Abstract
The pyrrolomycins are a family of polyketide antibiotics, some of which contain a nitro group. To gain insight into the nitration mechanism associated with the formation of these antibiotics, the pyrrolomycin biosynthetic gene cluster from Actinosporangium vitaminophilum was cloned. Sequencing of ca. 56 kb of A. vitaminophilum DNA revealed 35 open reading frames (ORFs). Sequence analysis revealed a clear relationship between some of these ORFs and the biosynthetic gene cluster for pyoluteorin, a structurally related antibiotic. Since a gene transfer system could not be devised for A. vitaminophilum, additional proof for the identity of the cloned gene cluster was sought by cloning the pyrrolomycin gene cluster from Streptomyces sp. strain UC 11065, a transformable pyrrolomycin producer. Sequencing of ca. 26 kb of UC 11065 DNA revealed the presence of 17 ORFs, 15 of which exhibit strong similarity to ORFs in the A. vitaminophilum cluster as well as a nearly identical organization. Single-crossover disruption of two genes in the UC 11065 cluster abolished pyrrolomycin production in both cases. These results confirm that the genetic locus cloned from UC 11065 is essential for pyrrolomycin production, and they also confirm that the highly similar locus in A. vitaminophilum encodes pyrrolomycin biosynthetic genes. Sequence analysis revealed that both clusters contain genes encoding the two components of an assimilatory nitrate reductase. This finding suggests that nitrite is required for the formation of the nitrated pyrrolomycins. However, sequence analysis did not provide additional insights into the nitration process, suggesting the operation of a novel nitration mechanism.
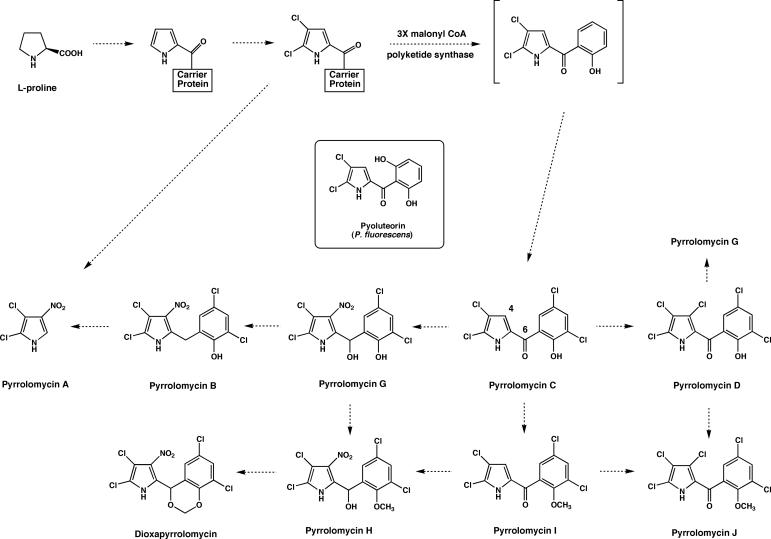
The pyrrolomycins are a family of antibiotics with high halogen content that are produced by Actinosporangium and Streptomyces species (3, 6, 8, 9, 19, 23, 24, 31, 40). Representative examples of the pyrrolomycins include dioxapyrrolomycin and pyrrolomycins A to D (Fig, 1). Most of the pyrrolomycins bear a close structural resemblance to pyoluteorin, a polyketide antibiotic produced by Pseudomonas species (32, 33) (Fig. 1). The pyrrolomycins exhibit potent antibiotic activity against gram-positive bacteria and inhibit substance P-induced release of myeloperoxidase from human polymorphonuclear leukocytes (29). Biosynthetic studies of dioxapyrrolomycin have shown that the skeleton of this antibiotic is derived from l-proline and three molecules of acetate, while the methylenedioxy group originates from the methyl group of l-methionine (5). These studies demonstrate that the biosynthetic pathway for the pyrrolomycins is similar to that for pyoluteorin. A hypothetical biosynthetic pathway showing the relationships of the known pyrrolomycin antibiotics is shown in Fig. 1.
FIG. 1.
Hypothetical biosynthetic pathway for the pyrrolomycins produced by A. vitaminophilum, Streptomyces fumanus, and Streptomyces sp. strain UC 11065.
The most interesting structural feature of the pyrrolomycins is the presence of a nitro group on the pyrrole ring in some of the antibiotics. Naturally occurring nitro compounds are rare in nature, and relatively little is known about the biochemical mechanisms used to introduce a nitro group. Examples of naturally occurring aryl nitro compounds include pyrrolnitrin (46), chloramphenicol (47), and aureothin (13), which are antibiotics, and the thaxtomins, which are phytotoxins (14). In the case of pyrrolnitrin and aureothin, the aryl nitro group has been shown to originate from the oxidation of an aryl amino group (12, 27, 41), while the biosynthesis of the 4-nitrotryptophan moiety of the thaxtomins has been shown to require the participation of a nitric oxide synthase (20). In order to gain insight into the nitration mechanism involved in the biosynthesis of the pyrrolomycins, we have undertaken experiments to clone and characterize the pyrrolomycin biosynthetic gene cluster.
MATERIALS AND METHODS
Bacterial strains and cloning vectors.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown and transformed as described previously by Sambrook et al. (37). Culture media were obtained from Becton Dickinson and Co. (Sparks, MD). Chemical reagents were purchased from Sigma-Aldrich Co. (Milwaukee, WI) unless otherwise noted. Actinosporangium vitaminophilum and Streptomyces sp. strain UC 11065 were cultured in seed medium (9) composed of 1% glucose, 1% soluble starch, 0.5% polypeptone, 0.3% yeast extract, 0.2% soy flour, 0.2% beef extract, and 0.2% CaCO3 (pH 7.0). For maintenance on solid medium, 2% agar was added to a modified seed medium in which CaCO3 was omitted and 2% soytone was used in place of soy flour.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Actinosporangium vitaminophilum (Streptomyces vitaminophilus) (ATCC 31673) | Producer of pyrrolomycins A to D, wild type | ATCC |
| Streptomyces sp. strain UC 11065 | Producer of dioxapyrrolomycin and pyrrolomycins B to D, wild type | Upjohn Laboratories |
| Streptomyces sp. strain UC 11065(dox8::pDOX8hx) | Streptomyces sp. strain UC 11065 with single-crossover disruption of dox8 | This work |
| Streptomyces sp. strain UC 11065(dox17::pDOX17hx) | Streptomyces sp. strain UC 11065 with single-crossover disruption of dox17 | This work |
| E. coli DH10B | General cloning host | Invitrogen |
| E. coli 12567 | dam-13::Tn9 dcm-6 hsdM | 28 |
| Plasmids | ||
| pBluescript II SK(+) | E. coli cloning vector; Ampr | Stratagene |
| pSP72 | E. coli cloning vector; Ampr | Promega |
| pGEM-T Easy | PCR cloning vector; Ampr | Promega |
| pOJ446 | E. coli-Streptomyces shuttle cosmid for conjugal transfer; Apr | 1 |
| pKC1139 | E. coli-Streptomyces shuttle plasmid for conjugal transfer with temperature-sensitive Streptomyces replicon; Apr | 1 |
| pGEM-T Easy 15001 | 1.5-kb PCR fragment spanning the C terminus of pyr16 and the N terminus of pyr17 in pGEM-T Easy | This work |
| pDOX8hx | 995-bp PCR internal fragment of dox8 in pKC1139 | This work |
| pDOX17hx | 1,211-bp PCR internal fragment of dox17 in pKC1139 | This work |
| pCAV2 | pOJ446 cosmid clone of A. vitaminophilum hybridizing to the pGEM-T Easy 15001 pyr16-pyr17 insert | This work |
| pCAV5 | pOJ446 cosmid clone of A. vitaminophilum hybridizing to the pGEM-T Easy 15001 pyr16-pyr17 insert | This work |
| pCAV2PX3 | pOJ446 cosmid clone of A. vitaminophilum overlapping CAV2 | This work |
| pCAV3PX1 | pOJ446 cosmid clone of A. vitaminophilum overlapping CAV5 | This work |
| pCU13 | pOJ446 cosmid clone of Streptomyces sp. strain UC 11065 hybridizing to the pGEM-T Easy 15001 pyr16-pyr17 insert | This work |
DNA manipulations and sequence analysis.
Plasmid and cosmid DNA were purified with a QIAprep Spin Miniprep kit (QIAGEN, Valencia, CA). PCR products were separated on agarose gels and purified from the gels using a Geneclean Spin kit (MP Biomedicals, Solon, OH). Digestion with restriction endonucleases and ligation experiments were carried out by standard procedures under conditions recommended by the manufacturers. Automated DNA sequencing was performed using universal and synthetic oligonucleotide primers. Cosmids were sequenced by the release of the inserts with XbaI and SpeI followed by further digestion with PstI to generate a pool of fragments that were then cloned into pBluescript II SK(+) (Stratagene, La Jolla, CA) or pSP72 (Promega Corp., Madison, WI) plasmid vectors. The clones were then sequenced with universal primers followed by primer walking with designed primers. DNA gaps between subclones were filled in by direct sequencing of the parent cosmid. Sequences were determined by complete sequencing of both DNA strands with multiple sequencing of some regions. Synthetic primers were obtained from Sigma-Genosys (St. Louis, MO) or Integrated DNA Technologies, Inc. (Coralville, IA). Sequencher, version 4.1 (Gene Codes Corp., Ann Arbor, MI), was used to compile DNA sequence data. Sequence alignments were performed using Gene Inspector software, version 1.5 (Textco, Inc., Lebanon, NH). Open reading frames (ORFs) were deduced from sequence data by using FramePlot 2.3.2 (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pI). The corresponding deduced proteins were compared with other proteins using BLAST methods (http://www.ncbi.nlm.nih.gov/BLAST/).
PCR amplification of halogenase genes from A. vitaminophilum.
Degenerate PCR primers were designed based upon the conserved domains of bacterial halogenases found in the pyrrolnitrin (11), pyoluteorin (32), teichoplanin (42), pentachloropseudilin (48), and dalbivancin (43) biosynthetic gene clusters. In addition, primers devised by Piraee and Vining (34) to amplify a halogenase gene from Streptomyces venezuelae were also used directly or in combination with other halogenase primers. PCRs were run for 30 cycles. The conditions for each cycle were 1 min at 94°C, 1 min of touchdown from 55°C to 40°C, and 2 min at 72°C. Each reaction mix contained 3 ng of genomic DNA, 12.5 pmol of each primer, 1 U of Ex Taq DNA polymerase (Takara Bio Inc., Mountain View, CA), and 10% dimethyl sulfoxide. The PCR products were purified with a Geneclean Spin kit (MP Biomedicals) after agarose gel separation and cloned into the pGEM-T Easy plasmid vector (Promega Corp.) for sequencing. One set of degenerate primers led to the successful amplification of a 1.5-kb halogenase gene fragment. The sequences of these primers were as follows: 5′-S IRS RSS TGG ITS KGS VWS ATC CCS-3′ (N-terminal primer) and 5′-AG GTG SAC VCC VKD SGA GAA CA-3′ (C-terminal primer), where I is inosine; D is A, G, or T; K is G or T; R is A or G; S is C or G; V is A, C, or G; and W is A or T.
Cosmid library construction and screening.
A. vitaminophilum and Streptomyces sp. strain UC 11065 were grown in yeast extract-malt extract medium (21), and genomic DNA was extracted as described previously (21). Purified genomic DNA was partially digested with Sau3AI so that most of the resulting fragments were between 40 and 60 kb in size as judged by pulsed-field gel electrophoresis using a FIGE mapper (Bio-Rad, Hercules, CA), and the partially digested DNA was then dephosphorylated using calf intestinal alkaline phosphatase (New England Biolabs, Ipswich, MA). Cosmid vector pOJ446 (1) was digested with HpaI, dephosphorylated using calf intestinal alkaline phosphatase, and further digested with BamHI to generate two vector arms. The cosmid arms and the partially digested genomic fragments were then ligated and packaged into lambda phage heads using Stratagene Gigapack III XL packaging extract (Stratagene), and the resulting particles were used to transduce E. coli strain DH10B, with selection of transductants occurring on Luria-Bertani (LB) agar containing 50 μg/ml apramycin (Ap). Southern hybridizations were carried out according to standard protocols (37). Denatured probes were radiolabeled with [α-32P]dCTP using the NEBlot kit (New England Biolabs). For colony hybridizations, the cosmid libraries were spread out onto LB agar containing 50 μg/ml apramycin, incubated for 20 h at 37°C, and transferred onto nitrocellulose membranes (Whatman Inc., Florham Park, NJ). Amplification of the libraries on LB agar containing chloramphenicol as well as subsequent lysis, denaturation, neutralization, and final hybridization were conducted according to standard protocols (37). Both the A. vitaminophilum and Streptomyces sp. strain UC 11065 cosmid libraries were screened with the 1.5-kb halogenase fragment amplified from A. vitaminophilum by PCR. The hybridizing cosmids were analyzed by restriction mapping, and two overlapping A. vitaminophilum cosmids, CAV2 and CAV5, were initially selected for sequencing. When it became clear that the entire A. vitaminophilum pyrrolomycin gene cluster was probably not contained on CAV2 and CAV5, a 798-bp N-terminal fragment of the CAV5 insert and a 702-bp C-terminal fragment from the CAV2 insert were amplified by PCR with primers 3px1F (5′-GATCGGCCTCCTGCTGTTC-3′) and 3px1R (5′-GGGCGGCGTCCTGCTCTC-3′) and with primers 2px3F (5′-GATCTGGAAGGAGCGCAG-3′) and 2px3R (5′-GAGTCCTACCTCGGCGAC-3′), respectively. Each PCR product was then used to probe the A. vitaminophilum cosmid library. In this way, cosmids CAV3PX1 and CAV2PX3 were recovered.
Fermentation of A. vitaminophilum and Streptomyces sp. strain UC11065 and isolation of pyrrolomycins.
Fermentations of A. vitaminophilum (ATCC 31673) and Streptomyces sp. strain UC11065 were carried out at 28°C and 250 rpm in seed medium. Since neither organism could be induced to sporulate, cultures were maintained as frozen mycelial suspensions in sterile 20% glycerol. Seed medium (100 ml) was inoculated from a frozen mycelial suspension and grown at 28°C for 48 h. Fresh seed medium (5 × 200 ml) was then inoculated with seed culture (5 × 10 ml), and the fermentation was allowed to proceed for 120 h. At the end of this time, the pH of the culture was adjusted to 7.0, and the mixture was centrifuged to separate the fermentation broth from the mycelium. The broth was extracted three times with ethyl acetate, while the mycelium was suspended in excess 50% aqueous acetone and stirred for 1 h. The aqueous acetone extract was filtered, and the acetone was removed in vacuo. The aqueous residue was then extracted three times with ethyl acetate, and the resulting extracts were combined with the ethyl acetate extracts of the fermentation broth. The combined ethyl acetate extracts were dried over anhydrous sodium sulfate and filtered, and the ethyl acetate was removed in vacuo. The resulting residue was dissolved in methanol for high-performance liquid chromatography (HPLC) analysis. Pyrrolomycins were isolated by preparative HPLC using an Agilent SB-C18 column (4.6 by 250 mm) (VWR International, West Chester, PA) at a flow rate of 1 ml per min using the conditions described below.
HPLC analysis of the pyrrolomycins.
HPLC analysis was carried out using a ThermoFinnigan P2000 HPLC pump equipped with a Varian ProStar 325 UV-visible detector set to 268 nm and an Agilent SB-C18 column (4.6 by 250 mm). Elution was initially carried out for 3 min at a flow rate of 1 ml/min with a solvent consisting of a 75:25 mixture of methanol-water containing 1% acetic acid. The solvent mixture was then changed to a mixture of 90:10 methanol-water containing 1% acetic acid over 1 min, and elution with the latter solvent continued for an additional 16 min. Under these conditions, pyrrolomycins A, B, C, and D had approximate retention times of 4.4 min, 9.8 min, 14.3 min, and 12.7 min, respectively, while dioxapyrrolomycin exhibited a retention time of ca. 12.1 min. Under the fermentation conditions described above, A. vitaminophilum produced pyrrolomycins A to D, while Streptomyces sp. strain UC 11065 produced pyrrolomycins B to D and dioxapyrrolomycin.
Inactivation of pyrrolomycin production by single-crossover gene disruptions.
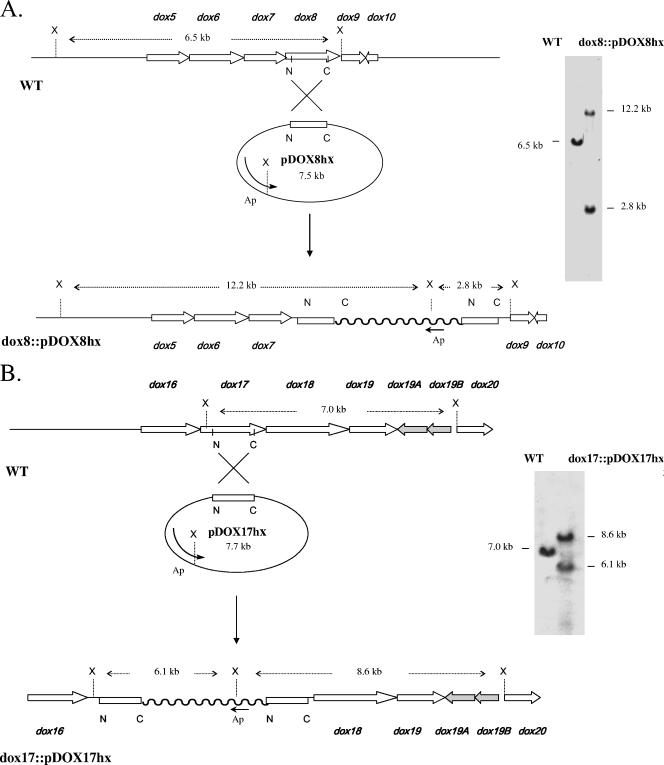
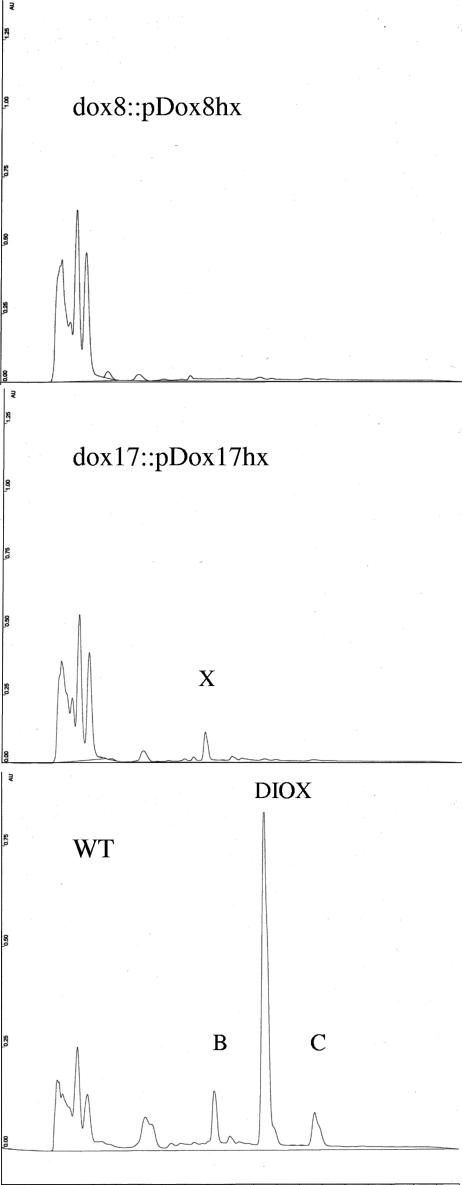
The dox8 and dox17 genes from the dioxapyrrolomycin gene cluster of Streptomyces sp. strain UC11065 were targeted for disruption. A 995-bp fragment of dox8 and a 1,211 bp fragment of dox17 were amplified by PCR using primers containing XbaI and HindIII sites (shown in boldface type): dox8-N-term (5′-TCTAGAAGAGCACCGAGGC-3′), dox8-C-term (5′-AAGCTTCACCATGTGGTCGC-3′), dox17-N-term (5′-TCTAGACCAACGAGTTCCAC-3′), and dox17-C-term (5′-AAGCTTCGGGCTCATGAAGTG-3′). The PCR products were cloned into the XbaI-HindIII sites of the temperature-sensitive shuttle vector pKC1139 (1). The resulting constructs, pDOX8hx and pDOX17hx, were passed through nonmethylating E. coli strain ET12567 (28). Streptomyces sp. strain UC 11065 was inoculated into liquid modified seed medium containing 17% sucrose and grown at 28°C and 250 rpm for 48 h. Protoplasts were generated by treatment with 1 mg/ml of lysozyme at 30°C for 1 h and purified by filtration through cotton wool. After polyethylene glycol-mediated transformation, the transformed protoplasts were regenerated on R2YE agar plates (21). After 20 h at 30°C, transformants were selected by flooding the R2YE agar plates with apramycin solution to a final concentration of 50 μg/ml. After 7 to 10 days, apramycin resistance transformants were transferred onto fresh modified seed medium agar plates containing 100 μg/ml of apramycin and then incubated at 37°C for 5 days to eliminate pKC1139-based plasmids. Actively growing clones were transferred onto fresh modified seed medium agar plates without apramycin and incubated for 5 days at 37°C. After two additional rounds of nonselective growth at 37°C on solid modified seed medium, individual clones were purified by inoculation into liquid modified seed medium containing 17% sucrose, followed by the formation and regeneration of protoplasts in the manner described above. Individual colonies obtained by protoplast regeneration were selected and grown in yeast extract-malt extract medium at 28°C for 48 h. Genomic DNA was then extracted from these cultures by standard methods, digested with XhoI, and analyzed by Southern hybridization using the PCR-amplified 995-bp fragment of dox8 or the PCR-amplified 1,211bp fragment of dox17 as a probe. Both mutants exhibited hybridization patterns that were completely consistent with the genetic alterations expected from single-crossover disruptions (Fig. 2). Single-crossover disruptants of the dox8 and dox17 genes identified by Southern hybridization were fermented under standard conditions, and the production of pyrrolomycins was analyzed by HPLC in the manner described above. Wild-type strain UC 11065 was used as a control. No pyrrolomycins were detected in the organic extracts obtained from either of the disruptants (Fig. 3).
FIG. 2.
(A) Southern blot analysis of XhoI-digested genomic DNA isolated from wild-type Streptomyces sp. strain UC 11065 (WT) and UC11065(dox8::pDOX8hx) using a 995-bp internal dox8 fragment as a probe. The sizes of the predicted and observed hybridizing fragments present in the wild type and in mutant dox8::pDOX8hx are shown. X, XhoI; Ap, apramycin resistance gene. (B) Southern blot analysis of XhoI-digested genomic DNA isolated from wild-type Streptomyces sp. strain UC 11065 (WT) and UC 11065(dox17::pDOX17hx) using a 1,211-bp internal dox17 fragment as a probe. The sizes of the predicted and observed hybridizing fragments present in the wild type and in mutant dox17::pDOX17hx are shown. X, XhoI; Ap, apramycin resistance gene.
FIG. 3.
HPLC analysis of fermentation broths from Streptomyces sp. strains UC 11065(dox8::pDox8hx), UC 11065(dox17::pDox17hx), and wild-type UC 11065 (WT) using HPLC conditions described in Materials and Methods. B, pyrrolomycin B; C, pyrrolomycin C; DIOX, dioxapyrrolomycin; X, unknown metabolite.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited into the GenBank database under the accession numbers EF140901, EF140902, and EF140903.
RESULTS
Cloning of the pyrrolomycin biosynthetic gene cluster of A. vitaminophilum.
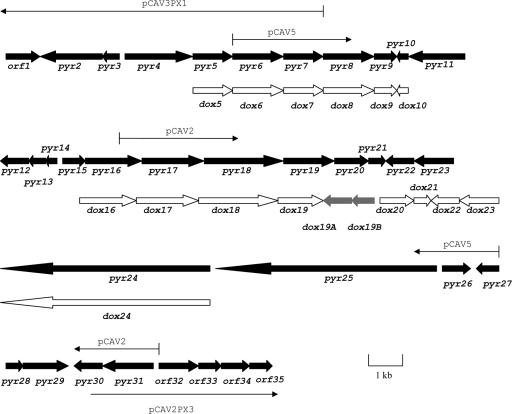
Investigations to clone the pyrrolomycin biosynthetic gene cluster were initially carried out with A. vitaminophilum, an organism reported to produce pyrrolomycins A to E (8, 9, 19, 24). HPLC analysis of the A. vitaminophilum fermentation carried out in our laboratory confirmed the production of pyrrolomycins A to D. Since halogenated natural products are relatively rare, degenerate PCR primers designed from conserved regions of known bacterial halogenases were used to successfully clone a halogenase gene fragment from A. vitaminophilum. The successful primer set amplified a 1.5-kb segment of DNA rather than the anticipated 0.6- to 0.8-kb fragment. Sequencing of this PCR product revealed that it spanned two adjacent halogenase genes (pyr16 and pyr17) that are present in A. vitaminophilum. A cosmid library from A. vitaminophilum was then constructed in the E. coli-Streptomyces shuttle cosmid pOJ446 and probed with the 1.5-kb PCR product. The hybridizing cosmids were analyzed by restriction mapping, and two overlapping cosmids, pCAV2 and pCAV5, were initially selected for sequencing. When it became clear that the entire pyrrolomycin gene cluster was probably not contained on pCAV2 and pCAV5, an N-terminal fragment of the pCAV5 insert and a C-terminal fragment from the pCAV2 insert were amplified by PCR and then used to probe the A. vitaminophilum cosmid library. In this way, cosmids pCAV2PX3 and pCAV3PX1 were recovered. Sequencing of the pCAV2 and pCAV5 inserts as well as portions of the pCAV2PX3 and pCAV3PX1 inserts identified 35 contiguous ORFs (Fig. 4). Sequence analysis of the genes encoded by these ORFs (see below) provided strong evidence the they constitute the pyrrolomycin biosynthetic gene cluster of A. vitaminophilum. To provide more conclusive proof for this assumption, extensive efforts were made to introduce foreign DNA into this organism in order to create gene disruptions. Unfortunately, all attempts to introduce DNA into A. vitaminophilum either by conjugation with E. coli or by transformation of protoplasts were completely unsuccessful (data not shown). To gain additional evidence that the pyrrolomycin gene cluster of A. vitaminophilum had been correctly identified, experiments to clone the pyrrolomycin gene cluster from the pyrrolomycin producer Streptomyces sp. strain UC11065 were undertaken.
FIG. 4.
Genetic organization of the pyr biosynthetic gene cluster from A. vitaminophilum ATCC 31673 and the partial dox gene cluster from Streptomyces sp. strain UC 11065. Arrows indicate positions of A. vitaminophilum cosmid inserts.
Cloning of the pyrrolomycin biosynthetic gene cluster of Streptomyces sp. strain UC 11065.
Streptomyces sp. strain UC 11065 was previously reported to produce the nitrated metabolite dioxapyrrolomycin as well as pyrrolomycins C and D (6). HPLC analysis of the UC 11065 fermentation carried out in our laboratory confirmed the production of these three pyrrolomycins as well as the production of pyrrolomycin B. Extensive experimentation with Streptomyces sp. strain UC 11065 revealed that small plasmids (<12 kb) can be introduced into this organism by protoplast transformation but not by conjugation with E. coli (data not shown). Although the efficiency of protoplast transformation proved to be quite low, the successful isolation of transformants indicated that genetic manipulations of Streptomyces sp. strain UC11065 should be possible. Therefore, genomic DNA of UC 11065 was digested separately with BamHI, BgIII, and PstI and probed by Southern hybridization using each of the four halogenase genes (pyr11, pyr16, pyr17, and pyr29) that had previously been cloned from A. vitaminophilum. Each probe gave a unique hybridization pattern, suggesting that homologs of all four halogenase genes are present in UC 11065 (data not shown). A pOJ446 cosmid library of Streptomyces sp. strain UC 11065 was then constructed and screened by Southern hybridization using the same 1.5-kb PCR product used to screen an A. vitaminophilum cosmid library. Eighteen positive cosmids were isolated by colony hybridization and analyzed by restriction mapping. The cosmid insert in one cosmid, pCU13, was subcloned and partially sequenced by the same methods used for A. vitaminophilum. Seventeen ORFs were identified, and 15 of these encoded genes that exhibit a high degree of similarity to the genes cloned from A. vitaminophilum (see Table 3) as well as similar organization (Fig. 4).
TABLE 3.
Organization and analysis of the partial pyrrolomycin biosynthetic gene cluster from Streptomyces sp. strain UC 11065
| Gene | Size (aa) | Protein homolog (accession number), similarity/identity (%) | Possible function (% identity to A. vitaminophilum protein homolog) |
|---|---|---|---|
| dox5 | 374 | SCO4594 (CAC08295), 86/80 | 2-Oxoglutarate ferredoxin oxidoreductase, β-subunit (77) |
| dox6 | 479 | GntQ (AAR98563), 68/49 | Membrane antiporter (76) |
| dox7 | 379 | PltE (AAD24879), 69/55 | Pyrrole synthesis (87) |
| dox8 | 489 | CloN4 (AAN65233), 64/52 | Adenylation of l-proline (86) |
| dox9 | 187 | AvinDRAFT_6224 (EAM07574), 60/43 | Flavin reductase (81) |
| dox10 | 98 | Ava_4741 (ABA24338), 56/36 | Acyl carrier protein (56) |
| dox16 | 583 | HalA (AAQ04684), 81/69 | Halogenase (87) |
| dox17 | 552 | HalB (AAQ04685), 75/59 | Halogenase (92) |
| dox18 | 735 | NasA (CAC19469), 76/66 | Assimilatory nitrate reductase, catalytic subunit (79) |
| dox19 | 477 | NasC (AAK77360), 59/49 | Assimilatory nitrate reductase, electron transfer subunit (68) |
| dox19A | 327 | med-Orf21 (BAC79034), 73/65 | Kinase (absent from A. vitaminophilum) |
| dox19B | 148 | MflvDRAFT_0738 (EAS08754), 55/41 | Methyltransferase (absent from A. vitaminophilum) |
| dox20 | 405 | NocL (AAT09797), 59/44 | Cytochrome P450 (65) |
| dox21 | 103 | Nwi_2139 (ABA05393), 62/47 | Monooxygenase (76) |
| dox22 | 226 | MAP_2796c (AAS05113), 60/46 | Methyl transferase (59) |
| dox23 | 353 | Tmn22 (BAE93743), 50/35 | Membrane protein (45) |
| dox24 | 2,067 | MxaE (AAK57189), 48/34 | Polyketide synthase (82) |
Confirmation that the cloned locus from UC 11065 is required for pyrrolomycin production.
To confirm that the cloned locus from Streptomyces. sp. strain UC 11065 contains genes that are required for pyrrolomycin biosynthesis, single-crossover disruptions were created in dox8 and dox17. The disruption plasmids pDOX8hx and pDOX17hx were created by the insertion of a 995-bp fragment of dox8 and a 1,211-bp fragment of dox17, respectively, into the temperature-sensitive E. coli-Streptomyces shuttle vector pKC1139. Each plasmid was transferred into wild-type Streptomyces. sp. strain UC 11065 by protoplast-mediated transformation. Single-crossover mutants were selected, and their genotypes were confirmed by Southern analysis. Genomic DNA from the wild-type strain and each of the two mutants was digested with XhoI and probed by Southern hybridization with the PCR-amplified 995-bp fragment of dox8 or the PCR-amplified 1,211-bp fragment of dox17. The hybridization pattern for each mutant was consistent with that expected for the insertion of the disruption plasmid at the target site by a single-crossover-mediated event (Fig. 2). The resulting mutants completely lost their ability to produce pyrrolomycins when grown under the conditions used for pyrrolomycin production by the wild-type strain, as evidenced by HPLC analysis (Fig. 3). Since single-crossover mutations are likely to exhibit polar effects that interfere with the transcription of downstream genes, complementation of the dox8 and dox17 mutants was not attempted.
DNA sequence analysis of A. vitaminophilum genes.
Sequencing of a total of ca. 54 kb of DNA surrounding the halogenase A and halogenase B homologs pyr16 and pyr17 revealed the presence of 33 additional ORFs (Fig. 4). The 35 sequenced genes and the actual or putative functions assigned to them are listed in Table 2. The genes can be divided into three groups: (i) biosynthetic genes with identifiable functions, (ii) genes encoding tailoring enzymes or proteins of unknown function, and (iii) genes encoding potential regulatory and resistance proteins.
TABLE 2.
Organization and analysis of pyrrolomycin biosynthetic gene cluster from A. vitaminophilum
| Gene | Size (aa) | Protein homolog(s) (GenBank accession number), similarity/identity (%)a | Possible function |
|---|---|---|---|
| orf1 | 280 | FdhD (Q82M23), 82/72 | Formate dehydrogenase |
| pyr2 | 536 | Nfa35380 (BAD58386), 49/37 | Flavin adenine dinucleotide-dependent monooxygenase |
| pyr3 | 206 | Nfa14550 (BAD56300), 51/37 | TetR family regulator |
| pyr4 | 645 | KorA (NP_826054), 90/82 | 2-Oxoglutarate ferredoxin oxidoreductase, α-subunit |
| pyr5 | 355 | KorB (BAC72588), 84/75 | 2-Oxoglutarate ferredoxin oxidoreductase, β-subunit |
| pyr6 | 479 | GntQ (AAR98563), 71/50 | Membrane antiporter |
| pyr7 | 379 | PltE (AAD24879), 70/56 | Pyrrole synthesis |
| pyr8 | 470 | STIAU_0873 (EAU66332), 64/51; PltF (AAD24881), 63/51 | adenylation of l-proline |
| pyr9 | 187 | Amb2577 (BAE51381), 63/44 | Flavin reductase |
| pyr10 | 99 | Bpro_5327 (ABE47183), 60/42 | Acyl carrier protein |
| pyr11 | 557 | HalA (AAQ04684), 53/37; PltD (AAD24878), 52/34 | Halogenase(?); FADH2 motifs absent |
| pyr12 | 289 | SAV3633 (NP_824810), 62/46 | Unknown |
| pyr13 | 260 | TTC1745 (AAS82087), 55/37 | Oxidoreductase |
| pyr14 | 172 | Mlr2550 (BAB49655), 63/46 | Unknown |
| pyr15 | 313 | PA3330 (AAG06718), 56/42 | Oxidoreductase |
| pyr16 | 584 | HalA (AAQ04684), 80/69 | Halogenase |
| pyr17 | 552 | HalB (AAQ04685), 75/59 | Halogenase |
| pyr18 | 694 | NasA (CAC19469), 76/66 | Assimilatory nitrate reductase, catalytic subunit |
| pyr19 | 473 | NasC (AAK77360), 62/49 | Assimilatory nitrate reductase, electron transfer subunit |
| pyr20 | 405 | NocL (AAT09797), 62/44 | Cytochrome P450 |
| pyr21 | 104 | TcmH (AAA67512), 71/51 | Monooxygenase |
| pyr22 | 232 | MAP0125c (NP_959059), 49/36 | Methyl transferase |
| pyr23 | 355 | Tmn22 (BAE93743), 54/37 | Membrane protein |
| pyr24 | 2,051 | PltC (AAC38076) 58/42 | Polyketide synthase |
| pyr25 | 2167 | PltB (AAC38075), 57/43 | Polyketide synthase |
| pyr26 | 302 | LmnN (AAN85527), 58/43; PltG (AAD24880), 49/31 | Thioesterase |
| pyr27 | 207 | SCO3346 (CAB42672), 60/41 | TetR family regulator |
| pyr28 | 90 | PltL (AAQ90170), 69/41 | Proline carrier protein |
| pyr29 | 447 | ChlB4 (AAZ77674), 82/69; PltA (AAD24884), 78/65 | Halogenase |
| pyr30 | 297 | Orf56 (BAC76514), 61/48 | PPTase |
| pyr31 | 594 | Orf57 (BAC76515), 54/44 | Acyl CoA dehydrogenase |
| orf32 | 480 | SCO1715 (CAB50930), 83/75 | Homogentisate 1,2-dioxygenase |
| orf33 | 250 | SCO1716 (CAB50931), 81/70 | GntR family regulatory protein |
| orf34 | 383 | MvanDRAFT_5628 | Acyl CoA dehydrogenase |
| orf35 | 387 | Bcep18194_C7158 (AAB06202), 69/56 | l-Carnitine dehydratase |
plt genes are from the pyoluteorin gene cluster of P. fluorescens.
Biosynthetic genes with identifiable functions.
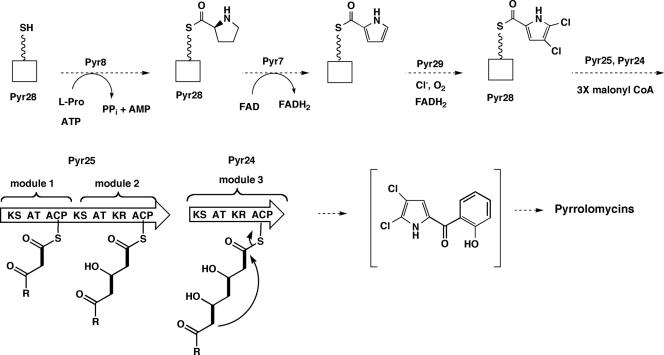
Seven genes with identifiable functions are present in the pyrrolomycin gene cluster of A. vitaminophilum. The translation products of six of these genes exhibit strong similarities to proteins encoded by the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. Pyr7, Pyr8, Pyr28, and Pyr29 exhibit identities of 56%, 45%, 44%, and 65% to PltE, PltF, PltL, and PltA, respectively. Investigations by Dorrestein et al. and Thomas and coworkers (7, 45) identified the roles played by each of these four Plt proteins in pyoluteorin biosynthesis. PltF catalyzes the reaction of l-proline with ATP to generate an amino acyl adenylate, which then reacts with the 4′-phosphopantethiene arm of the carrier protein PltL to produce a thioester. PltE catalyzes a flavin-dependent dehydrogenation of l-prolyl-PltL to pyrrolyl-PltL. Finally, PltA carries out a reduced flavin adenine dinucleotide (FADH2)-dependent double chlorination of pyrrolyl-PltL to produce 4,5-dichloropyrrolyl-PltL, which is then converted into pyoluteorin by the pyoluteorin polyketide synthase (PKS). The high degree of similarity between these four proteins from the pyoluteorin pathway and Pyr7, Pyr8, Pyr28, and Pyr29 suggests that the A. vitaminophilum proteins fulfill the same function in the pyrrolomycin pathway (Fig. 5). Additional support for the role assigned to Pyr8 is provided by a more detailed analysis of the amino acid sequence of this protein. Recent studies have identified the amino acid residues that determine the substrate specificities of enzymes or domains that catalyze the adenylation of α-amino acids (4, 44). PltF and RedM from the undecylprodiginine pathway of Streptomyces coelicolor have both been shown to utilize l-proline as a substrate (45). Analysis of the specificity codes of these two adenylation proteins revealed that the codes are DLLYLALV and DLFYLALV, respectively. Analysis of Pyr8 revealed that its specificity code is DLFYIAWV. The similarity of the Pyr8 specificity coded to the codes of PltF and RedM supports the hypothesis that Pyr8 catalyzes the adenylation of l-proline.
FIG. 5.
Predicted functions for some early pyr biosynthetic genes.
Two additional genes in the pyrrolomycin gene cluster that can be assigned functions on the basis of their similarities to pyoluteorin biosynthetic genes are pyr24 and pyr25. The translation products of these genes exhibit 42% and 44% identity to the pyoluteorin type I polyketide synthases PltC and PltB, respectively. Analysis of Pyr24 revealed that, like PltC, it consists of one PKS module that contains ketosynthase (KS), acyltransferase (AT), ketoreductase (KR), and acyl carrier protein (ACP) domains. Neither PltC nor Pyr24 appears to contain the active-site motif for a dehydratase domain. Analysis of Pyr25 shows that this polyketide synthase is similar in organization to PltB and contains two PKS modules. The first module of Pyr25 consists of KS, AT, and ACP domains, while the second module contains KS, AT, KR, and ACP domains. Both the AT and KR domains of the second module exhibit some alterations in the expected consensus motifs. Furthermore, the KR domain of the second module lacks the Lys residue of the Lys-Ser-Tyr catalytic triad found in the KR domains of modular PKSs (35). However, this Lys residue may not be required for ketoreductase activity since the mutation of the corresponding Lys residue in the KR domain in module 6 of 6-deoxyerythronolide B synthase leads to a 2:1 ratio of reduced to unreduced product (35). The presence of a KR domain in the second module of Pyr25 is consistent with the fact that only one phenolic hydroxyl group is attached to the benzene ring of the pyrrolomycins (Fig. 5). Pyr25 also appears to lack a dehydratase domain. This suggests that the dehydration reactions necessary to form the benzene ring of the pyrrolomycins probably occur spontaneously, after the release of the fully extended, cyclized polyketide. These dehydrations would be facilitated by the gain in resonance stabilization energy that accompanies the formation of the aromatic ring. Like PltB, Pyr25 lacks a thioesterase (TE) domain at its C terminus. The lack of a TE domain is consistent with the release of the extended polyketide chain by an intramolecular cyclization reaction (Fig. 5). The pyrrolomycin cluster contains a stand-alone thioesterase, Pyr26. A homolog of Pyr26, called PltG (33% identity), is present in the pyoluteorin cluster, but its role in pyoluteorin biosynthesis has not been clarified. A number of type I polyketide gene clusters contain stand-alone thioesterase genes. These have been called thioesterase II genes to distinguish them from the TE domains that are often are present at the C terminus of the final type I PKS module. Experimental evidence supports an editing role for the type II thioesterases (15, 16, 22).
The seventh gene with an identifiable function is pyr30. This gene appears to encode a 4′-phosphopantetheinyl transferase (PPTase) and shows 48% identity to putative PPtases from Streptomyces rochei and Streptomyces ambofaciens. The PPTases are a family of enzymes that catalyze the transfer of a 4′-phosphopantetheine residue from coenzyme A (CoA) to the active-site serine residue of carrier proteins or carrier protein domains, thereby converting the proteins from the apo to the holo form (25). The size of Pyr30 suggests that it is most closely related to PPTases in the Sfp subfamily (10). It appears likely that Pyr30 catalyzes the conversion of the carrier protein Pyr28 from the apo to the holo form. Pyr30 may also catalyze the conversion of the ACP domains of the polyketide synthases Pyr24 and Pyr25 to the holo form and of the putative ACP Pyr10 to its holo form.
Genes encoding tailoring enzymes or proteins of unknown function.
The pyrrolomycin gene cluster of A. vitaminophilum contains three halogenases (Pyr11, Pyr16, and Pyr17) in addition to the PltA homolog Pyr29. All FADH2-dependent halogenases appear to contain two motifs, one near the N terminus (GxGxxG) and one in the middle of the proteins (GWxWxIP) (49). These motifs are present in Pyr16, Pyr17, and Pyr29, but they are absent from Pyr11. This suggests that Pyr11 cannot function as a halogenase. It is noteworthy that Pyr11 exhibits homology to PyrD (32% identity), a putative halogenase from the pyoluteorin cluster that also lacks the characteristic flavin-dependent halogenase motifs. Since Pyr29 probably catalyzes the formation of the 4,5-dichloropyrrole moiety of the pyrrolomycins (Fig. 5), Pyr16 and Pyr17 may be required for the formation of the 2,4-dichlorophenol moiety that is present in all of the pyrrolomycins as well as for the introduction of a third chlorine atom into the pyrrole ring to produce pyrrolomycins D and J. The pyr9 gene appears to encode a flavin reductase. The function of Pyr9 may be to supply FADH2 to the halogenase enzymes.
A noteworthy feature of the A. vitaminophilum pyr cluster is the presence of two genes (pyr18 and pyr19) that appear to encode the two subunits of assimilatory nitrate reductase. Pyr18 and Pyr19 exhibit the highest identities (66% and 49%, respectively) to the putative assimilatory nitrate reductase components NasC and NasB of Amycolatopsis mediterranei. The function of Pyr18 and Pyr19 may be to reduce nitrate to nitrite in order to provide nitrite for use in the biosynthesis of the nitrated pyrrolomycins. The presence of an assimilatory nitrate reductase in the pyrrolomycin gene cluster may explain the presence of the pyr4 and pyr5 genes. The deduced products of these genes exhibit 82% and 75% identity to the two subunits of a putative 2-oxoglutarate ferredoxin oxidoreductase from Streptomyces avermitilis. In cyanobacteria, reduced ferredoxin is known to serve as the electron donor for assimilatory nitrate reductases (18), and so the function of Pyr4 and Pyr5 may be to provide reduced ferredoxin to the assimilatory nitrate reductase encoded by pyr18 and pyr19.
The translation products of three genes within the A. vitaminophilum pyr cluster exhibit signatures that suggest that they may be involved in oxidative tailoring processes. Pyr20 exhibits high similarity to cytochrome P450 enzymes (43% identity to OlmB from the gene cluster encoding the macrolide antibiotic oligomycin). Its function may be to catalyze oxidative cyclization of an aryl O-methyl group to produce the methylenedioxy group of dioxapyrrolomycin. The formation of the methylenedioxy group of the alkaloid berberine has been shown to involve a cytochrome P450-dependent oxidation (2). Interestingly, significant amounts of dioxapyrrolomycin are not produced by A. vitaminophilum, whereas it is the major pyrrolomycin antibiotic produced by Streptomyces sp. strain UC11065. The pyrrolomycin gene cluster of the latter species appears to be very similar to that of A. vitaminophilum, and it contains the pyr20 homolog dox20 (see below). The introduction of the aryl O-methyl group could possibly be catalyzed by the deduced product of pyr22, which exhibits 36% identity to a putative benzoquinone methyltransferase from Mycobacterium tuberculosis.
The other two genes possibly involved with oxidative tailoring are pyr2 and pyr21. Pyr2 exhibits strong similarity to flavin adenine dinucleotide-dependent monooxygenases (37% identity to putative rifampin monooxygenase from Nocardia). Pyr21 exhibits homology to a family of monooxygenases that catalyze the oxidation of some aromatic polyketide intermediates to the quinone level. This family includes TcmH (51% identity) from the tetracenomycin pathway and ActVA (Orf6) from the actinorhodin pathway (31% identity). These enzymes are unique since they carry out oxygenation without the aid of any prosthetic groups, metal ions, or cofactors that are normally associated with the activation of molecular oxygen (38, 39). It is conceivable that Pyr2 or Pyr21 might play a role in the introduction of a nitro group into the pyrrolomycin skeleton.
Two genes (pyr13 and pyr15) in the pyr cluster of A. vitaminophilum appear to be related to short-chain alcohol dehydrogenases. A possible function for one of the two enzymes encoded by these genes might be the reduction of the C-6 keto group that connects the pyrrole and benzene rings to the alcohol level that is found in dioxapyrrolomycin and in pyrrolomycins G and H. The second alcohol dehydrogenase homolog may be responsible for the further reduction of the resulting alcohol moiety to the methylene group that is found in pyrrolomycin B.
Four ORFs without identifiable functions exist within the apparent boundaries of the A. vitaminophilum pyrrolomycin cluster. The first of these ORFs is pyr10, which appears to encode an acyl carrier protein. The translation product of the second ORF, pyr14, shows significant similarity to proteins that may catalyze the carboxymethylation of S-isoprenylcysteine residues as well as similarity to RemK, a protein of unknown function that is found within the gene cluster for the aromatic polyketide resistomycin (17). The third ORF (pyr23) encodes a putative membrane protein, while the last of the four ORFs (pyr31) shows strong similarity to a number of acyl coenzyme A dehydrogenases.
Genes encoding potential regulatory and resistance proteins.
Two genes in the pyr cluster appear to encode regulatory proteins in the TetR family. Pyr3 shows similarity to putative TetR regulatory proteins in Nocardia and Bradyrhizobium, while Pyr27 exhibits similarity to putative TetR proteins of S. coelicolor and S. avermitilis. A pairwise alignment between Pyr3 and Pyr27 revealed that they are only 20% identical to one another. Either one or both of these proteins may play a role in the regulation of the pyr cluster.
Two putative transporter genes were identified within the pyr cluster. Pyr6 shows strong relationships to the membrane cation antiporter proteins GntQ and StaN (50% and 46% identities, respectively), while Pyr12 shows low similarity to the membrane transport protein DedA. The DedA protein of Ralstonia metallidurans has been implicated in the uptake of selenite (26). Either Pyr6 or Pyr12 might confer self-resistance to the pyrrolomycins by facilitating the export of the antibiotics from the producing organism.
DNA sequence analysis of Streptomyces sp. strain UC 11065 genes.
The pyrrolomycin gene cluster of Streptomyces sp. strain UC11065 was cloned by using a 1.5-kb halogenase PCR product amplified from A. vitaminophilum (see Materials and Methods). Two halogenase genes, dox16 and dox17, with deduced translation products that exhibited a high degree of identity to Pyr16 and Pyr17 were recovered by this cloning procedure. Sequencing of ca. 24 kb of DNA surrounding dox16 and dox17 disclosed the presence of 15 additional ORFs. These ORFs are listed in Table 3. The genes can be divided into two groups: (i) dox genes homologous to genes in the pyr biosynthetic gene cluster and (ii) genes unique to the dox cluster.
dox genes homologous to genes in the pyr biosynthetic gene cluster of A. vitaminophilum.
Fifteen of the ORFs in the sequenced region of UC 11065 are homologs of genes in the pyr cluster. Furthermore, the organization of the genes in the pyr and dox clusters appears to be highly conserved (Fig. 4). These findings provide strong evidence that both the A. vitaminophilum and the UC 11065 clusters are involved in the production of a similar set of metabolites. The functions of the 15 homologous dox genes are expected to be similar or identical to those of the corresponding pyr genes. The amino acid specificity code for the Pyr8 homolog Dox8 is DLFYLAWV, a result that is consistent with the hypothesis that Dox8 also catalyzes the adenylation of l-proline (see above).
Genes unique to the dox cluster.
Partial sequencing of the dox cluster has thus far disclosed the presence of two ORFs that appear to be absent from the pyr cluster. The first of these is dox19A. The deduced product of this ORF exhibits 65% identity to a putative carbohydrate kinase from Streptomyces sp. strain AM-7161. The potential function of this gene product is unknown. The second ORF unique to the dox cluster is dox19B. The deduced product of this gene exhibits about 40% identity to putative thiopurine S-methyltransferases from Mycobacterium. It is conceivable that this protein is required to produce a metabolic intermediate bearing an aryl O-methyl group that is then oxidized by the cytochrome P450 enzyme encoded by dox20 to produce the methylenedioxy group of dioxapyrrolomycin. If this interpretation were correct, then the genes pyr22 and dox22 that encode a methyltransferase that is common to both the pyr and dox gene clusters would play an alternative role in the biosynthetic pathways. The absence of a dox19B homolog from the pyr cluster might explain the observation that A. vitaminophilum produces very little, if any, dioxapyrrolomycin.
DISCUSSION
Using PCR primers designed from the conserved regions of bacterial halogenase genes, we amplified a halogenase DNA probe from A. vitaminophilum, a producer of the pyrrolomycin antibiotics. This probe was used to clone a gene cluster from A. vitaminophilum whose sequence analysis suggested that it was probably the pyrrolomycin biosynthetic gene cluster of this organism. To verify the identity of this gene cluster, numerous attempts were made to transform A. vitaminophilum in order to create gene disruptions. All of these efforts were unsuccessful. Consequently, investigations of a second pyrrolomycin-producing organism, Streptomyces sp. strain UC 11065, were undertaken. Once it had been established that DNA could be introduced into UC 11065 by protoplast-mediated transformation, the pyrrolomycin gene cluster was cloned from this organism using the same halogenase DNA probe that was employed to clone the A. vitaminophilum cluster. Sequencing of ca. 26 kb of the cloned UC 11065 DNA revealed the presence of 17 ORFs. Fifteen of these genes exhibit high degrees of similarity to genes within the A. vitaminophilum cluster as well as a nearly identical organizations (Fig. 4). Furthermore, both gene clusters contain a number of genes that exhibit strong similarities to genes in the pyoluteorin biosynthetic gene cluster. These observations support the hypothesis that both gene clusters are required for the biosynthesis of the pyrrolomycins in their respective organisms. To evaluate this hypothesis, the dox8 and dox17 genes of the UC 11065 cluster were inactivated by single-crossover disruptions. HPLC analysis of the metabolites isolated from each mutant indicated that the production of pyrrolomycins was abolished in both mutants (Fig. 3). These results confirm that the genetic locus cloned from UC 11065 is essential for pyrrolomycin production, and, by implication, they further confirm that the highly similar locus in A. vitaminophilum also encodes pyrrolomycin biosynthetic genes. Attempts to produce double-crossover gene disruptions in UC 11065 have not been successful thus far (data not shown). Although the entire pyrrolomycin cluster of UC 11065 has not yet been sequenced, the more extensive sequencing carried out on the A. vitaminophilum gene cluster appears to have reached the cluster boundaries in this organism. The 5′ end of the A. vitaminophilum cluster probably begins with pyr2 (Table 2 and Fig. 4), since the upstream ORF (orf1) exhibits a very high degree of similarity to formate dehydrogenase, an enzyme that is unlikely to be required for the pyrrolomycin biosynthetic pathway. The 3′ end of the pyrrolomycin cluster is probably defined by pyr31 since the ORF immediately downstream (orf32) exhibits a high degree of similarity to homogentisate 1,2-dioxygenase, an enzyme that should be irrelevant to the pyrrolomycin pathway. A precise determination of the boundaries of the A. vitaminophilum cluster will be accomplished only if the cluster can be expressed in a heterologous strain where the effect of systematic gene disruptions created outside of the native background can be assessed. Attempts to produce heterologous expression in S. coelicolor CH999 by the introduction of two cosmids with inserts spanning the entire pyrrolomycin gene cluster were unsuccessful (data not shown).
An important motivation for cloning the pyrrolomycin biosynthetic gene clusters has been to elucidate the mechanism for the formation of the nitrated pyrrolomycins. Unfortunately, the data obtained by sequence analysis of the cloned genes do not fully clarify the nature of the nitration process. The presence of genes encoding the two components of an assimilatory nitrate reductase in both clusters strongly suggests that nitrite is required for the nitration reaction. However, sequence analysis failed to reveal how nitrite is utilized to nitrate the pyrrole ring in this family of antibiotics. Evidence from precursor incorporation experiments supports the intermediacy of pyrrolomycin C in dioxapyrrolomycin biosynthesis (5). This observation suggests that nitration occurs at a relatively late stage of the pyrrolomycin biosynthetic pathway. Several mechanisms for the nitration reaction can be envisioned on the basis of currently available information. One possibility is a nucleophilic, aromatic substitution reaction that proceeds by the chlorination of pyrrolomycin C to pyrrolomycin D followed by the reaction of pyrrolomycin D with nitrite to generate a 2,3-dichloro-4-nitropyrrole that could be converted into pyrrolomycins A and B as well as dioxopyrrolomycin. A second possible mechanism would involve the nucleophilic addition of nitrite to C-4 of pyrrolomycin C followed by the oxidation of the resulting nitro dihydropyrrole to produce a 2,3-dichloro-4-nitropyrrole. A third possibility is an electrophilic aromatic substitution reaction that proceeds by the reaction of a pyrrolomycin C derivative with nitrosonium ion generated from nitrous acid to produce a 2,3-dichloro-4-nitrosopyrrole, which is then oxidized to the corresponding nitropyrrole. Lastly, nitration of a pyrrolomycin C derivative could proceed by the conversion of nitrite into an electrophilic, nitrating species by a reaction with some form of activated oxygen (30, 36), followed by nitration at the C-4 position of the pyrrole nucleus. Since no nitrated antibiotics with an intact C-6 keto group have been isolated, the reduction of this keto group may precede the nitration reaction. Additional investigations will clearly be required to determine whether any of the above-described hypotheses for the nitration mechanism are correct.
Acknowledgments
Support of this work by the Office of Naval Research (grant N00014-04-1-0285) and by the Robert A. Welch Foundation (grant C-0729) is gratefully acknowledged.
We thank John Frost for providing a culture of Streptomyces sp. strain UC 11065, Wensheng Li for the HPLC protocol used to separate the pyrrolomycins, and Phagun Garg for preparative HPLC experiments.
Footnotes
Published ahead of print on 11 December 2006.
REFERENCES
- 1.Bierman, M., R. Logan, K. Obrien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia-coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkland, J. A., T. Frenzel, M. Rueffer, M. Kobayashi, U. Mocek, C. Fox, J. M. Beale, S. Groger, M. H. Zenk, and H. G. Floss. 1995. Cryptic stereochemistry of berberine alkaloid biosynthesis. J. Am. Chem. Soc. 117:1533-1545. [Google Scholar]
- 3.Carter, G. T., J. A. Nietsche, J. J. Goodman, M. J. Torrey, T. S. Dunne, D. B. Borders, and R. T. Testa. 1987. LL-F42248α, a novel chlorinated pyrrole antibiotic. J. Antibiot. (Tokyo) 40:233-234. [DOI] [PubMed] [Google Scholar]
- 4.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 5.Charan, R. D., G. Schlingmann, V. S. Bernan, X. Feng, and G. T. Carter. 2006. Dioxapyrrolomycin biosynthesis in Streptomyces fumanus. J. Nat. Prod. 69:29-33. [DOI] [PubMed] [Google Scholar]
- 6.Conder, G. A., R. J. Zielinski, S. S. Johnson, M. S. Kuo, D. L. Cox, V. P. Marshall, C. L. Haber, P. J. DiRoma, S. J. Nelson, R. D. Conklin, et al. 1992. Anthelmintic activity of dioxapyrrolomycin. J. Antibiot. (Tokyo) 45:977-983. [DOI] [PubMed] [Google Scholar]
- 7.Dorrestein, P. C., E. Yeh, S. Garneau-Tsodikova, N. L. Kelleher, and C. T. Walsh. 2005. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc. Natl. Acad. Sci. USA 102:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ezaki, N., M. Koyama, T. Shomura, T. Tsuruoka, and S. Inouye. 1983. Pyrrolomycins C, D and E, new members of pyrrolomycins. J. Antibiot. (Tokyo) 36:1263-1267. [DOI] [PubMed] [Google Scholar]
- 9.Ezaki, N., T. Shomura, M. Koyama, T. Niwa, M. Kojima, S. Inouye, T. Ito, and T. Niida. 1981. New chlorinated nitro-pyrrole antibiotics, pyrrolomycin A and B (SF-2080 A and B). J. Antibiot. (Tokyo) 34:1363-1365. [DOI] [PubMed] [Google Scholar]
- 10.Finking, R., J. Solsbacher, D. Konz, M. Schobert, A. Schafer, D. Jahn, and M. A. Marahiel. 2002. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:50293-50302. [DOI] [PubMed] [Google Scholar]
- 11.Hammer, P. E., D. S. Hill, S. T. Lam, K.-H. van Pee, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, J., and C. Hertweck. 2004. Biosynthetic origin of the rare nitroaryl moiety of the polyketide antibiotic aureothin: involvement of an unprecedented N-oxygenase. J. Am. Chem. Soc. 126:3694-3695. [DOI] [PubMed] [Google Scholar]
- 13.He, J., and C. Hertweck. 2003. Iteration as programmed event during polyketide assembly; molecular analysis of the aureothin biosynthesis gene cluster. Chem. Biol. 10:1225-1232. [DOI] [PubMed] [Google Scholar]
- 14.Healy, F. G., M. Wach, S. B. Krasnoff, D. M. Gibson, and R. Loria. 2000. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38:794-804. [DOI] [PubMed] [Google Scholar]
- 15.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Z., B. A. Pfeifer, E. Chao, S. Murli, J. Kealey, J. R. Carney, G. Ashley, C. Khosla, and C. R. Hutchinson. 2003. A specific role of the Saccharopolyspora erythraea thioesterase II gene in the function of modular polyketide synthases. Microbiology 149:2213-2225. [DOI] [PubMed] [Google Scholar]
- 17.Jakobi, K., and C. Hertweck. 2004. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J. Am. Chem. Soc. 126:2298-2299. [DOI] [PubMed] [Google Scholar]
- 18.Jepson, B. J., L. J. Anderson, L. M. Rubio, C. J. Taylor, C. S. Butler, E. Flores, A. Herrero, J. N. Butt, and D. J. Richardson. 2004. Tuning a nitrate reductase for function. The first spectropotentiometric characterization of a bacterial assimilatory nitrate reductase reveals novel redox properties. J. Biol. Chem. 279:32212-32218. [DOI] [PubMed] [Google Scholar]
- 19.Kaneda, M., S. Nakamura, N. Ezaki, and Y. Iitaka. 1981. Structure of pyrrolomycin B, a chlorinated nitro-pyrrole antibiotic. J. Antibiot. (Tokyo) 34:1366-1368. [DOI] [PubMed] [Google Scholar]
- 20.Kers, J. A., M. J. Wach, S. B. Krasnoff, J. Widom, K. D. Cameron, R. A. Bukhalid, D. M. Gibson, B. R. Crane, and R. Loria. 2004. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429:79-82. [DOI] [PubMed] [Google Scholar]
- 21.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 22.Kim, B. S., T. A. Cropp, B. J. Beck, D. H. Sherman, and K. A. Reynolds. 2002. Biochemical evidence for an editing role of thioesterase II in the biosynthesis of the polyketide pikromycin. J. Biol. Chem. 277:48028-48034. [DOI] [PubMed] [Google Scholar]
- 23.Koyama, M., N. Ezaki, T. Tsuruoka, and S. Inouye. 1983. Structural studies on pyrrolomycins C, D and E. J. Antibiot. (Tokyo) 36:1483-1489. [DOI] [PubMed] [Google Scholar]
- 24.Koyama, M., Y. Kodama, T. Tsuruoka, N. Ezaki, T. Niwa, and S. Inouye. 1981. Structure and synthesis of pyrrolomycin A, a chlorinated nitro-pyrrole antibiotic. J. Antibiot. (Tokyo) 34:1569-1576. [DOI] [PubMed] [Google Scholar]
- 25.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 26.Ledgham, F., B. Quest, T. Vallaeys, M. Mergeay, and J. Coves. 2005. A probable link between the DedA protein and resistance to selenite. Res. Microbiol. 156:367-374. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J., M. Simurdiak, and H. Zhao. 2005. Reconstitution and characterization of aminopyrrolnitrin oxygenase, a Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 280:36719-36727. [DOI] [PubMed] [Google Scholar]
- 28.Macneil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. Macneil. 1992. Analysis of Streptomyces-avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Masuda, K., K. Suzuki, A. Ishida-Okawara, S. Mizuno, and K. Hotta. 1991. Pyrrolomycin group antibiotics inhibit substance P-induced release of myeloperoxidase from human polymorphonuclear leukocytes. J. Antibiot. (Tokyo) 44:533-540. [DOI] [PubMed] [Google Scholar]
- 30.Monzani, E., R. Roncone, M. Galliano, W. H. Koppenol, and L. Casella. 2004. Mechanistic insight into the peroxidase catalyzed nitration of tyrosine derivatives by nitrite and hydrogen peroxide. Eur. J. Biochem. 271:895-906. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, H., K. Shiomi, H. Iinuma, H. Naganawa, T. Obata, T. Takeuchi, and H. Umezawa. 1987. Isolation and characterization of a new antibiotic, dioxapyrrolomycin, related to pyrrolomycins. J. Antibiot. (Tokyo) 40:899-903. [DOI] [PubMed] [Google Scholar]
- 32.Nowak-Thompson, B., N. Chaney, J. S. Wing, S. J. Gould, and J. E. Loper. 1999. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 181:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowak-Thompson, B., S. J. Gould, and J. E. Loper. 1997. Identification and sequence analysis of the genes encoding a polyketide synthase required for pyoluteorin biosynthesis in Pseudomonas fluorescens Pf-5. Gene 204:17-24. [DOI] [PubMed] [Google Scholar]
- 34.Piraee, M., and L. C. Vining. 2002. Use of degenerate primers and touchdown PCR to amplify a halogenase gene fragment from Streptomyces venezuelae ISP5230. J. Ind. Microbiol. Biotechnol. 29:1-5. [DOI] [PubMed] [Google Scholar]
- 35.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 36.Sala, A., S. Nicolis, R. Roncone, L. Casella, and E. Monzani. 2004. Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical nitrating agents. Eur. J. Biochem. 271:2841-2852. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Sciara, G., S. G. Kendrew, A. E. Miele, N. G. Marsh, L. Federici, F. Malatesta, G. Schimperna, C. Savino, and B. Vallone. 2003. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, B., and C. R. Hutchinson. 1993. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32:6656-6663. [DOI] [PubMed] [Google Scholar]
- 40.Shomura, T., S. Amano, J. Yoshida, N. Ezaki, T. Ito, and T. Niida. 1983. Actinosporangium vitaminophilum sp. nov. Int. J. Syst. Bacteriol. 33:557-564. [Google Scholar]
- 41.Simurdiak, M., J. Lee, and H. Zhao. 2006. A new class of arylamine oxygenases: evidence that p-aminobenzoate N-oxygenase (AurF) is a di-iron enzyme and further mechanistic studies. Chem. Biol. Chem. 7:1169-1172. [DOI] [PubMed] [Google Scholar]
- 42.Sosio, M., H. Kloosterman, A. Bianchi, P. de Vreugd, L. Dijkhuizen, and S. Donadio. 2004. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 150:95-102. [DOI] [PubMed] [Google Scholar]
- 43.Sosio, M., S. Stinchi, F. Beltrametti, A. Lazzarini, and S. Donadio. 2003. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 10:541-549. [DOI] [PubMed] [Google Scholar]
- 44.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, M. G., M. D. Burkart, and C. T. Walsh. 2002. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 9:171-184. [DOI] [PubMed] [Google Scholar]
- 46.van Pee, K.-H., and J. M. Ligon. 2000. Biosynthesis of pyrrolnitrin and other phenylpyrrole derivatives by bacteria. Nat. Prod. Rep. 17:157-164. [DOI] [PubMed] [Google Scholar]
- 47.Vining, L. C., and C. Stuttard. 1994. Chloramphenicol, p. 505-530. In L. C. Vining and C. Stuttard (ed.), Genetics and biochemistry of antibiotic production. Butterworth-Heinemann, Boston, MA.
- 48.Wynands, I., and K. H. van Pee. 2004. A novel halogenase gene from the pentachloropseudilin producer Actinoplanes sp. ATCC 33002 and detection of in vitro halogenase activity. FEMS Microbiol. Lett. 237:363-367. [DOI] [PubMed] [Google Scholar]
- 49.Zehner, S., A. Kotzsch, B. Bister, R. D. Sussmuth, C. Mendez, J. A. Salas, and K. H. van Pee. 2005. A regioselective tryptophan 5-halogenase is involved in pyrroindomycin biosynthesis in Streptomyces rugosporus LL- 42D005. Chem. Biol. 12:445-452. [DOI] [PubMed] [Google Scholar]