Abstract
Mosaic tetracycline resistance genes comprising tet(O), tet(W), and tet(32) sequences were abundant in DNA extracted from pig and human fecal samples, accounting for 78% (50/64) and 46% (37/80) of genes amplified with a tet(O) primer set, respectively, in two samples. The nonmosaic tet(32) gene was isolated from a human saliva bacterium.
Tetracycline resistance is one of the most common bacterial antibiotic resistances. Tetracycline resistance genes whose products have ≤80% amino acid sequence identity fall into 40 different gene classes (1, 4, 9), conferring resistance by 4 different mechanisms (2). Various mosaic ribosomal protection genes, presumably arising by recombination between wild-type genes, have been discovered recently. Mosaic tet(O/W) genes were prevalent in Megasphaera elsdenii isolates from swine farms in the United States (11) and conferred a higher level of resistance than the original tet(O) or tet(W) gene (12). The tet(32) ribosomal protection gene also exhibits a high level of resistance to tetracycline (6). This gene was recently reclassified as tet(O/32/O) (13), since the novel tet(32) sequence, which shares only 60% identity with tet(O) sequences, is limited to the central portion of the gene product. The nomenclature for these hybrid tet genes reflects the regions of similarity to known genes (5). In this study we investigated whether such hybrid tet genes represent occasional variants or whether they account for a significant proportion of ribosomal protection tet genes.
Incidence of mosaic tetracycline resistance genes in a range of environmental samples.
Mixed animal fecal samples (8), mainly from pig herds reared either intensively or organically (Table 1), and mixed human fecal and saliva samples from several countries were used for DNA extraction with the FastDNA SPINKit for soil (Qbiogene, Irvine, CA). Sample Ab1 was a single sample from an individual with a history of long-term tetracycline therapy (10), from whom the bacterium carrying the tet(O/32/O) gene had previously been isolated (6, 13).
TABLE 1.
PCR results obtained with human fecal and saliva samples and animal fecal DNA samples from across Europea
| Sample | No. of individuals | PCR no. and amplicona
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1, full-length tet(O) | 2, full-length tet(W) | 3, 5′ tet(O/W) | 4, 5′ tet(O/32) | 5, internal tet(W/32) | 6, 5′ tet(W/32) | 7, internal tet(W/32) (nested) | ||
| Human fecal DNAb | ||||||||
| Finland | 20 | + | + | − | + | − | ND | − |
| France | 20 | + | + | − | + | − | ND | − |
| Norway | 20 | + | + | + | + | − | ND | − |
| Italy | 20 | + | + | − | − | − | ND | − |
| Scotland | 20 | + | + | − | + | − | ND | − |
| England | 20 | + | + | + | + | − | ND | − |
| Ab1c | 1 | + | + | − | + | − | ND | − |
| Human saliva DNA | ||||||||
| Scotland | 20 | + | − | − | − | − | ND | − |
| Norway | 20 | + | − | − | − | − | ND | − |
| England | 20 | − | − | − | − | − | ND | − |
| France | 20 | + | − | − | − | − | ND | − |
| Finland | 20 | + | − | − | − | − | ND | − |
| Italy | 20 | + | + | − | − | − | ND | − |
| Animal fecal DNAd | ||||||||
| Norway pig herd 1 (O) | 5 | + | + | − | + | − | ND | − |
| Norway pig herd 2 (O) | 5 | + | + | − | + | − | ND | − |
| Norway pig herd 3 (O) | 5 | + | + | − | + | − | ND | − |
| Norway pig herd 4 (O) | 5 | + | + | + | + | − | ND | − |
| Scottish pig herd 1 (I) | 5 | + | + | + | + | + | − | + |
| Scottish pig herd 2 (O) | 5 | + | + | − | + | + | + | + |
| Spanish pig herd 1 (I) | 2 | + | + | + | + | + | + | + |
| Spanish pig herd 2 (I) | 2 | + | + | − | + | + | − | + |
| Spanish piglets | 2 | + | + | + | + | + | − | + |
| Italian pig 1 (O) | 1 | + | + | + | + | + | + | + |
| Italian pig 2(O) | 1 | + | + | − | + | + | + | + |
| Scottish sheep | 1 | + | + | − | − | − | ND | − |
| Scottish cow | 1 | − | − | − | − | − | ND | − |
| English pig | 1 | + | + | − | + | − | − | + |
| English cow | 1 | − | − | − | − | − | ND | − |
| Gene controls | ||||||||
| tet(O) | + | − | − | − | ND | ND | − | |
| tet(W) | − | + | − | − | ND | ND | − | |
| tet(O/W/O) | + | − | − | + | ND | − | − | |
PCRs and primer positions are as shown in Fig. 1. Routine PCR amplifications were carried out using a Bio-Rad iCycler in 5-μl volumes with the following cycling conditions: 1 cycle at 95°C for 5 min; 30 cycles of 95°C, specific annealing temperature, and 72°C for 30 s each; and a final cycle of 72°C for 10 min. PCR 7 was a nested PCR amplifying the internal tet(W/32) product from the full-length tet(O) template (product of PCR 1). +, PCR amplifications where a product of the correct size was obtained; −, those where no product was obtained; ND, not determined.
Human volunteers had not received antibiotics in the preceding 3 months (except Ab1).
Sample from individual on long-term tetracycline therapy.
O, organic (no use of antibiotics); I, intensive (antibiotics used as growth promoters limited to those approved by the EU [EU directive 70/524/ECC]).
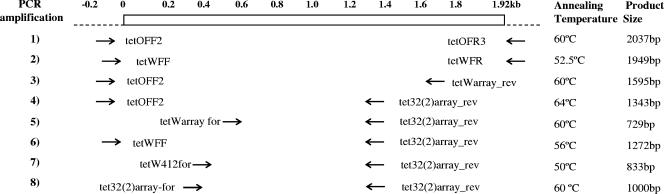
PCR was performed according to a standard protocol as detailed in Fig. 1, and the results are summarized in Table 1. tet(O) products were detected in all human fecal and pig fecal DNA samples and in all but one human saliva sample, while tet(W) was commonly detected in both sets of fecal DNA but in only one saliva sample. Mosaic genes were detected by specific PCR amplifications using mixed primer sets (Table 1, PCRs 3 to 7) that failed to amplify wild-type genes. Mosaic genes were not detected in saliva samples but were prevalent in fecal samples. Genes showing evidence of O/32 mosaicism within the first 1.2 kb were widespread in both human and animal fecal samples, and genes showing O/W mosaicism in the first 1.4 kb were detected in some samples. The internal W/32 mosaic product (Fig. 1, PCR 5) was detected in some animal fecal samples, but only four of these seven samples gave a product using the full-length tet(W) forward and internal tet(32) primers (Fig. 1, PCR 6), indicating that not all W/32 mosaic products are tet(W) at the 5′ end of the gene.
FIG. 1.
Schematic showing primer combinations, annealing temperatures, and expected product sizes for PCR amplification of tet(O), tet(W), and various mosaic Tcr genes. 1), full-length tet(O) product, tetOFF2 (5′-TTGTTTTGGGGCTATTGGAG-3′) plus tetOFR3 (5′-TATATGACTTTTGCAAGCTG-3′); 2), full-length tet(W) product, tetWFF (5′-TTGGGGCTGTAAAGGGAGGAC-3′) plus tetWFR (5′-CTTTACATTACCTTCTGA-3′); 3), tet(O/W) product, tetOFF2 plus tetWarray_rev (5′-AATCTTACAGTCCGTTACG-3′); 4), tet(O/32) product, tetOFF2 plus tet32(2)array_rev (5′ CTCTTTCATAGCCACGCC 3′); 5), tet(W/32) internal product, tetWarray for (5′-GGAGGAAAATACCGACATA-3′) plus tet32(2)array_rev; 6), tet(W/32) 5′ end, tetWFF plus tet32(2)array_rev; 7), tet(W/32) product from nested PCR using tet(O) full-length product as a template, tetW412for (5′-AGAGCGTGGTTCAGTCTGTT-3′) plus tet32(2)-array_rev; 8), tet32(2)array_for (5′-AACCGAAGCATACCGCTC-3′) plus tet32(2)array_rev.
Proportions of mosaic compared to wild-type tetracycline resistance genes.
DNA extracted from Spanish pig herd 1 and the human Ab1 sample was amplified using full-length tet(O) primers (Fig. 1, PCR 1), and the amplicons were ligated into the pGEM-T Easy Vector System I (Promega). Plasmids were purified from 64 or 80 transformant colonies, respectively, and inserts fully sequenced.
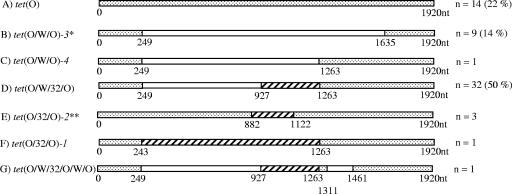
The majority of clones from the Spanish pigs carried mosaic genes (Fig. 2), and only 22% (14/64) contained the nonmosaic tet(O) gene. Crossover positions for each sequence were compared in order to identify hot spots for recombination. All 32 tet(O/W/32/O) amplicons had identical crossover points (Fig. 2C), and 9 tet(O/W/O) sequences were identical, with 1 changing back to tet(O) at position 1263 (Fig. 2B). None of these resembled tet(O/W/O) genes described previously (11). Three tet(O/32/O) genes were identical (Fig. 2E), while the other four were all different, with one the same as the previously described tet(O/32/O) gene (Fig. 2F) (13). One complex clone had an insert that aligned to tet(O/W/32/O/W/O) (Fig. 2G).
FIG. 2.
Schematic representation of mosaic tetracycline resistance genes from the fecal material of Spanish pig herd 1. Stippled regions indicate gene segments sharing high sequence identity with C. jejuni tet(O) (accession no. M18896). Open regions represent high sequence identity with B. fibrisolvens tet(W) (accession no. AJ222769). Hatched areas have a high level of similarity to the tet(32) region of the tet(O/32/O)-1 gene from Clostridium sp. strain K10 (accession no. AJ295238). Mosaic genes produced by recombination between the same tet classes at different crossover positions are differentiated by italicized numbers. *, tet(O/W/O)-1 and -2 were identified previously (11). **, three other tet(O/32/O) mosaics were also identified, tet(O/32/O)-3, -4, and -5, changing at positions 882 and 1263, 876 and 1248, and 867 and 1307, respectively.
The full-length amplicons from the Ab1 human fecal DNA were more evenly divided between mosaic and nonmosaic forms. Fifty-four percent of clones (43/80) carried nonmosaic tet(O) sequences. The remaining 46% (37/80) were all tet(O/32/O) genes identical to the original tet(O/32/O) gene (Fig. 2F) (6, 13).
Sequencing of Italian pig 1 clones amplified using the primer combination PCR 6 (Fig. 1) showed that other tetracycline resistance mosaic genes, including some derived from tet(W), remain to be discovered.
It has been suggested that a minimum stretch of 50 amino acids may be appropriate in defining a mosaic protein (5). However, the central tet(O) sequence in the tet(O/W/32/O/W/O) mosaic encodes 16 amino acids (Fig. 2G) and thus does not fit this description. The problems of nomenclature clearly increase with the complexity of the mosaic gene.
Tetracycline resistance mosaic genes can confer greater resistance than the wild-type genes (12). We attempted to determine the tetracycline resistance conferred by T-vector clones containing amplicons (Fig. 1, PCR 1) of each type of mosaic gene shown in Fig. 2 compared to that with the wild-type genes in an E. coli host. Only one of the clones gave any significant resistance: the tet(O/W/32/O/W/O) mosaic grew in 64 μg ml−1 tetracycline (results not shown). Upstream sequences of ∼300 nucleotides, absent here, have been shown to be essential for full expression of resistance for tet(O) (14) and tet(W) (7). The multiple mosaic may indeed confer a high level of resistance, but protein expression levels have not been determined.
Identification of tet(32).
During this work, a human oral bacterium (identified by full 16S rRNA sequencing as Streptococcus salivarius strain FStet12) was isolated that contained an internal tet(32) sequence (Fig. 1, PCR 8), but no product was amplified using the tet(O)-tet(32) primers (PCR 4). Genome walking (3) was used to complete the gene sequence, which was presumed to be the original, nonmosaic form of tet(32). The central region of the Tet(32) protein (amino acids 74 to 424) is >99% identical to the Tet(O/32/O) mosaic protein but otherwise the full-length protein has only 66% identity to its closest relatives, Tet(O) and Tet(W). The presence of tet(32) in S. salivarius FStet12 was confirmed by sequencing the full-length gene amplified from purified bacterial DNA. It is interesting that the tet(32) gene was identified in an environment that lacks mosaic genes (Table 1).
Conclusions.
In conclusion, this European study of humans and animals demonstrates that tetracycline resistance genes of the mosaic ribosome protection type, at least based on tet(O), are widespread and are as abundant as nonmosaics in some samples. These genes may influence the results of surveillance programs investigating the incidence of specific tetracycline resistance genes. In the case of tet(32), the abundance of mosaic combinations delayed identification of the full-length gene.
Nucleotide sequence accession numbers.
The sequence of tet(32) has been deposited in the GenBank database under accession number DQ647324. The sequence of a representative of each type of mosaic gene has been deposited in the GenBank database under accession numbers DQ679926 [tet(O/W/32/O/W/O)], EF065523 [tet(O/W/32/O)], and EF065524 [tet(O/W/O)-3].
Acknowledgments
We thank Pauline Young and Donna Henderson for DNA sequencing. We are indebted to Lorna Seville for providing the DNA from all the human samples except Ab1 and also for providing us with strain FStet12.
This study was carried out with financial support from the Commission of the European Communities, specific RTD program, “Quality of Life and Management of Living Resources”: QLK2-CT-2002-00843, “Antimicrobial resistance transfer from and between gram-positive bacteria of the digestive tract and consequences for virulence (ARTRADI),” and QLRT-2001-02056, “Gene mining of Metagenomes for novel enzymes and therapeutics (GEMINI).” RRI receives support from the Scottish Executive Environment and Rural Affairs Department (SEERAD).
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connell, S. R., D. M. Tracz, K. H. Nierhaus, and D. E. Taylor. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47:3675-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazimierczak, K., H. J. Flint, and K. P. Scott. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy, S. B., L. M. McMurry, and M. C. Roberts. 2005. Tet protein hybrids. Antimicrob. Agents Chemother. 49:3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melville, C. M., K. P. Scott, D. K. Mercer, and H. J. Flint. 2001. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob. Agents Chemother. 45:3246-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melville, C. M., R. Brunel, H. J. Flint, and K. P. Scott. 2004. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J. Bacteriol. 186:3656-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson, A. J., R. Colangeli, P. Spigaglia, and K. P. Scott. 6 December 2006. Distribution of specific tetracycline and erythromycin resistance genes in environmental samples assessed by macroarray detection. Environ. Microbiol. doi: 10.1111/j.1462-2920.2006.01190.x. [DOI] [PubMed]
- 9.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 10.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanton, T. B., and S. B. Humphrey. 2003. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69:3874-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanton, T. B., S. B. Humphrey, K. P. Scott, and H. J. Flint. 2005. Hybrid tet genes and tet gene nomenclature: request for opinion. Antimicrob. Agents Chemother. 49:1265-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Y., and D. E. Taylor. 1991. A DNA sequence upstream of the tet(O) gene is required for full expression of tetracycline resistance. Antimicrob. Agents Chemother. 35:2020-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]