Abstract
We tested 102 Campylobacter jejuni and 6 Campylobacter coli clinical isolates from Poland. All were susceptible to erythromycin. Among the tested C. jejuni isolates 55.9% and 13.7% were resistant to ciprofloxacin and tetracycline, respectively. Replacement of Thr86 with Ile in GyrA and a plasmid-borne tet(O) gene were the main resistance mechanisms for fluoroquinolones and tetracycline, respectively.
Campylobacteriosis is a significant public health problem in many developed countries. Campylobacter jejuni and Campylobacter coli are leading causes of food-borne gastroenteritis and enteritis in humans (7). The interest in campylobacteriosis in Poland started very recently, and its laboratory diagnostics, followed by recording, began in 2003. In 2004, only 24 confirmed cases of campylobacteriosis were recorded in Poland, whereas in the neighboring Czech Republic and Germany there were over 25,000 and 55,000 cases reported, respectively (http://www.efsa.europa.eu/en/science/monitoring_zoonoses/reports/1290.html). The low number of isolates recorded in Poland suggests that only a small number of infections are diagnosed and recorded.
In this work we present the first results on antimicrobial susceptibility measured by MIC assay of Campylobacter spp. isolated in Poland.
(A part of this report was presented at the Med-Vet-Net 2nd General Scientific Meeting in Malta, 3 to 6 May 2006).
In our study we tested all isolates of C. jejuni (n = 102) and C. coli (n = 6) collected from human diarrheal stool samples by the regional sanitary-epidemiological units, in four different districts in Poland, and the National Institute of Hygiene in Warsaw, Poland, between 2003 and 2005. All the isolates were epidemiologically unrelated. Bloody diarrhea, fever, vomiting, and abdominal pain were reported for 61, 56, 32, and 15% of cases, respectively. Isolates from children under 6 years of age predominated (84%).
The samples were spread onto CCDA plates (Oxoid Ltd., Basingstoke, United Kingdom) and incubated under microaerobic conditions at 37°C for 48 h. The MICs of tetracycline, ciprofloxacin, nalidixic acid, erythromycin, chloramphenicol, gentamicin, and amoxicillin-clavulanic acid were determined by the E-test method (AB Biodisk, Sweden) according to the manufacturer's instructions. C. jejuni ATCC 33560 was used as a control. The breakpoints were those recommended by CLSI for C. jejuni/C. coli (5), except for nalidixic acid, chloramphenicol, gentamicin, and amoxicillin-clavulanic acid, for which the breakpoints for Enterobacteriaceae were used (4).
All the tested isolates were susceptible to erythromycin, chloramphenicol, gentamicin, and amoxicillin-clavulanic acid, while 57 C. jejuni and 4 C. coli isolates were resistant to both ciprofloxacin and nalidixic acid. For all the ciprofloxacin-resistant isolates MICs were higher than 32 μg/ml. A similar percentage (45.1%) of ciprofloxacin resistance was detected in C. jejuni from Germany (19), whereas a higher resistance rate (81%) was reported in Spain (14).
To determine the mechanism of resistance to fluoroquinolones, we isolated genomic DNA as described previously (9) and performed the gyrA-restriction fragment length polymorphism (RFLP) analysis as described by Alonso et al. (1), with primers adapted for C. jejuni (20). Results showed that all the fluoroquinolone-resistant C. jejuni isolates carried a mutation in the gyrA gene resulting in the replacement of Thr86 with Ile. This substitution is known to be responsible for high-level resistance to fluoroquinolones (1, 13, 20). The relatively high fluoroquinolone resistance rates among Campylobacter isolates are most probably caused by the broad use of this class of antibiotics in veterinary medicine (especially in poultry) (10). This hypothesis is supported by the very low (2%) frequency of ciprofloxacin-resistant clinical isolates of C. jejuni observed in Australia, where the usage of fluoroquinolones in food-producing animals is prohibited (18). In Poland, such restriction was introduced in 2006, but fluoroquinolones are still allowed in veterinary medicine.
In our study 14 (13.7%) C. jejuni and 2 C. coli isolates resistant to tetracycline were observed (Table 1). The tetracycline resistance rate in Poland is lower than those reported in Canada (Alberta) (50%) and Spain (72%) (8, 14). Since the tet(O) gene is the most commonly reported determinant conferring resistance to tetracycline in these species, we analyzed its presence and localization in the tetracycline-resistant isolates. The tet(O) gene was amplified by PCR using primers and cycling conditions described by Bacon et al. (2). The C. jejuni 81-176 strain served as the tetracycline-resistant tet(O) reference strain (2). In agreement with previous findings (2, 3, 8, 11, 12, 15, 17), the tet(O) gene was detected in all the tetracycline-resistant isolates tested. Since tet(O) was reported to occur on plasmids (17), we carried out plasmid analyses and Southern hybridization with a tet(O) probe. Plasmids were extracted by the alkaline lysis method (16) with modification (15) or by a plasmid kit (A&A Biotechnology, Poland). The sizes of plasmids were calculated on the basis of the sum of fragment sizes obtained after ClaI (Bsu15I; Fermentas, Lithuania) digestions. Plasmids from isolates harboring multiple plasmids were compared to plasmids of known sizes harbored by E. coli V517. Southern blot analysis was carried out with DIG High Prime DNA labeling and detection starter kit I (Roche Diagnostics Gmbh, Germany) according to the manufacturer's instructions. Transfer of DNA to a nylon membrane (Serva, Germany) was performed as described by Sambrook et al. (16). A plasmid containing the tet(O) gene from the tetracycline-resistant strain C. jejuni 81-176 was used as a positive control, and bacteriophage lambda digested with HindIII was used as a negative control.
TABLE 1.
Characteristics of the C. jejuni and C. coli tetracycline-resistant strains
| Species and isolate | Tca MIC (μg/ml) | Plasmid size(s) (kb) | tet(O) on plasmidb | ClaI plasmid profile | Conjugation assay resultc |
|---|---|---|---|---|---|
| C. jejuni 47/05 | 128 | 38 | + | I | 8 × 10−6 |
| C. jejuni 44/05 | 256 | 38 | + | I | 3 × 10−7 |
| C. jejuni 23/05 | 256 | 38 | + | I | 10−8 |
| C. jejuni 45/03 | 32 | 38 | + | I | 8 × 10−6 |
| C. coli 202/04 | 64 | 38 | + | I | 1.8 × 10−6 |
| C. jejuni 42/05 | 64 | 38 | + | I | 2 × 10−8 |
| C. jejuni 90/04 | 64 | 40 | + | II | 2 × 10−6 |
| C. coli 317/04 | 64 | 43 | + | III | 1.2 × 10−7 |
| C. jejuni 75/04 | 64 | 46, 10, 5 | +e | IV | Kmr |
| C. jejuni 24/05 | 64 | 46 | + | IV | NC |
| C. jejuni 79/04 | 128 | 38 | + | V | Kmr |
| C. jejuni 229/04 | 64 | 38 | + | V | NC |
| C. jejuni 32/05 | 128 | 38 | + | VI | NC |
| C. jejuni 62/05 | 64 | 34 | + | VII | NC |
| C. jejuni 53/05 | 64 | 46 | + | VIII | 6 × 10−6 |
| C. jejuni 39/05 | 64 | NAd | NA | NA | NA |
Tc, tetracycline.
Determined by Southern blot assay.
Transfer frequency was calculated as the number of transconjugants per donor. Kmr, resistant to kanamycin; NC, no conjugation.
NA, not applicable.
On 46-kb plasmid.
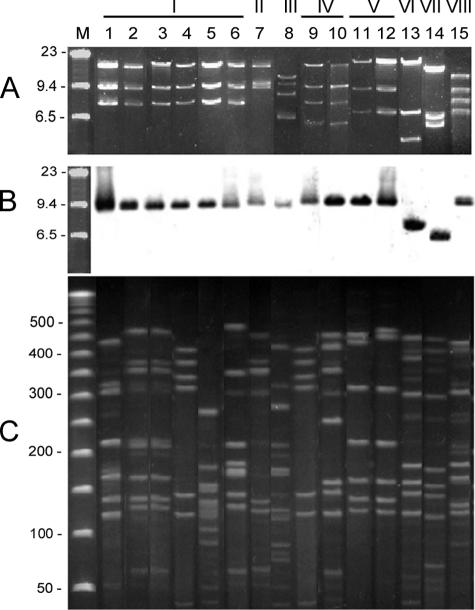
All the tetracycline-resistant isolates except a plasmidless one (39/05) harbored a large plasmid from 34 to 45 kb in size (Table 1; Fig. 1A) that carried the tet(O) gene as shown by the Southern hybridization assay (Fig. 1B). When compared to results by Lee et al. (11) and Gibreel et al. (8), who found that 47% and 67%, respectively, of tetracycline-resistant clinical isolates harbored a plasmid-borne tet(O) gene, our study revealed a surprisingly high frequency (94%) of plasmid-mediated tet(O) in Polish isolates of C. jejuni. To check the genetic diversity of the tet(O) plasmids, we performed ClaI-RFLP. Eight different profiles were noted, indicating heterogeneity of these plasmids. Nevertheless, five C. jejuni plasmids and one C. coli plasmid revealed the same predominating profile, I (Fig. 1A). These findings suggested that these isolates harbored the same horizontally transferred plasmid. To exclude a clonal dissemination of these plasmids, the tet(O)-positive isolates were genotyped by pulsed-field gel electrophoresis (PFGE) with SmaI according to the methodology described on the Campynet website (http://campynet.vetinst.dk/PFGE.html) (Fig. 1C). With the exception of isolates 44/05 and 23/05, no clonal structure was detected for isolates carrying plasmids of RFLP type I. Interestingly, isolates 79/04 and 229/04, carrying plasmids of RFLP type V, were closely related. However, we were unable to trace any direct epidemiological link between isolates 44/05 and 23/05 as well 79/04 and 229/04.
FIG. 1.
Results for the isolates harboring plasmids carrying the tet(O) gene. Lanes: 1, 47/05; 2, 44/05; 3, 23/05; 4, 45/03; 5, 202/04; 6, 42/05; 7, 90/04; 8, 317/04; 9, 75/04; 10, 24/05; 11, 79/04; 12, 229/04; 13, 32/05; 14, 62/05; 15, 53/05. (A) Patterns of the tet(O) gene-harboring plasmids digested by ClaI; (B) Southern hybridization of the tet(O) probe. Lane M, bacteriophage λ DNA HindIII molecular size marker (Sigma, Germany). (C) PFGE patterns of the respective isolates. Lane M, 50- to 1,000-kb pulse marker (Sigma, Germany). The size of DNA is shown in kilobases. The picture was electronically edited to support the lane compatibility in the all three parts of the picture.
Since tet(O) was reported to be often carried on conjugative plasmids (3, 8, 12, 15, 17), we carried out a conjugation assay as described by Taylor et al. (17) with modifications described by Pratt and Korolic (15). Streptomycin-resistant C. jejuni clinical isolates 367/04 and 375/04, obtained at the National Institute of Hygiene in Warsaw, Poland, and C. jejuni 81-116 mutant R1 strain with a kanamycin resistance gene inserted into flaA (21) were used as the recipient strains in conjugation experiments. As a control donor strain we used C. jejuni 81-176. The frequency of control conjugative transfer of tetracycline resistance between the C. jejuni 81-176 donor strain and the C. jejuni 81-116R1 recipient strain was 6 × 10−6. Conjugative transfer of tetracycline resistance was detected with 9 of 15 plasmid-possessing isolates (Table 1). It is noteworthy that all plasmids of the aforementioned ClaI-RFLP type I were conjugative, while generally being carried by genetically unrelated isolates (Fig. 1). We also observed differences in conjugation transfer ability of plasmids depending on the recipient strains. With C. jejuni 367/04 (Strr) or C. jejuni 375/04 (Strr) as recipients, conjugational transfer of tetracycline resistance was observed only from C. jejuni isolates 90/04, 42/05, 44/05, and 47/05 and frequency of transfer was much lower (<10−8). These differences might be explained by the presence of restriction-modification systems in Campylobacter spp. (6) that are capable of decreasing the efficiency of acquisition of DNA from other strains.
Taken together, results of the RFLP and PFGE experiments suggest that spread of tetracycline resistance mediated by tet(O) is mostly related to horizontal transfer of the resistance gene via conjugative plasmids rather than to the dissemination of specific clones in Poland.
Since 87% of the tetracycline-resistant isolates were obtained from children, for whom tetracycline administration is strictly limited, we may suppose that our results reflect the selection of resistant strains in food-producing animals, which are considered the main source of campylobacteriosis. To our knowledge, the usage of tetracyclines in animal medicine is relatively low in Poland in comparison to fluoroquinolones. Thus we may expect low selective pressure for the dissemination of the tet(O)-harboring plasmids in animal isolates in Poland.
Acknowledgments
This work was partially financed by Med-Vet-Net, CAMPYNET WPs 8 and 30.
We are grateful to G. Madajczak for kind help and advice. We thank E. K. Jagusztyn-Krynicka (University of Warsaw, Poland) and G. I. Perez-Perez (New York University School of Medicine) for providing the C. jejuni 81-176 strain and A. M. Ridley (VLA, Weybridge, United Kingdom) for providing the C. jejuni 81-116R1 strain.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Alonso, R., E. Mateo, C. Girban, E. Churruca, I. Martinez, and A. Fernández-Astorga. 2004. PCR-restriction fragment length polymorphism assay for detection of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli. Antimicrob. Agents Chemother. 48:4886-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor, R. A., B. M. Pearson, L. M. Friss, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. Informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Clinical and Laboratory Standards Institute. 2006. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Approved guideline M45-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Edmonds, P., B. Hall, W. Edwards, and K. M. Hartline. 1992. Presence of methylated adenine in GATC sequences in chromosomal DNAs from Campylobacter species. J. Bacteriol. 174:8156-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 8.Gibreel, A., D. M. Tracz, L. Nonaka, T. M. Ngo, S. R. Connell, and D. E. Taylor. 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48:3442-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gierczyński, R., S. Kałużewski, A. Rakin, M. Jagielski, A. Zasada, A. Jakubczak, B. Borkowska-Opacka, and W. Rastawicki. 2004. Intriguing diversity of Bacillus anthracis in eastern Poland—the molecular echoes of the past outbreaks. FEMS Microbiol. Lett. 239:235-240. [DOI] [PubMed] [Google Scholar]
- 10.Iovine, N. M., and M. J. Blaser. 2004. Antibiotics in animal feed and spread of resistant Campylobacter from poultry to humans. Emerg. Infect. Dis. 10:1158-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, C.-Y., C.-L. Tai, S.-C. Lin, and Y.-T. Chen. 1994. Occurrence of plasmids and tetracycline resistance among Campylobacter jejuni and Campylobacter coli isolated from whole market chickens and clinical samples. Int. J. Food Microbiol. 24:161-170. [DOI] [PubMed] [Google Scholar]
- 12.Nirdnoy, W., C. J. Mason, and P. Guerry. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piddock, L. J., V. Ricci, L. Pumbwe, M. J. Everett, and D. J. Griggs. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19-26. [DOI] [PubMed] [Google Scholar]
- 14.Prats, G., B. Mirelis, T. Llovet, C. Munoz, E. Miró, and F. Navarro. 2000. Antibiotic resistance trends in enteropathogenic bacteria isolated in 1985-1987 and 1995-1998 in Barcelona. Antimicrob. Agents Chemother. 44:1140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratt, A., and V. Korolic. 2005. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55:452-460. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Taylor, D. E., M. A. DeGrandis, D. A. Karmali, and P. C. Fleming. 1981. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 19:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unicomb, L. E., J. Ferguson, R. J. Stafford, R. Ashbolt, M. D. Kirk, N. G. Becker, M. S. Patel, G. L. Gilbert, M. Valcanis, and L. Mickan. 2006. Low level fluoroquinolone resistance among Campylobacter jejuni isolates in Australia. Clin. Infect. Dis. 42:1368-1374. [DOI] [PubMed] [Google Scholar]
- 19.Wagner, J., M. Jabbusch, M. Eisenblätter, H. Hahn, C. Wendt, and R. Ignatius. 2003. Susceptibilities of Campylobacter jejuni isolates from Germany to ciprofloxacin, moxifloxacin, erythromycin, clindamycin, and tetracycline. Antimicrob. Agents Chemother. 47:2358-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wardak, S., J. Szych, and A. Cieślik. 2005. Detection of mutation in the gyrA gene at position 86-Thr associated with fluoroquinolone resistance in Campylobacter jejuni using PCR-restriction fragment length polymorphism assay. Med. Dosw. Mikrobiol. 57:295-301. [PubMed] [Google Scholar]
- 21.Wassenaar, T. M., N. M. C. Bleumink-Pluym, and B. A. M. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]