Abstract
AcrAB-TolC is the major, constitutively expressed tripartite multidrug efflux system in Escherichia coli that recognizes various structurally unrelated molecules, including many antibiotics, dyes, and steroids. The AcrB inner membrane pump portion of the efflux system has been shown in recent structural studies to bind substrates at multiple sites, suggesting that particular substrate “sets” may compete for efflux by interfering with a certain binding site(s). However, our data indicate that the general structural class does not appear to dictate a particular substrate binding site that can be competitively inhibited in whole cells. In our study, substrate competition failed to increase cell-associated levels of steroids or dyes to levels characteristic of AcrB- or AcrB/EmrAB-deficient genomic mutants or achieved with the pump inhibitor carbonyl cyanide m-chlorophenylhydrazone. In addition, this general observation was sustained even with (i) a cocktail containing seven-pump substrates supplied slightly below their respective wild-type MIC levels, (ii) competing drug substrates of the same structural class (steroids or macrolides), and (iii) hyper-MIC levels of the exogenously supplied agents. Thus, this pump system (and possibly EmrAB-TolC) may have an extraordinary capacity to simultaneously handle multiple-drug substrates that is not necessarily reflected in MIC analyses. In addition, our study has extended the range of substrates recognized by the AcrAB- and EmrAB-TolC systems.
AcrAB-TolC is the major multidrug efflux system in Escherichia coli and has homologs in virtually every gram-negative organism sequenced to date (17). Similar to several other efflux systems encoded in the E. coli genome, it forms a tripartite complex composed of a resistance nodulation division (RND)-type cytosolic membrane pump proper (AcrB), a periplasmic membrane fusion protein (AcrA), and an outer membrane porin (TolC) to span both membranes of the gram-negative envelope (17, 20). Quantitatively and qualitatively, this system represents the strongest active intrinsic resistance mechanism known in this organism and is expressed constitutively (7). Another tripartite system of the major facilitator superfamily (MFS) type, EmrAB-TolC, is also expressed constitutively (19), but like other endogenous efflux systems (that are usually silent under normal laboratory conditions), its contribution to resistance is masked by the overwhelming effect and overlapping substrate repertoire of AcrAB-TolC (7, 9, 34).
AcrB is of considerable interest in the basic and clinical sciences for several reasons: (i) several genetic studies indicate that substrate specificity is determined by this component (5, 10, 21, 25, 35), (ii) the known substrate range is quite structurally diverse, with little rationale for selectivity, suggesting only a propensity for lipophiles (26, 28, 29) if aminoglycosides (which are substrates for at least one multidrug RND-type E. coli pump [32], AcrD) are excluded, (iii) crystal structures have shown that its architecture is large and complex (23, 39), fueling speculation on how substrates are recruited in a secondary active transport process (7, 24, 38), and (iv) the design of inhibitors is critically dependent on a fundamental understanding of this system that, once employed, could render E. coli (and potentially other gram-negative pathogens) susceptible to a wide range of older antibiotics, thus reinvigorating our current antibacterial drug arsenal (18).
Whether multidrug efflux pump inhibitors will prove clinically successful is currently unknown, albeit it is a subject of intense study (18). An associated issue involves substrate competition where in vivo efflux is measured in the presence of other drug substrates for inhibitory potential. Such studies have been performed with the mammalian P-glycoprotein (Pgp) efflux pump but have produced mixed results (14, 15), whereas this issue has not been adequately addressed in the literature for bacterial efflux systems. However, addressing this issue becomes difficult to interpret in light of structural studies with crystallized AcrB that identified at least four independent substrate binding sites within the large internal cavity (39) and, recently, another site deep within an external depression on the periplasmic face (37).
In light of this potentially complex substrate binding scheme, we tested whether certain tritium-labeled substrates could compete for efflux in a clinically relevant scenario with steady-state levels of cell-associated drug substrates in an E. coli whole-cell background. We previously demonstrated that steroid hormones are strong substrates for both AcrAB-TolC and EmrAB-TolC (9), and we employed them in this study as tritium-labeled competitors. In addition, we assessed resistance to and competition with various macrolide drugs which represent another structurally separate but likewise bulky substrate class.
(A preliminary account of this study was presented at the 50th annual Wind River Conference on Prokaryotic Biology, Estes Park, CO, 7 to 11 June 2006).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The E. coli strains used in this study were maintained as frozen stocks at −80°C in 10 to 15% glycerol and routinely cultured in Luria-Bertani Miller (LB) agar (1.5%) or broth medium (Fisher Biotech) at 37°C. The parent strain, AG102, was previously derived from AG100 (12) and is drug hyperresistant due to a mutation in MarR (marR1) which increases the expression of MarA, a global regulator, which in turn results in the overexpression of the AcrAB-TolC efflux system (31). AG102MB and HNCE4 are isogenic mutants derived from AG102 and AG102MB (9, 31), respectively, containing mutations at the acrB locus (kan; AG102MB) or at both the acrB (kan) and emrAB (cat; HNCE4) loci. These two derivatives were cultured with 25 μg/ml of either kanamycin (AG102MB) or chloramphenicol (HNCE4) to maintain selection.
Radioactivity and miscellaneous reagents.
[6,7-3H(N)]estradiol, [1,2-3H]hydrocortisone, [1,2-3H(N)]progesterone, [1α,2α-3H]testosterone, and [N-methyl-3H]erythromycin were obtained from American Radiolabeled Chemicals, Inc. in ethanol at a concentration of 1 mCi/ml and a specific activity of 50 Ci/mmol except for testosterone and erythromycin, which were obtained at specific activities of 40 mCi/mmol and 80 Ci/mmol, respectively. Unlabeled steroid hormones (Sigma-Aldrich Corporation), bile acids (Fluka), and detergents (Fisher Biotech) were prepared at 100-mg/ml stock concentrations in dimethyl sulfoxide (DMSO; Fisher Biotech) for use in competition analyses. Pump inhibitors carbonyl cyanide m-chlorophenylhydrazone (CCCP; Fluka) and reserpine (Sigma-Aldrich Corporation) were dissolved in DMSO at 10 mM and 2 mg/ml, respectively. Stock concentrations of various other drugs and dyes (Sigma-Aldrich Corporation) were dissolved in either water or ethanol at 50 mg/ml except for ciprofloxacin, ethidium bromide, and tetracycline, which dissolved at 0.5, 10, and 10 mg/ml, respectively. Substrate cocktail solutions were prepared as a 20× stock containing ampicillin (AMP; 40 μg/ml), chloramphenicol (CHL; 60 μg/ml), ciprofloxacin (CIP; 0.6 μg/ml), ethidium bromide (EtBr; 2.56 mg/ml), erythromycin (ERY; 2.56 mg/ml), novobiocin (NOV; 1.28 mg/ml), and tetracycline (TET; 20 μg/ml).
MIC analyses.
The MICs of several compounds were determined by preparing serial twofold microdilutions (150 μl) with LB medium in 96-well microtiter flat-bottom plates (Falcon; Becton Dickinson). The dilution series were inoculated with 5 μl of mid-log-phase cultures that were grown in 2× YT broth (1.6% tryptone, 1% yeast extract, and 0.5% NaCl) with selection (when appropriate) and normalized by turbidity for comparison between strains. Visible signs of bacterial growth were recorded after 24 h at 37°C. Those compounds requiring high initial concentrations in MIC analyses (taurocholic and cholic acids and Triton X-100; 256, 128, and 8 mg/ml, respectively) were dissolved directly in LB medium at the appropriate concentration to avoid any significant dilution of the growth medium.
Accumulation (uptake) assays.
An uptake assay for various tritium-labeled steroids and erythromycin was performed essentially as described previously (9, 11). In brief, the procedure involved concentrating mid-log-phase cells tenfold in fresh LB medium and normalizing by culture turbidity if uptake levels were compared between different strains. After a short preincubation (7.5 min) in a 37°C water bath (with or without different drug substrates and inhibitors), 200-μl aliquots of cells were exposed to tritium-labeled compounds (2.5 μl of commercial stock) for 3.5 min. Unbound radiolabel was washed from the cells in two successive, repeated steps with excess (1 ml) ice-cold 100 mM lithium chloride-100 mM potassium phosphate buffer (pH 7.0), followed by rapid cell pelleting in a 4°C microcentrifuge. The tritium-labeled cell pellet was digested with formamide (1 ml) at 65°C, and the remaining formamide-solubilized tritium label was quantified by liquid scintillation counting.
For dye uptake experiments, ethidium bromide was replaced by crystal violet in substrate cocktails at a final 1× concentration of 16 μg/ml. The above procedure was followed except quantization by liquid scintillation counting was replaced with absorption spectroscopy at 590 nm in formamide.
RESULTS
General drug resistance phenotypes of efflux-deficient strains.
The resistance profiles for AG102 and its mutant derivatives, AG102MB and HNCE4, were determined for various antimicrobial compounds with microdilution MIC analyses (Table 1). AG102MB displayed lower levels of resistance to every drug tested in this study than did AG102 when the drug-resistant marker phenotypes used in engineering the mutant strains were excluded (i.e., kanamycin, chloramphenicol, and cat-mediated fusidic acid cross-resistance [4, 10]). Except for ampicillin, the decreases in MICs were generally severalfold, which highlights the broad significance of the AcrAB-TolC system to intrinsic drug resistance in this organism. With the double-knockout strain HNCE4, further twofold decreases in resistance to cholic and taurocholic acids were apparent and in keeping with the known substrates of the EmrAB-TolC system (9), which also included a twofold decrease in resistance to the nonionic detergent Triton X-100. However, ethidium bromide is also a known substrate of this efflux system (17), yet MIC analysis revealed an eightfold-higher level of resistance to this molecule in HNCE4 than in AG102MB. Similarly, a higher level of resistance was also observed for crystal violet and erythromycin, which, taken collectively, may result from the induction of a separate efflux system. Nevertheless, the generally observed resistances in HNCE4 more closely resembled those of AG102MB in respective magnitudes than those of the parent strain AG102.
TABLE 1.
Drug, dye, and detergent susceptibilities of E. coli marR1 and its isogenic efflux-deficient derivatives
| Strain | MIC (μg/ml) for indicated compounda
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CHL | CAb | CIP | CV | ERY | EtBr | FA | KAN | NOV | SDS | TCAb | TET | TrX | |
| AG102 | 32 | 8 | 16 | >0.25 | 32 | 128 | 256 | >512 | 8 | 512 | 1,000 | 64 | 8 | 8,000 |
| AG102MB | 16 | 2 | 4 | 0.0625 | 0.5 | 4 | 4 | 16 | >512 | 4 | 62.5 | 8 | 2 | 125 |
| HNCE4 | 16 | >128 | 2 | 0.0625 | 8 | 8 | 32 | 128 | >512 | 4 | 125 | 4 | 2 | 62.5 |
Abbreviations: AMP, ampicillin; CHL, chloramphenicol; CA, cholic acid; CIP, ciprofloxacin; CV, crystal violet; ERY, erythromycin; EtBr, ethidium bromide; FA, fusidic acid; KAN, kanamycin; NOV, novobiocin; SDS, sodium dodecylsulphate; TCA, taurocholic acid; TET, tetracycline; TrX, Triton X-100.
MICs for bile acids are reported as milligrams per milliliter.
Competition studies with AcrAB-TolC substrates.
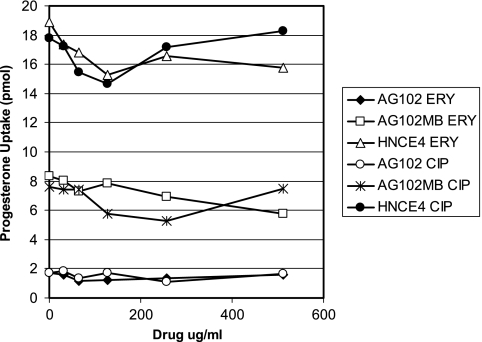
Fully efflux proficient AG102 cells were exposed independently to increasing concentrations of various AcrAB-TolC substrates (CHL, CIP, ERY, EtBr, NOV, and TET) and a nonsubstrate aminoglycoside (gentamicin) to determine their effects at steady state on steroid hormone accumulation. Low concentrations (0 to 16 μg/ml) of each compound were used in competition against tritium-labeled hydrocortisone, a substrate of AcrAB-TolC but not EmrAB-TolC (9), which produced only modest fluctuations (approximately twofold at most) in hydrocortisone levels (data not shown). This analysis was extended with erythromycin and ciprofloxacin at much higher concentrations (Fig. 1) and measured in all three E. coli strains (AG102, AG102MB, and HNCE4) using progesterone, which is a substrate of both AcrAB-TolC and EmrAB-TolC (9). As consistently observed, erythromycin and ciprofloxacin also failed to modulate progesterone levels in AG102 cells to those observed with AG102MB or HNCE4, even at concentrations well above the MICs of both drugs (Table 1).
FIG. 1.
Uptake of progesterone by marR1 strains exposed to increasing concentrations of exogenous ERY and CIP that exceed MICs for the respective strains (Table 1).
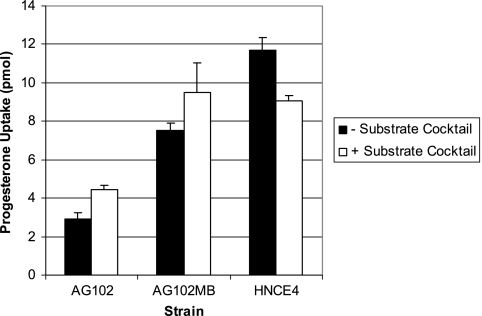
The three strains were also assessed for changes in steroid uptake when equilibrated with a substrate cocktail containing seven AcrAB-TolC substrates supplied slightly below the reported MICs for AG102 (Fig. 2). Progesterone levels (as well as estradiol and testosterone [data not shown]) for AG102 increased only marginally (at most 2.3-fold) by the cocktail. A similar and consistent slight increase was also observed in AG102MB but not with HNCE4. On the contrary, a consistent decrease in steroid uptake was observed with HNCE4 compared to that with an HNCE4 cocktail-free control. Upon further examination, a successive increase in ethidium bromide staining of the AG102, AG102MB, and HNCE4 cell pellets was observed by visual inspection in the cocktail-treated strains, demonstrating that the other substrates in the uptake assay milieu were responding predictably to the three efflux phenotypes. Therefore, we replaced ethidium bromide with crystal violet in the cocktails to assess whether its levels in the mutant strains would increase similar to those of ethidium bromide. Cocktail treatment had little effect on crystal violet levels for which absorbance values successively increased with the single (0.466 ± 0.02, untreated; 0.373 ± 0.04, treated) and double (0.547 ± 0.08, untreated; 0.502 ± 0.02, treated) efflux-mutant strains from levels observed with AG102 (0.307 ± 0.02, untreated; 0.297 ± 0.01, treated), suggesting that it too may serve as a substrate of EmrAB-TolC. Taken in aggregate, these data suggest that in the presence of multiple substrates, the major E. coli efflux systems can maintain consistently and simultaneously low levels of each substrate.
FIG. 2.
Uptake of progesterone by marR1 strains when exposed to substrate cocktails containing seven AcrAB-TolC pump substrates at or slightly below MICs for AG102. Cocktail treatment resulted in statistically significant changes in uptake (as assessed by the Wilcoxon rank sum test) over respective nontreated control levels. Error bars indicate standard deviations. −, absence of; +, presence of.
Competition with structurally similar substrates. (i) Macrolide studies.
Substrate competition may occur only with substrates of the same structural class, assuming that they are competing for identical binding sites. Using a group of six macrolide drugs, tritium-labeled erythromycin uptake was measured in AG102 cell aliquots equilibrated with one of these later-generation macrolides at increasing concentrations from 0 to 256 μg/ml. In each case, these drugs also failed to produce any significant fluctuation in erythromycin uptake, even with the structurally similar macrolides dirithromycin, oleandomycin, and roxithromycin containing, like erythromycin, 14-membered lactone rings (data not shown). In addition, MIC analyses revealed that these compounds serve as substrates for AcrAB-TolC since severalfold decreases in resistance to each of the six additional macrolides were observed in AG102MB compared to respective AG102 values (Table 2). Moreover, josamycin and tylosin MICs were lowest in the HNCE4 background, suggesting that, quite possibly, these two 16-membered lactone ring macrolides are also recognized by the EmrAB-TolC system.
TABLE 2.
Macrolide susceptibilities of E. coli marR1 and its isogenic efflux-deficient derivativesa
| Strain | Macrolide susceptibilityb
|
|||||
|---|---|---|---|---|---|---|
| DTM | JOS | OLEAN | ROX | SPIR | TYL | |
| AG102 | 256 | >512 | >512 | 512 | >512 | >512 |
| AG102MB | 16 | 32 | 128 | 64 | 64 | 128 |
| HNCE4 | 16 | 4 | 256 | 128 | 64 | 64 |
Susceptibilities are reported as MICs in micrograms per milliliter.
Abbreviations: DTM, dirithromycin; JOS, josamycin; OLEAN, oleandomycin; ROX, roxithromycin; SPIR, spiramycin; TYL, tylosin.
(ii) Steroid studies.
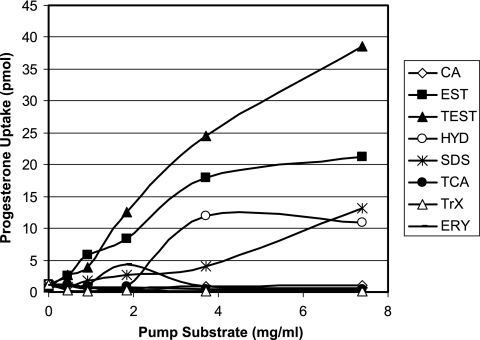
Substrate hierarchy or preference by the efflux pumps was also examined to eliminate any competitive advantage that steroids may have as preferred substrates. Various steroid hormones were employed in competition with each other, bile acid-type steroids (which have amphipathic, detergent-like properties), nonionic (Triton X-100) and ionic (sodium dodecyl sulfate [SDS]) detergents, and erythromycin at high (mg/ml) concentrations. Using this approach, we were able to produce significant modulations in progesterone uptake (and that of hydrocortisone and testosterone [data not shown]) with SDS and other steroid hormones (Fig. 3). However, these compounds exhibited reduced solubility, especially at such high concentrations, resulting in their precipitation with the cells during the ice-cold buffer washes of the uptake assay. Indeed, when assessed in a cell-free milieu, tritium-labeled testosterone associated with estradiol precipitates in a linear relationship with concentration (R2 = 0.956 [data not shown]), indicating that the steroid-steroid competition may also have failed to inhibit the efflux phenotype. In support of these findings, more-soluble compounds, such as Triton X-100 and, as previously observed with estradiol uptake (9), the bile acids cholic and taurocholic acids, uniformly failed in this capacity.
FIG. 3.
Effect of high concentrations (mg/ml) of steroid molecules, ionic and nonionic detergents, and erythromycin on progesterone uptake profile of AG102 cells. Substrate abbreviations are defined in Table 1; EST, estradiol; TEST, testosterone; HYD, hydrocortisone.
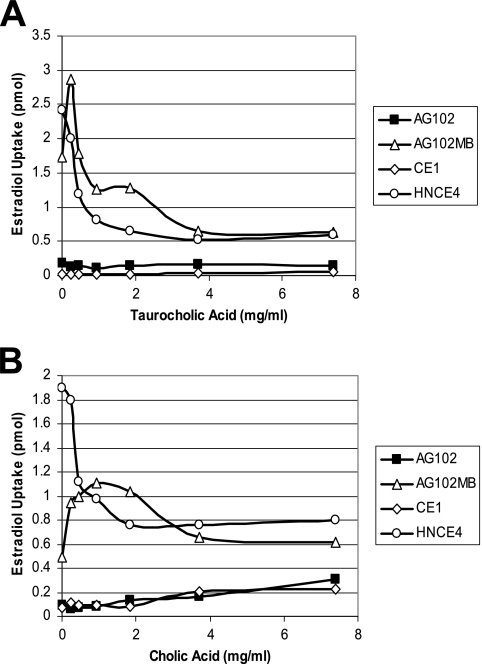
To investigate steroid competition for the EmrAB-TolC system apart from the overwhelming effect of the AcrAB-TolC phenotype, the three marR1 strains were employed together with a single isogenic emrAB mutant strain, CE1 (9), to monitor the uptake of estradiol at increasing concentrations of either conjugated (taurocholic) or deconjugated (cholic) bile acids within gastrointestinal tract physiological levels of <10 mg/ml (approximately 18 to 20 mM [8]). At every concentration tested, estradiol uptake was unaffected by exogenous taurocholic acid (Fig. 4A) and increased marginally by cholic acid (Fig. 4B) in strains containing a functional AcrAB-TolC system (AG102 and CE1), whereas strains containing knockouts of this system (AG102MB and HNCE4) were quite sensitive to bile acids (Fig. 4A and B). AG102MB cells, which contain a functional EmrAB-TolC system, demonstrated less initial uptake of estradiol than did HNCE4 cells and, in addition, exhibited some apparent modest bile acid-mediated inhibition of EmrAB-TolC activity, indicated by the initial increase in estradiol uptake at low concentrations of either bile acid. However, this initial inhibition became dominated presumably by the diffusion of high concentrations of bile acids (>2 mg/ml) at the exclusion of estradiol association, resulting in uptake levels similar to those of HNCE4 (Fig. 4A and B). Interestingly, these data corresponded well with the general observations previously made with substrate cocktails, especially regarding the decrease in steroid hormone uptake of cocktail-treated HNCE4 cells (Fig. 2).
FIG. 4.
Steroid competition studies on uptake of tritium-labeled estradiol by E. coli marR1 strains. (A) Effect of exogenous, unlabeled taurocholic acid competition on estradiol uptake. (B) Effect of cholic acid competition on estradiol uptake.
Impact of pump inhibitors as substrates.
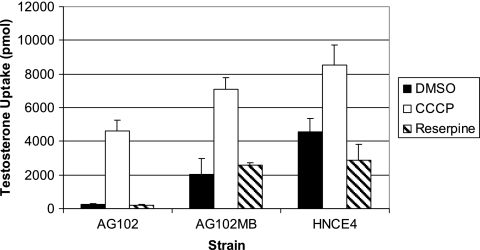
Thus far in the study, substrate competition was expected to result in pump inhibition to levels approaching those for pump-deficient strains. To demonstrate comparable inhibition both within and between strains, steroid uptake was assessed using classical inhibitors, such as CCCP (ionophore and proton motive force uncoupler) and reserpine (general efflux pump inhibitor). During the course of the study, testosterone was indeed confirmed to be an additional substrate of the AcrAB-TolC and EmrAB-TolC efflux systems since its uptake levels repeatedly increased approximately 8- and 19-fold from AG102 levels in DMSO-treated AG102MB and HNCE4 backgrounds, respectively (Fig. 5). When exposed to CCCP at 100 μM, levels of this steroid increased dramatically for each strain that reached levels in AG102 cells approximating those of untreated HNCE4, potentially representing maximal pump inhibition (AG102 MIC for CCCP, 128 μM). On the contrary, 100 μg/ml reserpine had no statistically significant effect on testosterone uptake when assessed with the Wilcoxon rank sum test (Fig. 5). Accordingly, MIC analyses with reserpine at 100 μg/ml failed to demonstrate any repeatable significant difference in the MICs of cholic acid, CIP, crystal violet, ERY, EtBr, NOV, SDS, and TET for AG102 cells from the values reported in Table 1 but, interestingly, these drug resistances fell below concentration thresholds measured for AG102MB cells, indicating that reserpine itself may affect cell viability. Indeed, the MIC of reserpine decreased from 256 μg/ml for AG102 cells to 128 μg/ml for both AG102MB and HNCE4 cells, which suggests that this inhibitor is a potentially weak substrate of the AcrAB-TolC system. In addition, the MICs of CCCP for AG102MB and HNCE4 cells decreased to 32 and 16 μM, respectively, suggesting that CCCP may also be recognized by AcrAB-TolC as has previously been shown for EmrAB-TolC (19). Thus, these inhibitors may act in concert as substrates and inhibitors to confound the issue especially in whole-cell backgrounds. At the very least, exogenous reserpine as a potential substrate of AcrAB-TolC failed to modulate the uptake of testosterone.
FIG. 5.
Effect of the ionophore, CCCP, and the general efflux pump inhibitor, reserpine, on testosterone uptake by marR1 strains. Error bars indicate standard deviations.
DISCUSSION
The universal inability to compete substrates of either major efflux system in E. coli to produce an inhibitory effect on macrolide, steroid, or dye accumulation in whole-cell assays is an interesting finding that may be a common property for efflux systems in general. Although this is the first report for bacterial systems, this phenomenon has been reported for mammalian Pgp, MDR1, with which several drug substrates failed to compete with daunomycin when this multidrug efflux system was overexpressed in breast cancer cells, leading the authors to conclude, as one possibility, that distinct substrate binding sites exist on MDR1 (14). Interestingly, the modulation of this same system in Calu-3 subbronchial cells was achieved with particularly hydrophobic β-ligand substrates but also with steroid hormones as measured with rhodamine 123. However, a dichotomy existed where only non-Pgp steroid substrates, such as testosterone and progesterone, affected rhodamine accumulation, whereas several known Pgp steroidal substrates were ineffective (15). Since AcrAB-TolC serves as a model to study multidrug efflux and antibiotic resistance in bacterial systems (17) and, likewise, Pgp serves accordingly for mammalian multidrug efflux pumps that contribute to intrinsic resistance to many chemotherapeutic agents (1), our study reinforces the functional similarities that exist between bacterial and mammalian multidrug efflux systems.
The relationship between MICs and the observed efflux of steroids in the presence of hyper-MIC levels of other supplied drug substrates provides one of the most striking observations in this study. In several instances, the concentrations of drug substrates supplied in the uptake system (CHL and TET, up to 16 μg/ml; CIP and ERY, up to 512 μg/ml) (Fig. 1) were higher than the respective MICs for those drugs (Table 1), yet steroid (hydrocortisone or progesterone) and, in one instance, erythromycin accumulation (versus dirithromycin) remained virtually unaffected, indicating an efflux capacity that may far exceed observed MIC levels. Other possible explanations for these data may involve the metabolism of the labeled compound by E. coli or the involvement of some impurity in the commercial stock preparation of the labeled substrates, the latter of which seems unlikely considering that several different tritium-labeled steroid substrates and erythromycin were employed in the study. Although it is impossible to eliminate the effect of steroid metabolism from our analyses using whole-cell backgrounds, such activity is generally associated with members of the gram-positive commensal microbiota and involves ring transformation and deconjugation (3). E. coli has been associated with only two such mechanisms: hydrolysis of steroid glucuronides (13) and 7α-dehydroxylation of bile acids (36). Thus, we chose to employ free steroid hormones to eliminate the impact of potential hydrolysis of steroid conjugates that would impact solubility and undoubtedly hydrophobic properties of the parent compound. Furthermore, we speculate that the effects of endogenous metabolism on our data are negligible at best since the labeled steroid substrates were exposed to E. coli for only a short period of time (3.5 min).
In substrate cocktail studies, dye and steroid levels in the same uptake system responded in tandem to the three genetic backgrounds. These data indicate that the efflux pump systems may also have the capability to handle multiple substrates simultaneously. During the submission of our work, two papers describing crystal structures of AcrB with and without substrates were published (22, 33). In these most recent developments, both groups were able to discriminate different conformations of each protomer within the homotrimeric organization of AcrB, leading them to advance a three-step, functionally rotating mechanism that allows multisite substrate binding. Taken in aggregate with previous structural studies, such a mechanism is in agreement with our inability to compete substrates and, potentially, our independent conclusion that simultaneous entry (and subsequent binding) of multiple different substrates may indeed occur initially in the “access” protomer conformation (22). Although significant efflux inhibition was not achieved with the substrate cocktail used in our study, it is important to address whether refinement of cocktail components and their respective concentrations may alter this property and potentially prove informative for inhibiting multisite and multisubstrate binding.
A caveat of our study involves an assumption that the exogenously supplied levels of drugs equilibrate across the membranes, thereby exposing them to the cytoplasmic pumps at or near such levels. This assumption is difficult to make since the outer membrane provides a barrier to retard lipophile influx. The pathway of such influx, whether through porins or membrane intercalation, is difficult to assess individually, much less in combination (27). It is conceivable that some of the observed effect may result from this molecular “sieving” capability of the outer membrane, which may bias exposure of the efflux pump to particular drugs in effect at concentrations different from those initially supplied. Nonetheless, with MIC levels as a guide, the influence of outer membrane diffusion on our data is fundamentally limited but was tested in several instances at exceedingly high (mM) concentrations (Fig. 3 and 4). This is especially relevant considering that the various tritium-labeled substrates were supplied at concentrations several orders of magnitude lower (approximately 125 nM) than those of the exogenously supplied and preincubated competitors at steady state.
Another potential criticism involves our methodology in assessing substrate competition for efflux. Thus, in our study, changes in the accumulation of labeled substrate were measured as an indicator of potential efflux inhibition rather than efflux itself. The interpretation of this methodology becomes problematic under conditions where significant nonspecific membrane association of substrates, especially lipophiles, occurs, whereby changes mediated by efflux become diluted if not insignificant. We offer two observations that indicate the contrary: (i) in each case where uptake was measured with efflux-proficient AG102 cells, levels of cell-associated substrate were low, and (ii) whether uptake was compared with AG102 and efflux-deficient strains or with AG102 exposed to CCCP, the differences in accumulation were severalfold, which we failed to achieve with any substrate combination in this study. Thus, measuring steady-state levels becomes pertinent since whole-cell drug association is important for assessing activity in vivo. Nevertheless, substrate accumulation is best described as net cellular levels involving interplay between influx (diffusion or facilitated entry), efflux, and nonspecific association which, collectively, is complex and undoubtedly dynamic.
This report also describes characteristic drug resistances in E. coli where two constitutively expressed tripartite systems composed of pumps from different phylogenetic families have been mutated and characterized. In one other related study, efflux pumps were deleted by groups according to phylogenetic family, which failed to reveal changes in drug susceptibilities attributable to an emrAB deletion undoubtedly because of the overwhelming effect of the AcrAB-TolC system (34). Such effect was demonstrated here with the CE1 strain by using substrates recognized by both pump systems (Fig. 4). In our study, we found that crystal violet and possibly certain 16-membered lactone ring macrolides, josamycin and tylosin, may serve as substrates for the EmrAB-TolC efflux system. However, MIC analyses for HNCE4 revealed higher levels of resistance to ethidium bromide, crystal violet, erythromycin, (potentially) SDS (Table 1), oleandomycin, and roxithromycin (Table 2) than for AG102MB. With regards to this observation, we can hypothesize that one or more of several other efflux systems in the E. coli genome may be induced by these drugs during overnight growth for the MIC analyses by the efflux-crippled strain. Such systems are under tight regulatory control by various local and global regulatory genes that can be induced by drugs (2, 6, 17, 30), whereas on the contrary, any potential induction could not occur in the uptake assays. Interestingly, a recent study identified an increase in MdtEF-dependent drug tolerance as a function of growth phase that conferred resistance (with respect to compounds used in our study) to erythromycin, crystal violet, ethidium bromide, and SDS in a ΔacrB background (16). Whether this system is responsible for the results observed in our study with HNCE4 appears likely but is unknown at this point.
Acknowledgments
We thank Carl Cerniglia, Huizhong Chen, and Fatemeh Rafii for critically reviewing the manuscript and providing several helpful suggestions. We also thank an anonymous reviewer for several insightful comments that helped ensure correct interpretation of our results.
This study was funded by the United States Food and Drug Administration and the National Center for Toxicological Research under protocol E0718001.
The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Ambudkar, S. V., I. W. Kim, and Z. E. Sauna. 2006. The power of the pump: mechanisms of action of P-glycoprotein (ABCB1). Eur. J. Pharm. Sci. 27:392-400. [DOI] [PubMed] [Google Scholar]
- 2.Baranova, N., and H. Nikaido. 2002. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, S. F., and P. B. Hylemon. 1997. Biotransformation of bile acids, cholesterol, and steroid hormones, p. 470-510. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology, vol. I. Gastrointestinal ecosystems and fermentations. International Thomson Publishing, New York, NY. [Google Scholar]
- 4.Bennett, A. D., and W. V. Shaw. 1983. Resistance to fusidic acid in Escherichia coli mediated by the type I variant of chloramphenicol acetyltransferase. A plasmid-encoded mechanism involving antibiotic binding. Biochem. J. 215:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eda, S., H. Maseda, and T. Nakae. 2003. An elegant means of self-protection in gram-negative bacteria by recognizing and extruding xenobiotics from the periplasmic space. J. Biol. Chem. 278:2085-2088. [DOI] [PubMed] [Google Scholar]
- 6.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 7.Elkins, C. A., and K. E. Beenken. 2005. Modeling the tripartite drug efflux pump archetype: structural and functional studies of the macromolecular constituents reveal more than their names imply. J. Chemother. 17:581-592. [DOI] [PubMed] [Google Scholar]
- 8.Elkins, C. A., and L. B. Mullis. 2004. Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 70:7200-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elkins, C. A., and L. B. Mullis. 2006. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J. Bacteriol. 188:1191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graef, V., E. Furuya, and O. Nishikaze. 1977. Hydrolysis of steroid glucuronides with β-glucuronidase from bovine liver, Helix pomatia and Escherichia coli. Clin. Chem. 23:532-535. [PubMed] [Google Scholar]
- 14.Hall, J. G., A. H. Cory, and J. G. Cory. 1999. Lack of competition of substrates for P-glycoprotein in MCF-7 breast cancer cells overexpressing MDR1. Adv. Enzyme Regul. 39:113-128. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton, K. O., M. A. Yazdanian, and K. L. Audus. 2001. Modulation of P-glycoprotein activity in Calu-3 cells using steroids and β-ligands. Int. J. Pharm. 228:171-179. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, A., H. Hirakawa, T. Hirata, K. Nishino, and A. Yamaguchi. 2006. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J. Bacteriol. 188:5693-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 18.Lomovskaya, O., and K. A. Bostian. 2006. Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem. Pharmacol. 71:910-918. [DOI] [PubMed] [Google Scholar]
- 19.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomovskaya, O., H. I. Zgurskaya, and H. Nikaido. 2002. It takes three to tango. Nat. Biotechnol. 20:1210-1212. [DOI] [PubMed] [Google Scholar]
- 21.Mao, W., M. S. Warren, D. S. Black, T. Satou, T. Murata, T. Nishino, N. Gotoh, and O. Lomovskaya. 2002. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 46:889-901. [DOI] [PubMed] [Google Scholar]
- 22.Murakami, S., R. Nakashima, E. Yamashita, T. Matsumoto, and A. Yamaguchi. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173-179. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 24.Murakami, S., and A. Yamaguchi. 2003. Multidrug-exporting secondary transporters. Curr. Opin. Struct. Biol. 13:443-452. [DOI] [PubMed] [Google Scholar]
- 25.Murata, T., M. Kuwagaki, T. Shin, N. Gotoh, and T. Nishino. 2002. The substrate specificity of tripartite efflux systems of Pseudomonas aeruginosa is determined by the RND component. Biochem. Biophys. Res. Commun. 299:247-251. [DOI] [PubMed] [Google Scholar]
- 26.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikaido, H., and H. Zgurskaya. 1999. Antibiotic efflux mechanisms. Curr. Opin. Infect. Dis. 12:529-536. [DOI] [PubMed] [Google Scholar]
- 30.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295-1298. [DOI] [PubMed] [Google Scholar]
- 34.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto, T., H. Higashi, A. Kanatani, X. S. Lin, H. Nagai, H. Oyama, K. Kurazono, and D. Tsuru. 1991. Cloning and sequencing of the 7α-hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. J. Bacteriol. 173:2173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, E. W., J. R. Aires, G. McDermott, and H. Nikaido. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 187:6804-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]