Abstract
Klebsiella pneumoniae causes common and severe hospital- and community-acquired infections with a high incidence of multidrug resistance. The emergence and spread of β-lactamase-producing K. pneumoniae strains highlight the need to develop new therapeutic strategies. In this study, we developed antisense peptide nucleic acids (PNAs) conjugated to the (KFF)3K peptide and investigated whether they could mediate gene-specific antisense effects in K. pneumoniae. No outer membrane permeabilization was observed with antisense PNAs when used alone. Antisense peptide-PNAs targeted at two essential genes, gyrA and ompA, were found to be growth inhibitory at concentrations of 20 μM and 40 μM, respectively. Mismatched antisense peptide-PNAs with sequence variations of the gyrA and ompA genes when used as controls were not growth inhibitory. Bactericidal effects exerted by peptide-anti-gyrA PNA and peptide-anti-ompA PNA on cells were observed within 6 h of treatment. The antisense peptide-PNAs specifically inhibited expression of DNA gyrase subunit A and OmpA from the respective targeted genes in a dose-dependent manner. Both antisense peptide-PNAs cured IMR90 cell cultures that were infected with K. pneumoniae (104 CFU) in a dose-dependent manner without any noticeable toxicity to the human cells.
Klebsiella pneumoniae has emerged as a common cause of serious epidemic and nosocomial infections in hospitals, resulting in high morbidity and mortality (2). K. pneumoniae infections occur in almost all age groups, with urinary and respiratory tract infections being most commonly encountered. K. pneumoniae accounts for a significant proportion of hospital-acquired pneumonia, bacteremia, meningitis, septicemia, and soft tissue infections. Klebsiella neonatal infections are becoming a major concern of pediatricians, as septicemia and meningitis in newborns are now often caused by multidrug-resistant strains.
In addition to host factors, bacterial virulence determinants, such as production of capsular polysaccharides and the aerobactin-mediated iron uptake system contribute to the outcome of K. pneumoniae infections (20). K. pneumoniae is resistant to penicillins and expanded-spectrum cephalosporins through the production of β-lactamases that are encoded mainly by the blaSHV, blaTEM, and ampC genes. β-lactams such as imipenem and meropenem, which are highly resistant to hydrolysis by TEM, SHV, and AmpC β-lactamases remain effective antibiotic options (18). However, the emergence of carbapenem-resistant K. pneumoniae strains will have a serious impact on remaining therapeutic options (22). β-Lactamase-producing strains are clinically significant, as they are difficult to treat. Furthermore, these strains often acquire additional mechanisms of resistance, such as mutations in the gyrA gene and/or expression of efflux pumps (13).
The emergence of antibiotic-resistant bacteria and the slow progress in identifying new classes of antimicrobial agents call for research that will uncover novel therapeutic strategies. Peptide nucleic acid (PNA) is designed to incorporate the predictable recognition properties of nucleic acids and the chemical stability of peptides. The backbone of PNA carries 2-aminoethyl-glycine linkages in place of the regular phosphodiester backbone of DNA, and the nucleotide bases are connected to this backbone at the amino nitrogens through a methylene carbonyl linker (3, 16). PNA oligomers are found to form exceptionally strong complexes with complementary strands of DNA or RNA (4, 5). In vitro studies indicate that PNA could inhibit both transcription and translation of genes to which it has been targeted and holds promise as an antigene or antisense therapy (8, 15). However, as with other high-molecular-weight drugs, the delivery of PNA appears to be a general problem. The lipid bilayer, lipopolysaccharide, and peptidoglycan of the gram-negative bacteria act as major barriers to the entry of PNAs. When PNAs were conjugated with a cell wall-permeabilizing peptide such as KFFKFFKFFK, their target-specific antisense effects were found to improve (7, 9, 21).
Sequence-specific PNAs can bind to complementary template DNA or mRNA and give rise to gene-specific silencing through blocking transcription or translation processes (17). In this study, we demonstrated that antisense peptide-PNAs targeted at genes essential for growth carried in the chromosomal DNA were able to inhibit the growth of β-lactamase-producing K. pneumoniae.
MATERIALS AND METHODS
K. pneumoniae cell preparation.
The β-lactamase-producing K. pneumoniae strain obtained from a hospital was grown in Muller-Hinton broth (MHB) or Luria-Bertani (LB) broth at 37°C on a shaker overnight. The β-lactamase-producing K. pneumoniae strain used in this study was resistant to amikacin, ampicillin, aztreonam, ceftazidime, ceftriaxone, cefuroxime, ciprofloxacin, tazobactam, cotrimoxazole, levofloxacin, and minocycline. This strain contained TEM-1, SHV-11, and AmpC type (DHA-I) β-lactamases that were confirmed by phenotypic tests and sequence analysis of the respective β-lactamase genes.
Inhibition of bacterial growth.
PNAs were synthesized by Eurogentec and listed in Table 1. Overnight grown β-lactamase-producing K. pneumoniae cell cultures were diluted to ∼105 CFU/ml in 100 μl MHB in a low-binding 96-well microtiter plate (Costar 7424), and antisense peptide-PNA, antisense PNA, or mismatched peptide-PNA were each added to a final concentration of 0, 2, 10, 20, 40, or 80 μM in the well. Microtiter plates were incubated at 37°C in a GENios spectrophotometer (TECAN Austria GmbH) which was set to shake the plate for 5 s at 3-min intervals for 12 h. Turbidities of the cultures were recorded at 550 nm every 3 min. Viable cell counts were determined at different time intervals on LB agar plates in triplicates. The plates were then incubated overnight at 37°C, and the colonies were enumerated by visual inspection.
TABLE 1.
Antisense PNAs used to investigate K. pneumoniae gene expression and growth
| PNA type | PNA designationa | Sequenceb |
|---|---|---|
| Anti-reporter gene peptide-PNAs | Peptide-Fluor | Fluorescein-H(KFF)3K-L-NH2 |
| Peptide-anti-lacZ | H(KFF)3K-L-CATAGCTGTTTC-NH2 | |
| Mismatched-anti-lacZ | H(KFF)3K-L-ACATGTCGTCTT-NH2 | |
| Anti-essential gene peptide-PNAs or PNAs | ||
| Anti-gyrA gene | Anti-gyrA | H-GCCATCTCGGACATC-Lys* |
| Peptide-anti-gyrA | H(KFF)3K-L-GCCATCTCGGACATC-NH2 | |
| Mismatched-anti-gyrA | H(KFF)3K-L-GCCATGTCGGAGATC-NH2 | |
| Anti-ompA gene | Anti-ompA | H-ATACCAGGTGTTATCT-Lys* |
| Peptide-anti-ompA | H(KFF)3K-L-ATACCAGGTGTTATCT-NH2 | |
| Peptide-anti-ompA-Fluor | Fluorescein-H(KFF)3K-L-ATACCAGGTGTTATCT-NH2 | |
| Mismatched-anti-ompA | H(KFF)3K-L-ATACCACGTGATATCT-NH2 |
Anti, antisense PNAs (complementary oligonucleotide sequence).
The PNAs are written from their amino to carboxy termini. All PNA sequences except two (*) were attached to a common peptide sequence KFFKFFKFFK.
β-Galactosidase activity assay.
β-Galactosidase activity in liquid cultures was measured by using the chromogenic LacZ substrate o-nitrophenyl-β-d-galactopyranoside (ONPG), as described by Miller (14).
Nitrocefin outer cell barrier permeabilization assay.
Outer membrane permeability was determined as described previously (1). Briefly, the β-lactamase-producing K. pneumoniae strain was prepared as described above. Permeabilization assays were carried out using 96-well microtiter plates with wells containing 100 μl of 5 mM HEPES (pH 7.4), 5 mM carbonyl cyanide m-chlorophenylhydrazone, and 20 μg/ml nitrocefin, and cells were added to reach an absorbance reading of 0.1 at 550 nm. Following the addition of the permeabilizing compounds at various concentrations, ranging from 0.1 μg to 1 μg, nitrocefin cleavage was monitored by absorption measurements at 500 nm. In view of future therapeutic applications, lower concentrations of PNAs were tested to prevent toxicity to cells. Nitrocefin generally diffuses into the cell at a rate similar to other β-lactams, and it can be cleaved by β-lactamase localized within the periplasmic space. Cleavage of nitrocefin results in a color change from yellow to red, and this can be used to monitor outer membrane permeabilization.
Cell cultures.
Normal human epithelial fibroblasts (IMR90) were cultured routinely in minimal essential medium (MEM; GIBCO, Invitrogen) supplemented with 10% heat-inactivated (30 min, 56°C) fetal calf serum (Seromed, Berlin, Germany), 1% l-glutamine (Boehringer Mannheim, Germany), 1% nonessential amino acids (Boehringer Mannheim, Germany), 10,000 U of penicillin/ml, and 10 mg of streptomycin/ml and in an atmosphere of 5% CO2. IMR90 cells were used for all assays. In some experiments, the exact number of IMR90 cells per well was determined by detaching the cells using trypsin-EDTA treatment and counting them in a Malassez chamber.
K. pneumoniae infection and PNA treatment.
For the infection assays, IMR90 cells were washed three times with phosphate-buffered saline and incubated for 1 to 6 h or overnight at 37°C in 5% CO2 with a suspension of 5 × 107 bacterial cells in MEM. After incubation, wells were washed three times with phosphate-buffered saline, and any adhering bacteria were released by the addition of 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO) and quantified by plating appropriate dilutions on LB agar plates. Control wells without mammalian cells were prepared in a similar manner to quantitate nonspecific bacterial adherence to plastic. Specific adhesion to IMR90 cells was expressed as the total number of CFU minus the number of CFU adhering to wells without IMR90 cells per cm2 of well. Cytotoxicity was estimated by assessing the ability of IMR90 cells from duplicate wells to exclude trypan blue. For peptide-PNA treatment assay, IMR90 cells were suspended by trypsin-EDTA treatment, transferred to 96-well Falcon 3872 plates, and grown to approximately 30% confluence. Freshly grown K. pneumoniae cells were added at 104 CFU/ml. After 1 h of infection, antisense peptide-PNA (10, 20, or 40 μM), antisense PNA solution with a similar dose range, or an equivalent volume of water was added, and the plates were incubated overnight at 37°C in 5% CO2.
RESULTS AND DISCUSSION
Reporter gene expression.
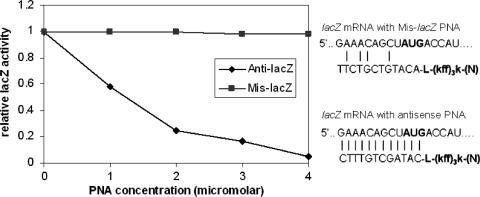
To test the antisense effects of PNA in K. pneumoniae, a reporter system based on the expression of the β-galactosidase gene (lacZ) was constructed and evaluated. For antisense inhibition, the lacZ recombinant plasmid was transferred into K. pneumoniae by conjugation, and the expression of the lacZ gene was induced with 50 μM isopropyl β-d-thiogalactopyranoside (IPTG) and assayed at A420 with the substrate ONPG (Fig. 1). An antisense peptide-PNA targeted at the β-galactosidase reporter gene was able to inhibit gene expression with sequence specificity (Fig. 1). The Mis-lacZ control designed with eight mismatched sequences showed no lacZ inhibition compared with the fully complementary construct (Anti-lacZ) of similar size and composition (Fig. 1).
FIG. 1.
LacZ expression and inhibition with antisense peptide-PNA conjugate in K. pneumoniae. The values indicate relative enzyme activities in K. pneumoniae cultures growing in MHB. For antisense inhibition, the lacZ gene in K. pneumoniae was induced with 50 μM IPTG and assayed at A420 with the substrate ONPG. Anti-lacZ, antisense peptide-PNA targeted at lacZ gene; Mis-lacZ, mismatched peptide-PNA targeted at lacZ gene.
Outer membrane permeabilization assay.
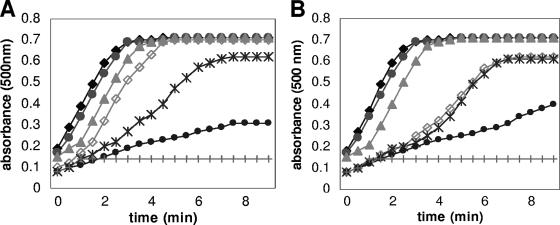
Uptake of antisense PNA alone and antisense peptide-PNA conjugates by the β-lactamase-producing K. pneumoniae strain was evaluated by the outer membrane permeabilization assay. The antisense PNA or antisense peptide-PNA against gyrA and ompA genes are noninhibitory to cell growth when used at 0.1 μM to 1 μM concentrations. Permeabilization of the outer membrane was monitored using the chromogenic reporter molecule nitrocefin (6). Free antisense PNAs targeted against gyrA and ompA genes when added to cells in the presence of nitrocefin did not permeabilize the membrane, whereas the respective antisense peptide-PNA conjugates permeabilized the outer membrane rapidly. As seen in the nitrocefin assay, membrane permeabilization was much more rapid in the presence of the antisense peptide-PNA conjugates than when antisense PNA was used alone. Permeabilization of the antisense peptide-PNA was found to be dose dependent, as reflected by the conversion of nitrocefins. When used at concentrations of 0.4 μM onwards, the rates of permeabilization of antisense peptide-PNA conjugates against gyrA and ompA through the outer membrane was exponential and reached a plateau within 3 min of addition (Fig. 2A and B, respectively). The results show that antisense PNAs when conjugated to the peptide were good membrane-permeabilizing agents.
FIG. 2.
Kinetics of outer membrane permeabilization by different concentrations of antisense peptide-PNAs. (A) Peptide-anti-gyrA PNA was included in the reactions at 0.1 μM (•), 0.2 μM (*), 0.3 μM (⋄), 0.4 μM (▴), 0.5 μM (•), and 1 μM (⧫) concentrations. Anti-gyrA PNA alone without peptide conjugation was included at 1 μM concentration (+). (B) Peptide-anti-ompA PNA was included in the reactions at 0.1 μM (•), 0.2 μM (*), 0.3 μM (⋄), 0.4 μM (▴), 0.5 μM (•), and 1 μM (⧫) concentrations. Anti-ompA PNA alone without peptide conjugation was included at 1 μM concentration (+).
Effect of growth exerted by antisense peptide-PNAs.
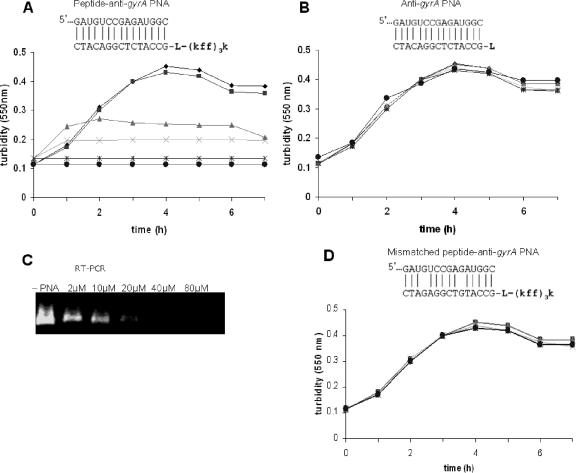
The antisense potency of the peptide-PNA targeted at the essential gyrA gene encoding the DNA gyrase subunit A was evaluated. Growth of the β-lactamase-producing K. pneumoniae was evaluated by examining the inhibitory effects of various concentrations of peptide-anti-gyrA PNA compared to treatment with antisense PNA (anti-gyrA) or with the mismatched antisense peptide-PNA (Mis-peptide-anti-gyrA PNA). Growth was totally inhibited by a 15-mer anti-gyrA PNA when it was attached to the carrier peptide (KFF)3K and added to cells at 20 μM concentration (Fig. 3A). No growth inhibition was observed in cultures treated with anti-gyrA PNA which was not conjugated to the peptide (Fig. 3B). The effect of peptide-anti-gyrA PNA on gyrA transcripts in K. pneumoniae was observed by electrophoresis of amplicons after real-time reverse transcription-PCR (RT-PCR) (Fig. 3C). A dose-dependent gradual reduction of gyrA gene expression in K. pneumoniae cells was observed, and no expression was detectable when the antisense peptide-PNA was used above 20 μM. The mismatched peptide-anti-gyrA PNA with two nucleotide differences did not inhibit cell growth (Fig. 3D).
FIG. 3.
Effect of peptide-anti-gyrA PNA on growth and gyrA gene expression of K. pneumoniae. K. pneumoniae growth was indicated by turbidity measurements at 550 nm. (A) Dose-dependent growth inhibition of K. pneumoniae by peptide-anti-gyrA PNA was observed. (B) No growth inhibition of K. pneumoniae was observed with unconjugated anti-gyrA PNA. Antisense peptide-PNA, PNA alone, or mismatched peptide-PNA was added to K. pneumoniae cultures at 2 (▪), 10 (▴), 20 (×), 40 (*), and 80 (•) μM concentrations, and compared to cultures lacking PNA (−PNA, ♦). (C) RNA was extracted from the peptide-anti-gyrA PNA-treated cultures and subjected to RT-PCR and gel electrophoresis in Tris-acetate-EDTA buffer. RT-PCR was carried out to show the effect of various doses of anti-gyrA peptide PNA on the levels of gyrA transcripts. (D) No growth inhibition of K. pneumoniae was observed with mismatched control PNA (Mismatched peptide-anti-gyrA PNA).
The primary target of fluoroquinolones in gram-negative bacteria is DNA gyrase, a type II topoisomerase required for DNA replication and transcription. DNA gyrase is a tetrameric enzyme composed of two A and two B subunits. It catalyzes the negative supercoiling of DNA and is therefore essential for maintenance of DNA topology. In the gram-negative organisms, resistance to fluoroquinolones has been shown to be associated most frequently with alterations in gyrA (12). In this study, the PNA which was designed to be complementary to the gyrA gene present in the coding strand will bind directly to the mRNA, leading to degradation of the message and subsequent inhibition of translation. The failure to synthesize DNA gyrase subunit A, which is needed for DNA replication, resulted in inhibition of growth.
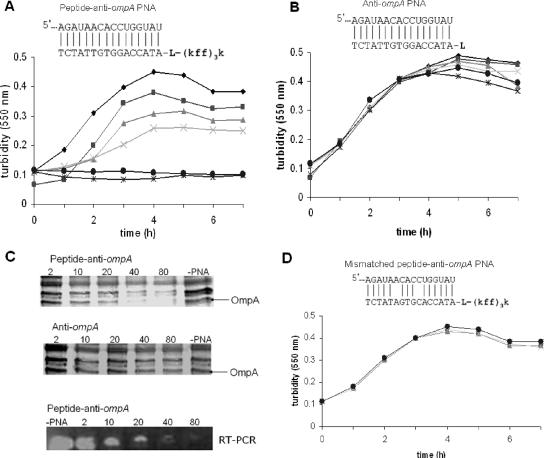
A 16-mer peptide-anti-ompA PNA conjugate targeted at the major outer membrane protein A (OmpA) prevented cell growth when it was used at a 40 μM concentration (Fig. 4A). When anti-ompA PNA was not conjugated to a peptide, growth was not inhibited and no change in OmpA protein expression was observed (Fig. 4B and C). Total inhibition of ompA gene and protein expression was observed by real-time RT-PCR and sodium dodecyl sulfate-polyacrylamide gel electrophoresis, respectively (Fig. 4C). Characterization of the OmpA protein was conducted using matrix-assisted laser desorption ionization-time of flight (mass spectrometry) analysis, and the data were compared to the NCBI database for sequence matches. The mismatched peptide-anti-ompA PNA which differed in 2 nucleotides from the antisense peptide-anti-ompA PNA was found not to be growth inhibitory (Fig. 4D). These results indicate that the (KFF)3K peptide was needed to carry the two antisense PNAs into K. pneumoniae. Such peptide-PNA conjugates open up new possibilities for anti-infective drug development. OmpA is highly represented in the bacterial cell wall, is conserved among the Enterobacteriaceae, and is involved in bacterial virulence and growth. OmpA is one of the major outer membrane proteins that assembles into the outer membrane via an N-terminal eight-transmembrane amphipathic β-barrel region, with the C-terminal region being retained in the periplasm (19). Functions attributed to OmpA include maintenance of structural cell integrity and a role in bacterial conjugation, bacteriophage binding, and cell growth (11). It also contributes to the ability of the gram-negative bacteria to invade mammalian cells (10).
FIG. 4.
Effect of peptide-anti-ompA PNA on K. pneumoniae growth and OmpA protein expression. K. pneumoniae growth was indicated by turbidity measurements at 550 nm. (A) Dose-dependent growth inhibition of K. pneumoniae by peptide-anti-ompA PNA. (B) No growth inhibition of K. pneumoniae was observed with unconjugated anti-ompA PNA. Peptide-PNA, PNA alone, or mismatched peptide-PNA was added to K. pneumoniae cultures at 2 (▪), 10 (▴), 20 (×), 40 (*), and 80 (•) μM concentrations and compared to cultures lacking PNA (−PNA, ♦). (C) Total protein was extracted from cells treated with the peptide-anti-ompA PNA and anti-ompA PNA alone before being subjected to one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis; RNA was extracted from the peptide-anti-ompA PNA-treated cultures and subjected to RT-PCR and gel electrophoresis in TAE buffer. (D) No growth inhibition of K. pneumoniae was observed with mismatched control PNA (Mismatched peptide-anti-ompA PNA).
Effect of peptide-PNA conjugate on cell viability.
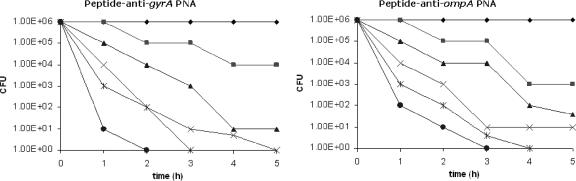
In Escherichia coli, the cationic peptide with the sequence KFFKFFKFFK was previously shown to be effective in carrying PNA (as peptide-PNA conjugate) across the membrane barrier (9). The antisense peptide-PNA conjugates targeting the gyrA and ompA genes were evaluated for their antibacterial efficacy against the multiresistant β-lactamase-producing K. pneumoniae strain in liquid culture. The bacteriostatic versus bactericidal effects of gene inhibition were studied by determining the number of CFU in cultures treated with antisense peptide-PNAs. To do this, aliquots were taken at multiple time points during antisense peptide-PNA treatment, and the number of viable cells in the sample was indicated by determining the number of CFU.
After incubation for 3 h, the numbers of viable cells in the culture were significantly reduced in the presence of the antisense peptide-PNA conjugates compared with cell cultures treated with PNA alone. After 6 h of incubation, numbers of CFU were determined by dilution and plating on LB agar. No CFU was observed when cultures were treated with antisense gyrA and ompA peptide-PNAs at concentrations above 40 μM. This inhibition was not observed in cell cultures treated with antisense mismatched PNA conjugates targeted at the respective genes (data not shown). When the respective antisense PNAs which were not conjugated to peptides were used, no bactericidal effect was observed (Fig. 5).
FIG. 5.
Bactericidal effects of antisense peptide-PNAs targeting gyrA and ompA. The number of CFU was calculated at different time points. Antisense peptide-PNAs were added to K. pneumoniae cultures at 2 (▪), 10 (▴), 20 (×), 40 (*), and 80 (•) μM concentrations and compared to the culture containing PNA alone at 80 μM (♦).
The antisense PNAs described in this paper are very promising compounds for antimicrobial development against β-lactamase-producing K. pneumoniae strains. Antisense PNAs targeted against essential genes are potential targets for new antimicrobial development and this approach enables one to find out which genes in K. pneumoniae are susceptible targets for more conventional antimicrobial development.
Treatment of K. pneumoniae-infected epithelial cells with antisense peptide-PNAs.
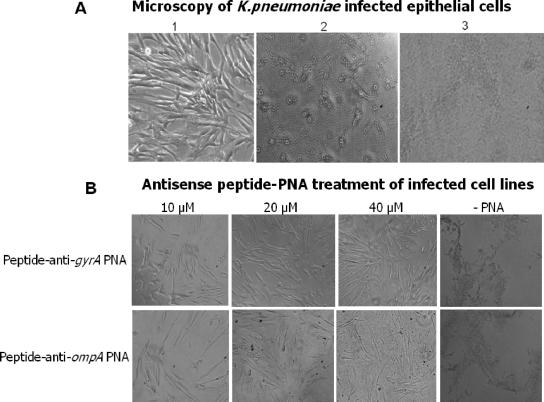
To investigate an in vivo model of K. pneumoniae infection, IMR90 monolayer cells were infected with a β-lactamase-producing K. pneumoniae strain. The morphology of the IMR90 monolayer cells was monitored. At 6 h postinfection, cells in the monolayer were observed to round up (Fig. 6A). Complete cell death was observed after overnight infection (Fig. 6A).
FIG. 6.
(A) Microscopy of epithelial cells infected with K. pneumoniae. (1) Normal healthy IMR90 cells in monolayer; (2) IMR90 cell morphology was changed (rounded cells) after 6 h of infection with bacteria; (3) after overnight infection, complete cell death was observed. (B) Antisense peptide PNA treatment of IMR90 cell lines infected with a β-lactamase-producing K. pneumoniae strain. The images show IMR90 cell cultures grown in MEM and 10% fetal calf serum. IMR90 cell cultures were treated with different concentrations of different antisense peptide-PNAs (10, 20, or 40 μM) after K. pneumoniae infection was allowed to proceed for 1 h. Complete cell death of the IMR90 monolayer was observed for the cultures not treated with PNA after overnight incubation.
To evaluate the antibacterial potential of antisense ompA and gyrA peptide-PNAs against the growth of K. pneumoniae in the presence of eukaryotic cells, IMR90 cell cultures were infected with 104 CFU of the β-lactamase-producing K. pneumoniae strain, and antisense peptide-PNAs were added 1 h postinfection. This system can be viewed as a very simple model for the growth of an extracellular pathogen in eukaryotic cells in the presence of antisense peptide-PNAs. The antisense peptide-PNA did not visibly affect IMR90 cell growth at the highest concentration tested (80 μM) (data not shown). When antisense peptide-PNAs at 20 μM or higher were added to K. pneumoniae-infected cells, no change in cell morphology was observed and the cells remained healthy after overnight incubation (Fig. 6B). No live bacteria were detected from the PNA-treated samples in an agar CFU test, confirming that these antisense peptide-PNA targets are bactericidal and serve as valuable targets for antigene therapeutic drugs against multiresistant K. pneumoniae strains. The observed antimicrobial effects of antisense peptide-PNA within a cell culture system broaden the potential scope of the approach and raise the prospects for PNA-based antibacterial drugs. However, of concern are delivery, potency, toxicity, and pharmacokinetics of antisense peptide-PNA conjugates in humans infected with antibiotic-resistant bacteria.
In conclusion, antisense peptide-PNAs targeted to essential genes of the human pathogen, a β-lactamase-producing K. pneumoniae strain, were shown to inhibit growth of the bacteria in vitro and in infected IMR90 epithelial cells. Treatment of K. pneumoniae cells with 20 μM peptide-anti-gyrA PNA was shown to be bactericidal, as no viable cells were detected after 6 h of infection. Higher concentrations of antisense peptide-PNA (at 40 μM) targeted at the ompA gene were able to inhibit K. pneumoniae growth. From this study, DNA gyrase subunit A and OmpA were established to be essential for bacterial growth and thus could serve as good anti-infective targets. Dose-dependent growth inhibition of K. pneumoniae was observed, with both of the antisense peptide-PNAs targeting the respective genes. No inhibition of growth was observed when control antisense PNAs with sequence variations or PNAs without the peptide were administered. Future experimentation with peptide-conjugated PNAs will need to be carried out with mice and primates to evaluate the potency, toxicity, and pharmacokinetics of PNAs for prevention and treatment of β-lactamase-producing K. pneumoniae infections.
Acknowledgments
This research was supported by a Microbiology Vaccine Initiative grant (no. R-182-006-067-731) awarded to C.L.P.
P.K. would like to acknowledge the receipt of a postgraduate research scholarship from the National University of Singapore.
Footnotes
Published ahead of print on 11 December 2006.
REFERENCES
- 1.Angus, B. L., A. M. Carey, D. A. Caron, A. M. Kropinski, and R. E. Hancock. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiPersio, J. R., L. M. Deshpande, D. J. Biedenbach, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2005. Evolution and dissemination of extended-spectrum β-lactamase-producing Klebsiella pneumoniae: epidemiology and molecular report from the SENTRY Antimicrobial Surveillance Program (1997-2003). Diagn. Microbiol. Infect. Dis. 51:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Egholm, M., O. Buchardt, P. E. Nielsen, and R. H. Berg. 1992. Peptide nucleic acids (PNA) oligonucleotide analogues with an achiral peptide backbone. J. Am. Chem. Soc. 114:1895-1897. [Google Scholar]
- 4.Egholm, M., P. E. Nielsen, O. Buchardt, and R. H. Berg. 1992. Recognition of guanine and adenine in DNA by thymine and cytosine containing peptide nucleic acids. J. Am. Chem. Soc. 114:9677-9678. [Google Scholar]
- 5.Egholm, M., O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, et al. 1993. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen bonding rules. Nature 365:566-568. [DOI] [PubMed] [Google Scholar]
- 6.Eriksson, M., P. E. Nielsen, and L. Good. 2002. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277:7144-7147. [DOI] [PubMed] [Google Scholar]
- 7.Good, L., and P. E. Nielsen. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355-358. [DOI] [PubMed] [Google Scholar]
- 8.Good, L., and P. E. Nielsen. 1998. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. USA 95:2073-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good, L., S. K. Awasthi, R. Dryselius, O. Larsson, and P. E. Nielsen. 2001. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360-364. [DOI] [PubMed] [Google Scholar]
- 10.Jeannin, P., et al. 2000. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat. Immunol. 1:502-509. [DOI] [PubMed] [Google Scholar]
- 11.Jeannin, P., et al. 2002. Outer membrane protein A (OmpA): a new pathogen-associated molecular pattern that interacts with antigen presenting cells-impact on vaccine strategies. Vaccine 20:A23-27. [DOI] [PubMed] [Google Scholar]
- 12.Kampranis, S. C., and A. Maxwell. 1996. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. USA 93:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Martínez, L., I. García, S. Ballesta, V. J. Benedí, S. Hernández-Allés, and A. Pascual. 1998. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 42:1850-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Nekhotiaeva, N., S. K. Awasthi, P. E. Nielsen, and L. Good. 2004. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10:652-659. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen, P. E., M. Egholm, R. H. Berg, and O. Buchardt. 1991. Sequence selective recognition of DNA by strand displacement with a thymine substituted polyamide. Science 254:1497-1500. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, P. E. 1999. Applications of peptide nucleic acids. Curr. Opin. Biotechnol. 10:71-75. [DOI] [PubMed] [Google Scholar]
- 18.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 20.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11:589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaara, M., and M. Porro. 1996. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yigit, H., A. M. Queenan, J. G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]