of the Pfmdr1 gene, we present genetic analyses of samples from a randomized four-arm antimalarial effectiveness trial in Muheza, Tanzania (10). In this trial, AQ monotherapy exhibited a high treatment failure rate of 76%, whereas AL was efficacious but permitted the establishment of new infections in 18% of subjects toward the end of the 28-day follow-up period. The prevalence of polymorphisms at Pfcrt codon positions 72 to 76 and Pfmdr1 changes N86Y, Y184F, S1034C, N1042D, and D1246Y were found in pretreatment parasite isolates and in a matched pair analysis of pre- and posttreatment samples from the AQ monotherapy and AL treatment arms of the trial.
MATERIALS AND METHODS
Subjects.
Briefly, a randomized four-arm drug effectiveness trial was carried out between September 2002 and October 2004. A total of 1,811 children (ages 4 to 59 months) were recruited from the Muheza district, Tanzania, and followed for 28 days. The drug groups consisted of those undergoing AQ monotherapy (terminated after 1 year due to a high level of treatment failure), AQ+sulfadoxine-pyrimethamine (SP), AQ+AS, and AL. Apart from the first dose, drug administration was not supervised, in order to provide an estimation of drug effectiveness in a more “real-world” setting. Full details of the trial, including recruitment, randomization, and follow-up have been published previously (10). After PCR detection of the polymorphic merozoite surface protein 2 gene (msp2 genotyping) to distinguish recrudescences from new infections, the percentage of parasitological treatment failures in each drug group was AQ, 48.5%; AQ+SP, 34.8%; AQ+AS, 12.1%; and AL, 2.7% (10).
Molecular analysis of filter paper-preserved blood samples collected during year 1 of the trial was carried out with two groups, as follows: (i) a block-randomized pretreatment subset of samples selected to provide a baseline estimate of allele prevalences, irrespective of drug group or treatment outcome (n = 300); and (ii) in patients who had parasitological treatment failure (i.e., those with recurrent parasites, with or without symptoms, during the 28-day follow-up) after AQ monotherapy or AL, samples were tested for the selection of alleles between day 0 (D0; pretreatment) and day of treatment failure (Dfail; from day 3 up to day 28). A total of 182 (AQ) and 54 (AL) treatment failures were analyzed as matched pairs, D0/Dfail.
Genotyping.
DNA was extracted from blood spots on filter paper by using the Chelex method (16).
To distinguish recrudescent from new infections in parasitological treatment failures, the block 3 region of msp2, which exists in three allelic forms, was amplified using allele-specific primers, and size polymorphisms were detected by agarose gel electrophoresis (26). Where 50% or more of the alleles present in the Dfail sample were new, compared to the D0 sample, this was classed as a new infection.
SNPs that resulted in an amino acid change at codons in the Pfcrt and Pfmdr1 genes were determined by PCR amplification, followed by a sequence-specific oligonucleotide probe assay adapted from Pearce et al. (14). The Pfmdr1 gene codons tested were N86Y (i.e., denoting an encoded amino acid change from asparagine to tyrosine at codon 86), F184Y, S1034C, D1042N, D1246Y, and in Pfcrt the codons tested were C72S, M74I, N75E, and K76T.
Two fragments from the Pfmdr1 gene and one from the Pfcrt gene, designed to encompass all SNP sites under investigation, were amplified using the primers and conditions shown in Table 1. All fragments were amplified by nested PCR in a 96-well plate format. The 25-μl PCR mixes contained primers at 200 nM final concentration, 1.5 mM MgCl2, 250 μM each deoxynucleotide triphosphate, and 1 U Bioline TaqI polymerase (Bioline, London, United Kingdom). Extracted DNA (5 μl) was added to each first-round PCR mixture. One microliter of the first-round product was then used as a template in a 25-μl nested amplification. DNA samples extracted from parasite lines 3D7, W2, and 7G8 were used as positive controls, as these lines carry all the Pfcrt and Pfmdr1 mutations under examination.
TABLE 1.
PCR primer sequences and reaction conditions for the nested amplification of Pfcrt and Pfmdr1 fragments
| Gene fragment | Primer name | Primer sequence | PCR product size (bp) | PCR cycling conditionsa |
|---|---|---|---|---|
| Pfcrt (for SNPs at codons 72 to 76) | Outer forward P1 | 5′-CCGTTAATAATAAATACACGCAG | 546 | 35 cycles of 94°C for 30 s; 56°C |
| Outer reverse P2 | 5′-CGGATGTTACAAAACTATAGTTACC | for 30 s; and 62°C for 1 min; then 62°C for 5 min | ||
| Nested forward D3 | 5′-AGGTTCTTGTCTTGGTAAATTTGC | 164 | 30 cycles of 94°C for 30 s; 56°C | |
| Nested reverse D2 | 5′-CAAAACTATAGTTACCAATTTTG | for 30 s; and 65°C for 1 min; then 65°C for 5 min | ||
| Pfmdr1 fragment 1 (for SNPs at codons 86 and 184) | Outer forward FN1/1 | 5′-AGGTTGAAAAAGAGTTGAAC | 578 | 30 cycles of 94°C for 30 s; 55°C |
| Outer reverse REV/C1 | 5′-ATGACACCACAAACATAAAT | for 30 s; and 65°C for 1 min; then 65°C for 5 min | ||
| Nested forward MDR2/1 | 5′-ACAAAAAGAGTACCGCTGAAT | 534 | 30 cycles of 94°C 30 s; 60°C | |
| Nested reverse NEWREV1 | 5′-AAACGCAAGTAATACATAAAGTC | for 30 s; and 65°C for 1 min; then 65°C for 5 min | ||
| Pfmdr1 fragment 2, long (for SNPs at codons 1034, 1042, and 1246) | Outer forward MDRFR2F1 | 5′-GTGTATTTGCTGTAAGAGCT | 958 | 34 cycles of 94°C for 30 s; 55°C |
| Outer reverse MDRFR2R1 | 5′-GACATATTAAATAACATGGGTTC | for 1 min; and 65°C for 1.5 min; then 65°C for 5 min | ||
| Nested forward MDRFR2F2 | 5′ CAGATGATGAAATGTTTAAAGATC | 864 | 29 cycles of 94°C for 30 s; 60°C | |
| Nested reverse MDRFR2R2 | 5′-TAAATAACATGGGTTCTTGACT | for 30 s; and 65°C for 1 min; then 65°C for 5 min |
Each PCR program was preceded by an initial denaturation step of 94°C for 3 min.
The denatured, nested PCR products were UV cross-linked to nylon membranes. After blocking, one membrane was exposed to an 18-bp 3′-digoxigenin-labeled oligonucleotide probe specific to the wild-type sequence. A second identical membrane was exposed to a similar probe bearing the mutant sequence (Table 2). High-stringency washing in 3 M tetramethylammonium chloride ensured that only probes with the exact complementary sequence remained hybridized to the immobilized PCR product; those with a single nucleotide mismatch at the SNP site were washed off (14). Probes that remained bound were detected by incubation with antidigoxigenin Fab fragments conjugated to alkaline phosphatase, followed by exposure to the substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.3.13,7]decan}-4-yl) phenyl phosphate. The resulting chemiluminescence was detected by autoradiography and scored visually by two readers.
TABLE 2.
Oligonucleotide probes used to detect sequence polymorphisms in genes Pfcrt and Pfmdr1
| Probe namea | Probe sequence |
|---|---|
| Pfcrt | |
| 72C | 5′-TAT TTA AGT GTA TGT GTA |
| 72S | 5′-TAT TTA AGT GTA AGT GTA |
| 74-76 IET | 5′-TA ATT GAA ACA ATT TTT G |
| 74-76 MNK | 5′-TA ATG AAT AAA ATT TTT G |
| 74-76 MNT | 5′-TA ATG AAT ACA ATT TTT G |
| Pfmdr1 | |
| 86 N | 5′-AG AAC ATG AAT TTA GGT G |
| 86 Y | 5′-AG AAC ATG TAT TTA GGT G |
| 184 Y | 5′-A GGT TTA TAT ATT TGG TC |
| 184 F | 5′-A GGT TTA TTT ATT TGG TC |
| 1034 S | 5′-A TGG GGA TTC AGT CAA AG |
| 1034 C | 5′-A TGG GGA TTC TGT CAA AG |
| 1042 N | 5′-TA TTT ATT AAT AGT TTT G |
| 1042 D | 5′-TA TTT ATT GAT AGT TTT G |
| 1246 D | 5′-AC TTA AGA GAT CTT AGA A |
| 1246 Y | 5′-AC TTA AGA TAT CTT AGA A |
The probe name indicates the polymorphic codon position and encoded amino acid. Nucleotide substitutions associated with drug resistance are in boldface type.
Statistical analysis.
Statistical analysis was carried out using STATA (version 9) software. A nonparametric test for matched pair data (McNemar's test) was used to analyze the selection of SNPs at each locus for D0 and the day of recurrent parasitemia (Dfail).
RESULTS
Baseline allele prevalence.
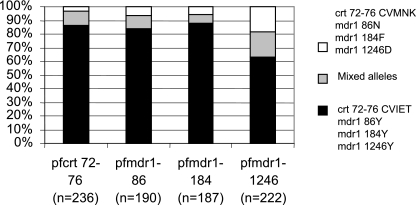
A baseline sample of 300 isolates for genotyping was generated by block randomization of pretreatment samples from all four arms of the trial, representing 27.3% of all participants in year 1 of the trial. Only the CVIET and CVMNK alleles of Pfcrt at codon positions 72 to 76 were identified in the pretreatment baseline. All of the analyzed samples contained the 1034S or 1042N alleles of the Pfmdr1 gene. Where both wild-type and mutant alleles were detected in the same sample, they were scored as mixed infections and were included in subsequent analyses as described. The prevalence of each allele at the three polymorphic loci in Pfmdr1 is shown in Fig. 1. The number of successful PCR amplifications is shown for each locus on the x axis.
FIG. 1.
Pretreatment random baseline prevalence of Pfcrt and Pfmdr1 alleles. The white portion of the bars shows the prevalence of the alleles associated with CQ sensitivity. The black portion shows the prevalence of alleles associated with CQ resistance. The gray portion shows the prevalence of samples where a mixed infection containing both alleles was detected.
Presentation alleles and treatment outcomes.
Using parasitological failure by day 28 as the outcome measure, we analyzed the baseline samples stratified by treatment group. Within the subset of subjects who had received AQ monotherapy, a significant association with parasitological failure was found, with the presence on day 0 of either the 86Y allele (Fisher's exact test, P = 0.032; n = 40) or the 184Y allele (P = 0.0054; n = 42). Conversely, pretreatment carriage of either the 86N allele (P = 0.005; n = 52) or the 184F allele (P = 0.0262; n = 52) was associated with treatment failure in the AL treatment group.
Allele prevalence among treatment failures.
Pfcrt genotypes were examined in the AQ treatment group only. All infections persisting after AQ treatment harbored the CVIET allele at amino acid positions 72 to 76, and 5% also carried mixed infections with the CVMNK allele.
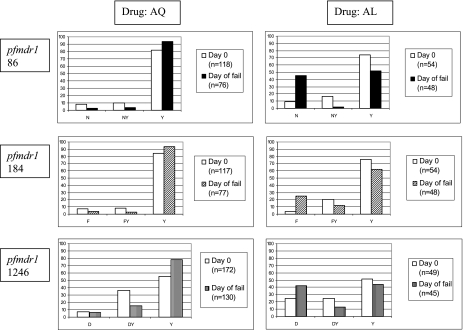
Within both the AQ and AL treatment groups, all the children who failed treatment by day 28 had both their pre- and posttreatment samples analyzed, and the allele prevalence at each locus, generated from all available genotyping results at each time point, is shown in Fig. 2. The number of samples involved in each calculation is shown in the legend for each graph. The 86Y allele prevalence increased after treatment with AQ from 81.4% to 93.4% and decreased after treatment with AL from 74.1% to 52.1%. Similarly, the 184Y allele was present in 93.5% of posttreatment infections from children who failed AQ treatment and in only 62.5% of those from children who failed AL treatment. For the 1246 locus, the posttreatment proportions of Y alleles were 78.5% and 44.4% in the AQ and AL arms, respectively. The pattern of Pfmdr1 allele prevalence change observed across the AL group is very similar to that reported recently by Dokomajilar et al. (2) from their study of Ugandan children, confirming that the alleles of interest in this group are Pfmdr1 86N, 184F, and 1246D. In contrast, the alleles of interest in the AQ treatment groups are Pfmdr1 86Y, 184Y, and 1246Y.
FIG. 2.
Prevalence of pre- and posttreatment Pfmdr1 alleles following therapy with either AQ or AL. Mixed infections are shown as separate columns in the middle of each graph. The number of samples involved in each prevalence calculation is shown in the legend in each graph.
Matched pair analysis of AQ monotherapy.
There were a total of 182 treatment failures in the AQ monotherapy arm of the trial. Of the 137 for which msp2 genotyping data were obtained, 45 (33%) were classed as new infections after PCR correction. Matched pre- and posttreatment data for the same individual were analyzed at Pfmdr1 loci 86, 184, and 1246 using a stringent two-by-two approach, with the omission of mixed infections harboring both alleles. After treatment with AQ, there was no evidence of directional selection for Pfmdr1 86Y or 184Y, but there was significant directional selection of Pfmdr1 1246Y (P = 0.0253) (Table 3).
TABLE 3.
Analysis of directional selection acting on the single locus alleles at Pfmdr1 positions 86, 184, and 1246 from children with parasites following therapy with either AQ or ALa
| AQ treatment group
|
AL treatment group
|
||||
|---|---|---|---|---|---|
| Allele present at D0 (n) | No. of alleles changed on Dfail | McNemar's χ2 test (P value) | Allele present at D0 (n) | No. of alleles changed on Dfail | McNemar's χ2 test (P value) |
| Pfmdr1 86Y (43) | 1 | 0.1797 | Pfmdr1 86Y (43) | 17 | 0.0017 |
| Pfmdr1 86N (5) | 4 | Pfmdr1 86N (5) | 3 | ||
| Pfmdr1 184Y (52) | 0 | 0.1573 | Pfmdr1 184Y (30) | 8 | 0.0196 |
| Pfmdr1 184F (4) | 2 | Pfmdr1 184F (1) | 1 | ||
| Pfmdr1 1246Y (62) | 0 | 0.0253 | Pfmdr1 1246Y (15) | 8 | 0.2482 |
| Pfmdr1 1246D (9) | 5 | Pfmdr1 1246D (10) | 4 | ||
The numbers of samples tested with each allele present on D0 (n) are shown with the allele name in columns one and four. The number of samples in which the other allele was detected (i.e., allele changed) in the Dfail sample is shown in columns two and five. The direction of the allele changes from D0 to Dfail were assessed using McNemar's χ2 test for paired samples. Significant P values are in boldface. Mixed infections harboring both alleles were omitted for this analysis.
Matched pair analysis of AL treatment.
There were a total of 54 treatment failures in the AL combination treatment arm of the trial (year 1 only). By msp2 genotyping, 39 of 45 interpretable treatment failure cases were classed as new infections (87%). Using the same stringent two-by-two tables as for the AQ arm, we found evidence for significant directional selection of Pfmdr1 86N (P = 0.0017) and 184F (P = 0.0196) after treatment with AL (Table 3).
Haplotype analysis.
Genotyping data for all three Pfmdr1 loci were combined to determine whether the 86Y/184Y/1246Y haplotype was present in each pre- and posttreatment sample in both treatment groups. If a sample contained one mixed allele (e.g., if Pfmdr1 86Y, 184Y, 1246Y, and 1246D were detected), we assumed that the haplotype of interest, YYY, must have occurred in one parasite genome within this sample (along with haplotype YYD) and therefore we included it in the analysis. If two or all three alleles were mixed, we have included these also, but it should be noted that there is a possibility that the YYY haplotype did not actually occur within a single genome in these samples. This effectively collapses each isolate into one of two states at each locus.
Pre- and posttreatment genotype data were available at all three Pfmdr1 loci (86, 184, and 1246) for 47 of those children who failed treatment in the AQ treatment group. Of those, 39 individuals harbored the YYY haplotype. In six of these isolates, mixed alleles were present at more than one of the three loci, but these were kept in the analysis. In 38 of these 39 individuals, the YYY haplotype remained in the treatment failure sample (Table 4). However, among AL treatment failures with available matched genotype data at all three loci, only 16 out of 30 samples that carried YYY at presentation harbored the YYY haplotype after treatment (Table 4). Therefore, children presenting with the 86Y/184Y/1246Y Pfmdr1 haplotype and treated with AQ were significantly more likely to retain this haplotype if they were found to be parasite positive during posttreatment follow-up than were children treated with AL (OR, 33.25; 95% CI, 4.17 to 1441; P, <0.001). The converse analysis, that is, analysis of the NFD haplotype, did not indicate a difference between the two treatment groups in persistence of infections harboring this genotype. However, as the NFD haplotype was rare in presentation infections, comprising only 2.4% of infections in the random baseline (total n = 164), this analysis lacked sufficient power to detect even moderate directional selection. Interestingly, the NFD haplotype was more common among pretreatment isolates of AL patients who later failed treatment (8 out of 49) than among pretreatment isolates of AQ-treated patients who later failed treatment (7 out of 107; OR, 2.79; 95% CI, 0.818 to 9.61; P = 0.0543). This is consistent with the single-locus analyses presented above.
TABLE 4.
Analysis of directional selection acting on the three-locus haplotype YYY at Pfmdr1 positions 86, 184, and 1246 from children with parasites following therapy with either AQ or ALa
| Drug | Haplotype on D0 | Haplotype on Dfail
|
P value | Comment | |||
|---|---|---|---|---|---|---|---|
| Any YYY | No YYY | Any NFD | No NFD | ||||
| AQ | Any YYY | 38 | 1 | 0.0339 | Directional selection for YYY | ||
| No YYY | 7 | 1 | |||||
| Any NFD | 0 | 3 | 0.3173 | No evidence of directional selection | |||
| No NFD | 1 | 44 | |||||
| AL | Any YYY | 16 | 14 | 0.0389 | Directional selection against YYY | ||
| No YYY | 5 | 6 | |||||
| Any NFD | 2 | 6 | 0.5930 | No evidence of directional selection | |||
| No NFD | 8 | 25 | |||||
The direction of the haplotype changes from D0 to Dfail were assessed using McNemar's χ2 test for paired samples. Only the samples where a change occurs (in bold type) contribute to this analysis. Significant P values are in bold type. Mixed alleles YN, YF, and YD are coded as “Y” in this analysis.
Linkage tests.
The YYY and NFD haplotypes of Pfmdr1 may be maintained in the parasite population by linkage disequilibrium. To investigate linkage disequilibrium in the population, pairwise associations between loci were examined in the baseline sample. Loci 86 and 184 of Pfmdr1 were significantly associated (n = 165; OR, 35.8; 95% CI, 6.30 to 201; P, <0.0001), as were loci 86 and 1246 (n = 129; OR, 11.6; 95% CI, 2.47 to 71.6; P, <0.0001), but positions 184 and 1246 were not strongly associated (n = 127; OR, 2.83; 95% CI, 0.518 to 14.1; P = 0.127). Only samples where single alleles were detected at both loci were included in the test. However, to confirm our estimations of haplotype and linkage analysis, full gene sequence data would be required.
Copy number of Pfmdr1.
The copy number of Pfmdr1 was determined in paired samples (day 0, day of treatment failure) from the AQ treatment group (n = 8 pairs) and the AL group (n = 15 pairs). The estimated gene copy number was close to 1.0 in all isolates tested (data not shown). The method used was adapted from Price et al. (19) for use on a Rotorgene 3000 (Corbett, Australia).
DISCUSSION
In this study, we examined the carriage of alleles of the Pfmdr1 gene in Tanzanian children with uncomplicated falciparum malaria before and after treatment with AQ or AL. Analysis of within-host selection at each locus and of the survival of specific three-locus haplotypes after treatment provided clear evidence that different Pfmdr1 alleles were selected in the two treatment groups. Our data suggest that AQ treatment positively selects for parasites harboring the Pfmdr1 86Y/184Y/1246Y haplotype, whereas AL treatment selects against such parasites, leading to enhanced survival of parasites with the Pfmdr1 86N/184F/1246D haplotype. We note that, as shown by the msp2 genotype results for the matched pair analysis, each treatment appears to exert its selective effect in different ways; AQ treatment failures are predominantly recrudescent infections in which AQ-tolerant parasites present in the D0 sample have survived treatment, whereas AL treatment failures are predominantly new infections, indicating that while most malaria parasites are cleared by AL, drug-tolerant genotypes have been able to reinfect hosts soon after treatment.
Among AL-treated children, the YYY Pfmdr1 haplotype tended to be replaced by the NFD haplotype in posttreatment infections, although the latter form of the gene was uncommon in the population as a whole. Our results are consistent with those from a recent study in Zanzibar (25), where the prevalence of 86N increased from 15% at day 0 to 41% at day of failure in children who failed AL treatment, and with the results of Dokomajilar et al. (2), who observed significant accumulation of Pfmdr1 alleles 86N, 184F, and 1246D among Ugandan children who were parasite positive after treatment with AL. We have extended the findings of these studies with regard to AL-treated cases by examining within-host selection and presenting new evidence that AL selects for the three-locus NFD haplotype. Furthermore, in contrast to the findings of Dokomajilar et al. (2), we found that the Pfmdr1 alleles carried at baseline were associated with different treatment outcomes in the two treatment groups.
This is the first study to systematically examine the role of Pfmdr1 haplotypes in AQ treatment failures in vivo. It is clear that multiple genes are responsible for AQ resistance. Previous studies of Pfcrt alleles in AQ treatment failures have found an incomplete association with the chloroquine-resistant K76T mutation (8, 11), in that this mutation alone is clearly not sufficient to confer in vivo resistance to AQ. In vitro studies show that the Pfcrt codon 72-to-76 SVMNT haplotype is associated with greatly reduced sensitivity to CQ and AQ in cultured parasites (23, 29). In contrast, parasite lines with the “African” Pfcrt CQ-resistant haplotype of CVIET show a range of in vitro sensitivities to AQ (23). We hypothesize that in African parasites carrying Pfcrt CVIET, such as the majority of isolates studied here, Pfmdr1 mutations also contribute to AQ resistance.
The prevalence of the 184Y allele of Pfmdr1 observed in this study is very different from that which we observed in West Africa, where the baseline prevalence of 184Y was 26% (9). The role of the 184 mutation in drug resistance remains unclear, suggesting that the use of CQ has selected different haplotypes of Pfmdr1 in different locations, depending on the predominant alleles of Pfcrt and other genes present in these populations. Analysis of the genetic diversity around the Pfmdr1 gene may provide evidence that a selective sweep has favored the spread of these genes, as has been shown for the alleles of Pfdhfr and Pfdhps associated with resistance to SP (22).
The increase in prevalence of alleles 86N, 184F, and 1246D after AL treatment can be explained in a number of ways. First, in vitro studies suggest that those parasites carrying “wild-type” 86N alleles have a higher fitness after removal of the selective drug pressure of CQ (7). Thus, the increase in prevalence may reflect a rebound in fitter, CQ-sensitive parasites. Second, in AL-treated patients, extended exposure of parasites to lumefantrine after clearance of artemether occurs and this may select for new infections emerging from the liver, with parasites carrying alleles 86N, 184F, and 1246D. The latter hypothesis is supported by in vitro studies demonstrating the increased tolerance of laboratory parasite clones to the related drugs mefloquine and halofantrine, if they carry the 86N allele (4) or the 1034S/1042N/1246D allele (24). If correct, this hypothesis might predict that in African settings where high transmission occurs and reinfection is common and these Pfmdr1 alleles are circulating, ACT based on AL may not have the success it has enjoyed in Southeast Asia. We and others have found no evidence that copy number of the Pfmdr1 gene contributes to either AL treatment failure or to AQ resistance, which suggests that sequence alteration rather than amplification is currently the predominant Pfmdr1-dependent mechanism of drug evasion in African parasite populations.
A third explanation for our observations for AL-treated patients is that posttreatment parasitemia was caused by recrudescence of parasite subpopulations that were at very low densities in the pretreatment population and thus not identified in the msp2 PCR genotyping assay. These may be remnants of the starting parasite population that survived the artemether concentrations present in the host in the first few days of treatment, and therefore, these genotypes have not only survived lumefantrine but may also bear a signal of artemisinin selection. Evidence to support this comes from our previous in vivo study of the combination CQ plus artesunate (CQ+AS) (27). Molecular analysis of the transmission potential of different genotypes demonstrated a fourfold reduction in transmissibility to mosquitoes of parasites harboring the 86Y allele compared to those with the 86N allele among children treated with CQ+AS. In contrast, among children receiving CQ monotherapy, the 86Y allele was associated with a 14-fold advantage (6). Thus the artesunate component was responsible for a 56-fold reduction of transmissibility of parasites carrying the 86Y allele compared to those carrying the 86N allele. This is also consistent with in vitro studies that show artemisinin sensitivity increasing in Pfmdr1 86Y allele-carrying parasites (4, 21, 24). We therefore hypothesize that part of the selective effect of AL is derived from the artemisinin component, which acts more strongly against parasites carrying Pfmdr1 86Y alleles than against those with 86N alleles.
There is now growing evidence for allelic selection after treatment with AL, demonstrating the importance of monitoring this combination closely in high transmission areas. The NFD haplotype at codon positions 86, 184, and 1246, as well as Pfmdr1 copy number (20), should be considered in future drug resistance studies with AL. Furthermore, we have shown that AQ monotherapy can select for the Pfmdr1 YYY haplotype at these codons. Thus, AQ and AL exert opposing selective effects, and the possibility of a balanced deployment of AL treatment for clinical malaria in children, together with AQ-containing combinations such as AQ+AS or AQ+SP for cases in older patients, should be considered in areas where AQ remains efficacious, such as the West African Sahel.
Acknowledgments
This research was funded by the Gates Malaria Partnership. C.J.S. is supported by the United Kingdom Health Protection Agency.
None of the authors have any commercial or other association that might pose a conflict of interest.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourte, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, C. V. Plowe, and D. Coulibaly. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257-263. [DOI] [PubMed] [Google Scholar]
- 2.Dokomajilar, C., S. L. Nsobya, B. Greenhouse, P. J. Rosenthal, and G. Dorsey. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Duraisingh, M. T., P. Jones, I. Sambou, L. von Seidlein, M. Pinder, and D. C. Warhurst. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13-23. [DOI] [PubMed] [Google Scholar]
- 5.Famin, O., and H. Ginsburg. 2002. Differential effects of 4-aminoquinoline-containing antimalarial drugs on hemoglobin digestion in Plasmodium falciparum-infected erythrocytes. Biochem. Pharmacol. 63:393-398. [DOI] [PubMed] [Google Scholar]
- 6.Hallett, R. L., C. J. Sutherland, N. Alexander, R. Ord, M. Jawara, C. J. Drakeley, M. Pinder, G. Walraven, G. A. T. Targett, and A. Alloueche. 2004. Combination therapy counteracts the enhanced transmission of drug-resistant malaria parasites to mosquitoes. Antimicrob. Agents Chemother. 48:3940-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayward, R., K. J. Saliba, and K. Kirk. 2005. Pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol. Microbiol. 55:1285-1295. [DOI] [PubMed] [Google Scholar]
- 8.Holmgren, G., J. P. Gil, P. M. Ferreira, M. I. Veiga, C. O. Obonyo, and A. Björkman. 2006. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect. Genet. Evol. 6:309-314. [DOI] [PubMed] [Google Scholar]
- 9.Meerman, L., R. Ord, J. T. Bousema, M. van Niekerk, E. Osman, R. Hallett, M. Pinder, G. Walraven, and C. J. Sutherland. 2005. Carriage of chloroquine-resistant parasites and delay of effective treatment increase the risk of severe malaria in Gambian children. J. Infect. Dis. 192:1651-1657. [DOI] [PubMed] [Google Scholar]
- 10.Mutabingwa, T. K., D. Anthony, A. Heller, R. Hallett, J. Ahmed, C. Drakeley, B. M. Greenwood, and C. J. M. Whitty. 2005. Amodiaquine alone, amodiaquine+sulfadoxine-pyrimethamine, amodiaquine+artesunate, and artemether-lumefantrine for outpatient treatment of malaria in Tanzanian children: a four-arm randomised effectiveness trial. Lancet 365:1474-1480. [DOI] [PubMed] [Google Scholar]
- 11.Ochong, E. O., I. V. van den Broek, K. Keus, and A. Nzila. 2003. Short report: association between chloroquine and amodiaquine resistance and allelic variation in the Plasmodium falciparum multiple drug resistance 1 gene and the chloroquine resistance transporter gene in isolates from the upper Nile in southern Sudan. Am. J. Trop. Med. Hyg. 69:184-187. [PubMed] [Google Scholar]
- 12.Olliaro, P., and P. Mussano. 2003. Amodiaquine for treating malaria. Cochrane Database Syst. Rev. 2:CD000016. [DOI] [PubMed] [Google Scholar]
- 13.Olliaro, P. L., and W. R. Taylor. 2004. Developing artemisinin based drug combinations for the treatment of drug resistant falciparum malaria: a review. J. Postgrad. Med. 50:40-44. [PubMed] [Google Scholar]
- 14.Pearce, R. J., C. Drakeley, D. Chandramohan, F. Mosha, and C. Roper. 2003. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob. Agents Chemother. 47:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. Scott Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plowe, C. V., A. Djimde, M. Bouare, O. Doumbo, and T. E. Wellems. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 52:565-568. [DOI] [PubMed] [Google Scholar]
- 17.Price, R., J. A. Simpson, P. Teja-Isavatharm, M. M. Than, C. Luxemburger, D. G. Heppner, T. Chongsuphajaisiddhi, F. Nosten, and N. J. White. 1999. Pharmacokinetics of mefloquine combined with artesunate in children with acute falciparum malaria. Antimicrob. Agents Chemother. 43:341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. van Vugt, N. J. White, F. Nosten, and S. Krishna. 1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, R. Patel, K. Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price, R. N., A.-C. Uhlemann, M. van Vugt, A. Brockman, R. Hutagalung, S. Nair, D. Nash, P. Singhasivanon, T. J. C. Anderson, S. Krishna, N. J. White, and F. Nosten. 2006. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin. Infect. Dis. 42:1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 22.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 23.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidhu, A. B., S. G. Valderramos, and D. A. Fidock. 2005. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 57:913-926. [DOI] [PubMed] [Google Scholar]
- 25.Sisowath, C., J. Strömberg, A. Mårtensson, M. Msellem, C. Obondo, A. Björkman, and J. P. Gil. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J. Infect. Dis. 191:1014-1017. [DOI] [PubMed] [Google Scholar]
- 26.Snounou, G., X. Zhu, N. Siripoon, W. Jarra, S. Thaithong, K. N. Brown, and S. Viriyakosol. 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 93:369-374. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland, C. J., C. J. Drakeley, U. Obisike, R. Coleman, M. Jawara, G. A. T. Targett, P. Milligan, M. Pinder, and G. Walraven. 2003. The addition of artesunate to chloroquine for the treatment of Plasmodium falciparum malaria in Gambian children delays, but does not prevent treatment failure. Am. J. Trop. Med. Hyg. 69:19-25. [PubMed] [Google Scholar]
- 28.Sutherland, C. J., R. Ord, S. Dunyo, M. Jawara, C. J. Drakeley, N. Alexander, R. Coleman, M. Pinder, G. Walraven, and G. A. T. Targett. 2005. Reduction of malaria transmission to Anopheles mosquitoes with a six-dose regimen of co-artememether. PLoS Med. 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warhurst, D. C. 2003. Polymorphism in the Plasmodium falciparum chloroquine-resistance transporter protein links verapamil enhancement of chloroquine sensitivity with the clinical efficacy of amodiaquine. Malaria J. 2:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, N. 1999. Antimalarial drug resistance and combination chemotherapy. Philos. Trans. R. Soc. Lond. B. 354:739-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. 2001. Antimalarial drug combination therapy: report of a WHO technical consultation. WHO, Geneva, Switzerland. www.rbm.who.int/cmc_upload/0/000/015/082/use_of_antimalarials2.