Abstract
Food-borne pathogens are responsible for most cases of food poisoning in developed countries and are often associated with poultry products, including chicken. Little is known about the role of β-defensins in the chicken digestive tract and their efficacy. In this study, the expression of chicken β-defensin gallinacin-6 (Gal-6) and its antimicrobial activity against food-borne pathogens were investigated. Reverse transcription-PCR analysis showed high expression of Gal-6 mRNA in the esophagus and crop, moderate expression in the glandular stomach, and low expression throughout the intestinal tract. Putative transcription factor binding sites for nuclear factor kappa beta, activator protein 1, and nuclear factor interleukin-6 were found in the Gal-6 gene upstream region, which suggests a possible inducible nature of the Gal-6 gene. In colony-counting assays, strong bactericidal and fungicidal activity was observed, including bactericidal activity against food-borne pathogens Campylobacter jejuni, Salmonella enterica serovar Typhimurium, Clostridium perfringens, and Escherichia coli. Treatment with 16 μg/ml synthetic Gal-6 resulted in a 3 log unit reduction in Clostridium perfringens survival within 60 min, indicating fast killing kinetics. Transmission electron microscopy examination of synthetic-Gal-6-treated Clostridium perfringens cells showed dose-dependent changes in morphology after 30 min, including intracellular granulation, cytoplasm retraction, irregular septum formation in dividing cells, and cell lysis. The high expression in the proximal digestive tract and broad antimicrobial activity suggest that chicken β-defensin gallinacin-6 plays an important role in chicken innate host defense.
Since the 1950s, subtherapeutic doses of antibiotics were added to feed to promote growth in chicken broilers (45). Increasing concern about the development of antibiotic resistance and prevalence of its transmission to human pathogens has led to a European ban on the use of antibiotics in animal feeds to promote growth. Hence, alternative methods to suppress microbial outgrowth in poultry are needed. An alternative strategy could be to stimulate the expression of endogenous antimicrobial proteins at mucosal surfaces of the chicken digestive tract by dietary modulation.
Currently, several chicken antimicrobial peptides, belonging to the cathelicidin, liver-expressed antimicrobial peptide (LEAP), and β-defensin families have been discovered (24, 39, 41, 46), but little is known about their roles in the chicken digestive tract.
Chicken cathelicidin 1 is expressed at moderate levels in the gizzard, small intestine, and large intestine (24), whereas low levels of chicken myeloid antimicrobial peptide 27 were found throughout the intestinal tract (41). High levels of chicken LEAP-2 expression were observed in the small intestine and liver (24) and upregulated in these tissues when challenged with Salmonella enterica serovar Enteritidis (39). Apart from gallinacin 11 (Gal-11), which is highly expressed in the small intestine, liver, gallbladder, and spleen, and Gal-13, which is found in colon, no significant β-defensin levels have been detected in the digestive tract (18, 24, 46).
Little is known about the antimicrobial properties of antimicrobial peptides in the chicken digestive tract. Recombinant chicken LEAP-2 was effective at microgram amounts against Salmonella enterica serovar Typhimurium SL1344, but not against Salmonella enterica serovar Typhimurium C5 and Salmonella enterica serovar Enteritidis (39). Chicken myeloid β-defensins gallinacin 1, 1α, and 2, isolated from chicken heterophils, showed activity against gram-positive, gram-negative bacteria and yeast (10). Although Salmonella enterica serovar Typhimurium and Listeria monocytogenes were inhibited by synthetic Gal-11, complete killing was only achieved at 500 μg/ml (18), suggesting a different role in the chicken gut.
Here we report the expression of β-defensin gallinacin-6 in the chicken digestive tract. The antimicrobial properties of synthetic and recombinant gallinacin-6 peptides were tested against gram-positive and gram-negative bacteria and yeasts using colony-counting and broth microdilution assays. Additionally, kill-curve studies and transmission electron microscopy were used to investigate the killing mechanism(s) involved.
MATERIALS AND METHODS
Tissue distribution of gallinacin-6.
Three healthy 6-week-old Ross 308 broiler chickens were sacrificed, and 23 tissue samples were taken within 30 min for RNA isolation. Samples were rinsed with cold, sterile saline, frozen immediately in liquid nitrogen, and stored at −80°C. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol, diluted to 2 μg RNA/μl, pooled per tissue, and DNase I treated (Invitrogen) before first-strand cDNA synthesis. To investigate the variability of expression in the upper part of the digestive tract, crop tissue samples were also collected from 13-day-old (n = 9) and 85-week-old (n = 5) Ross 308 chickens and 21-day-old Hybro chicken broilers (n = 3) and individually analyzed.
The tentative consensus sequence for chicken gallinacin-6 was retrieved from The Institute for Genomic Research chicken expressed sequence tag database (http://compbio.dfci.harvard.edu/tgi/; TC82510). Primer sequences were designed corresponding to the regions flanking the Gal-6 start and stop codon (Gal-6 forward and reverse primers) (Table 1). The β-actin forward and reverse primers were used to assess the quality and quantity of the chicken mRNA samples. All PCRs were performed with Faststart DNA Taq polymerase (Roche Diagnostics Gmbh, Mannheim, Germany) as follows: after an initial denaturing step of 5 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at 53°C, and 45 s at 72°C for Gal-6, and 33 cycles of 30 s at 95°C, 30 s at 61°C, and 45 s at 72°C for β-actin, followed by a final elongation step at 72°C for 7 min.
TABLE 1.
Vector construction, recombinant peptide, and RT-PCR primers for gallinacin-6
| Primer type and name | Primer sequence (5′-3′) |
|---|---|
| RT-PCRa primers | |
| Gal-6 (forward) | TCCACAGCAGAGGACAATC |
| Gal-6 (reverse) | AACTGCGTGGTCAGTGAGG |
| β-Actin (forward) | ACCCTGTCCTGCTTACTGAGG |
| β-Actin (reverse) | TCCCAATGGTGATCACCTGCC |
| Protein expression vector construction primers | |
| Oligo-1 (forward) | GTTTAAACCCGGGCCGCCACCATGGGATCCCCTCCTTCACTCGGACACACACC |
| Oligo-1 (reverse) | GAGATCTGCCCGGGCTATGCGGCCGCGCTAGCTATTTTGATGAAATGAAGAGTTTCGCCAAGGCTTTC |
| Oligo-2 (forward) | GTTTAAACCAAGGCCCAACTCCCCGAACCACTC |
| Oligo-2 (reverse) | CTGGATCAGAATTCAAGCATGCCCGCGGG |
| Oligo-3 (forward) | CGGAGATCTAAGCTTGGCTGTGGAATGTGTGTCAGTTAG |
| Oligo-3 (reverse) | GTTTAAACGAGCTTGGATCTGTAACGGCGCAG |
| Recombinant Gal-6 primers | |
| rGal-6 (forward) | GGATCCACCTTAGCATGCAGGCAG |
| rGal-6 (reverse) | GCGGCCGCTTAGGAGCTAGGTGCCCAT |
RT, reverse transcription.
Promoter analysis.
The published Gal-6 gene cDNA sequence (National Center for Biotechnology Information [GenBank], http://www.ncbi.nlm.nih.gov, AY621324), containing a 4,480-bp 5′ flanking sequence was investigated for the presence of putative transcription factor binding sites using the JASPAR (http://jaspar.cgb.ki.se/cgi-bin/jaspar_db.pl) and MatInspector software. A putative transcription start site (TSS) was predicted using the neural network promoter prediction software (Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/promoter.html).
Prediction of mature gallinacin-6 peptide sequence.
A protein Basic Local Alignment Search Tool (pBLAST) search (http://www.ncbi.nlm.nih.gov/BLAST/) using the Gal-6 precursor amino acid sequence was performed to investigate similarities with other β-defensins. The prepropeptide amino acid sequence of chicken gallinacin-6 was aligned with other avian and mammalian β-defensins using Clustal X (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). Signal peptide sequences were predicted using SignalP software (http://www.cbs.dtu.dk/services/SignalP/, version 3.0). The mature Gal-6 peptide sequence was deduced from comparison of the propiece sequences and cleavage sites of known mature β-defensins. Precursor and known mature peptide sequences were retrieved from the GenBank or EMBL Nucleotide Sequence Submission (EMBL; http://www.ebi.ac.uk) database. GenBank/EMBL accession numbers are as follows: mgBD-1, AAG09213; Gal-3, Q9DG58; Sphe-1, P83429; Gal-8, AAS99319; Gal-6, AAT48933; ptBD-1, AAK61462; hBD-1, AAC51728; hcBD-1, AAK61464; pBD-2, AAR90346; mBD-37, CAD33899; SBD-2, AAB61996; chBD-1, O97946; bbEBD, AAP57565; LAP, AAB33727; TAP, AAB61757; btEBD, AAC48804; and BNBD-1, AAB25864. The ostricacin 1 amino acid sequence was retrieved from the original citation (48).
Peptide synthesis.
A 41-amino-acid-residue peptide corresponding to the deduced mature Gal-6 peptide was synthesized by Genosphere Biotechnologies (Paris, France) using Fmoc solid-phase synthesis on a Symphony synthesizer (Protein Technology Inc., Tucson, AZ). Disulfide bridge formation was accomplished by the air-oxidation method as described by Hidaka et al. (17). Synthesized Gal-6 was dissolved in 100 mM Tris-HCl (pH 8.0) in the presence of reduced and oxidized glutathione (2 mM glutathione, 1 mM glutathione disulfide) and incubated under an N2 atmosphere at room temperature for 3 days. The folded Gal-6 peptide was purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Zorbax C8 column (Agilent Technologies, Palo Alto, CA) eluted with a 20-min linear gradient of 0 to 100% acetonitrile in 0.1% (wt/vol) trifluoroacetic acid. Finally, the RP-HPLC-purified folded peptide was dissolved in Milli Q and characterized by mass spectrometry, amino acid analysis, and electrophoresis on Tris-Tricine polyacrylamide gel electrophoresis (PAGE) gels.
Protein expression vector construction.
Plasmids pTT3 (8), pSGHV(0) (21), and pNUT-VWFcas (40) were used as templates for the construction of the pTT3-SRα-hGH-his8-TEV secreted protein expression vector. PCR fragment 1, containing the BamHI-NheI-NotI multiple cloning site, was generated using oligo 1 forward and reverse primers (Table 1) and plasmid pNUT-VWFcas as a template. PCR fragment 2, containing the human growth hormone gene (HGH), a His8 tag, and a tobacco etch virus (TEV) protease cleavage site, was generated using oligo 2 forward and reverse primers and plasmid pSGHV0 as a template. PCR fragment 3, containing the SRα promoter, was generated using oligo 3 forward and reverse primers and plasmid pSGHV0 as a template. The empty expression plasmid was constructed as follows. PmeI-BglII-digested PCR fragment 1 was ligated into PmeI-BamHI-digested pTT3, yielding pTT3a. PmeI-BamHI-digested PCR fragment 2 was ligated into PmeI-BamHI-digested pTT3a, yielding pTT3b. BglII (blunted)-PmeI-digested PCR fragment 3 was ligated into SalI (blunted)-PmeI-digested pTT3b, yielding pTT3-SRα-hGH-his8-TEV.
Production of rGal-6.
Prepro-Gal-6 cDNA was produced from chicken liver mRNA as described above. The sequence coding for the putative mature peptide was amplified using the recombinant Gal-6 (rGal-6) forward and reverse primers (Table 1). PCRs were performed using the following cycling protocol: 5 min at 94°C, 40 cycles of 94, 58, and 72°C for 30, 30, and 45 s. The PCR oligonucleotides introduced a BamHI site immediately for the codon coding for the N-terminal threonine residue of the mature peptide and a NotI restriction site after the stop codon. The amplified construct was cloned in the pGEM-T-Easy vector (Promega Co., Madison, WI) and subsequently sequenced in both directions to confirm that it contained no errors. The construct was digested using BamHI and NotI and ligated into digested pTT3-SRα-hGH-his8-TEV expression vector. HEK293-EBNA cells (ATCC CRL10852) were grown in 90% FreeStyle (Invitrogen) and 10% Ca2+ free Dulbecco's modified Eagle's medium (Invitrogen) containing 5% fetal calf serum (Invitrogen), 1% pluronic (Sigma-Aldrich, St. Louis, MO), 10 mM HEPES, 4 mM l-glutamine, 200 U/liter penicillin G, 0.1 mg/liter streptomycin, and 50 μg/ml Geneticin. Cells were maintained in exponential growth using Erlenmeyer flasks at 120 rpm on an orbital shaker mounted in a Reach-In CO2 incubator (Clean Air Techniek, Woerden, The Netherlands). HEK293-EBNA cells were transfected using DNA-polyethyleneimine (PEI; Polysciences, Warrington, PA) according to the method of Durocher et al. (8). Briefly, 24 h before transfection, cells were seeded at 2.5 × 105/ml in medium without fetal calf serum by dilution. The next day, DNA-PEI complexes were formed by a 10-min incubation of DNA at 20 μg/ml with PEI at 40 μg/ml in Optimem (Invitrogen); 25 μl of this mixture was used for each ml of cell culture to be transfected. Small-scale transfections (4 ml) were performed in six-well plates; large-scale transfections were performed in a Bioreactor (New Brunswick Scientific).
Purification of recombinant Gal-6.
The supernatant of transfected HEK293-EBNA cells was collected after 6 days and concentrated using a hollow fiber column (molecular mass cutoff, 10,000 Da; Amersham Biosciences, Uppsala, Sweden). Purification of the fusion protein HGH-His8-TEV-rGAL-6 was performed by affinity chromatography (Histrap HP, 1 ml; Amersham Biosciences). The supernatant was applied to the column, which was subsequently washed three times with buffer A (25 mM Tris, 300 mM NaCl, pH 8.2) containing 0, 20, and 50 mM imidazole, respectively. Bound protein was eluted with 250 mM imidazole in buffer A, collected, and dialyzed overnight (molecular mass cutoff, 1,000 Da; Spectrum Laboratories Inc., Rancho Domigues, CA) against 25 mM Tris-HCl, 150 mM NaCl, pH 8.2. After dialysis, the fusion protein was cleaved by TEV proteolysis for 2 h at 30°C (fusion protein:TEV = 10:1 mol/mol) and reapplied to the affinity column. The cleaved mature rGal-6 eluted at 50 mM imidazole in buffer A and was dialyzed against 25 mM ammonium formate. The purified protein was lyophilized and resuspended in 10 mM sodium phosphate buffer, pH 7.0, for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis and amino acid analysis.
Anti-His immunostaining of purified recombinant Gal-6.
Protein samples were analyzed by one-dimensional Tricine-sodium dodecyl sulfate (SDS)-PAGE (32), and protein bands were visualized by silver staining (Bio-Rad Laboratories, Richmond, CA). For immunoblot analysis, the proteins were transferred electrophoretically from the gels onto nitrocellulose membranes. Immunostaining was performed using mouse monoclonal anti-His6 (Roche) as primary antibodies and goat anti-mouse immunoglobulin G containing a peroxidase conjugate as the second antibody. Visualization was achieved using 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich).
Characterization of recombinant Gal-6.
Protein concentration and amino acid composition were determined by quantitative amino acid analysis on an HP 1090 Aminoquant, using a two-step precolumn derivatization with o-phthalaldehyde-3-mercaptopropionic acid for primary amino acids and 9-fluorenylmethylchloroformate for secondary amino acids (33). N-terminal sequence analysis was performed using an Applied Biosystems-Perkin Elmer sequencer model 476A. MALDI-TOF (mass spectrometry) analysis was performed by Eurosequence b.v. (Groningen, The Netherlands) on a Voyager-DE PRO (Applied Biosystems, Foster City, CA) in a positive linear mode, using alpha-cyano-4-hydrocinnamic acid as the matrix (16).
Quantitative determination of sulfhydryl groups in recombinant Gal-6.
Free thiol groups in rGal-6 were determined using the standard Ellman's Test (9) with small modifications. Briefly, a stock solution containing 100 mM Tris, pH 8.0, 0.1 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) and 2.5 mM sodium acetate was prepared. To 90 μl of this stock solution, 10 μl (256 μg/ml) rGal-6 was added, and the mixture was incubated at room temperature for 5 min. After incubation, the absorbance at 412 nm was determined as a measure of free thiols. Calibration was performed using l-cysteine solutions in the range of 0 to 100 μM.
Antimicrobial activity.
Bacillus cereus ATCC 9193, Campylobacter jejuni ATCC 33291, Candida albicans ATCC 10231, Clostridium perfringens ATCC 12915, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Saccharomyces cerevisiae CBS2978, Salmonella enterica serovar Typhimurium ATCC 14028, Staphylococcus aureus ATCC 29213, and Streptococcus pyogenes ATCC 19615 were used for antimicrobial assays. C. albicans at 37°C and S. cerevisiae at 25°C were grown overnight in yeast maltose broth (YMB; Oxoid Limited, Hampshire, United Kingdom). C. jejuni was grown overnight (37°C, 5% CO2) to stationary phase in heart infusion broth (Biotrading Benelux b.v., Mijdrecht, The Netherlands). S. pyogenes (37°C, 5% CO2) and C. perfringens (37°C, anaerobe) were grown overnight to stationary phase in Trypticase soy broth (TSB; Oxoid Limited). All other bacteria were maintained in TSB medium at 37°C. Bacteria and C. albicans were cultured to mid-logarithmic phase by transferring 100 μl of stationary-phase suspension into TSB or YMB medium followed by incubation and shaking for 4 h at 37°C. A hundred microliters of S. cerevisiae stationary-phase culture was transferred into YMB medium, incubated, and shaken for 10 h at 25°C. Mid-logarithmic phase cultures were centrifuged for 10 min at 4°C at 900 × g, and the bacterial and yeast pellets were diluted in minimal medium (TSB or YMB medium diluted 1,000-fold in distilled water for bacterial and yeast pellets, respectively). Initial concentrations of bacteria and yeasts were determined by measuring the optical density at 620 nm. To determine cell viability, 100 μl of 10-fold serial dilutions in peptone physiological salt solution were transferred on to Trypticase soy agar (TSA; Oxoid Limited) or yeast maltose agar (YMA; Oxoid Limited) plates, and colonies were counted after 24 h of incubation. C. jejuni viability was determined by transferring the serial dilutions onto saponin agar (SA; Oxoid Limited) plates, and colonies were counted after overnight incubation at 37°C and 5% CO2. Final dilutions were prepared in minimal TSB or minimal YMB media to reach a cell density of ∼2.5 × 106 CFU/ml for synthetic mature Gal-6 (sGal-6) assays and ∼2.5 × 105 CFU/ml for rGal-6 assays.
The antimicrobial activity of sGal-6 was determined using colony-counting assays. Twenty-five microliters of bacterial or yeast culture was mixed with 25 μl of 0 to 256 μg/ml sGal-6 in polypropylene microtiter plates and preincubated for 3 h at conditions suited to the investigated strain. After a 3-h incubation period, 200 μl of minimal medium was added, further diluted 10- to 1,000-fold in minimal medium, and transferred onto TSA, SA, or YMA plates, and colonies were counted after 24 h of incubation.
To investigate the effect of increased nutrient concentration and thereby higher metabolic activity of the bacteria on sGal-6 antimicrobial activity, additional colony-counting assays were performed in 100-fold-diluted TSB minimal medium. Minimal medium containing 100-fold-diluted TSB was buffered with 10 mM sodium phosphate (pH 6.5) to prevent acidification resulting from increased bacterial metabolism. Bacterial pellets were washed in phosphate-buffered 100-fold-diluted TSB minimal medium instead of 1,000-fold-diluted TSB in water, and the antibacterial activity of sGal-6 was determined as described above.
The influence of ionic strength on sGal-6 microbicidal activity was studied in 100-fold-diluted TSB medium containing 10 mM sodium phosphate (pH 6.5) supplemented with 20 or 150 mM NaCl. Bacterial pellets, washed in phosphate-buffered 100-fold-diluted TSB minimal medium, were subdivided for further dilution in 0, 20, or 150 mM NaCl in the same medium. The antibacterial activity of sGal-6 was assessed as described above.
Additionally, MIC and minimal bactericidal concentration (MBC) of the sGal-6 and rGal-6 peptides against bacterial and yeast strains were assessed using broth microdilution assays. Bacterial and yeast suspensions were exposed to sGal-6 and rGal-6 concentrations ranging from 0 to 256 μg/ml in 50 μl of minimal medium as described for the colony-counting assays. After 3 h of preincubation, 180 μl of TSB, heart infusion broth, or YMB medium was added to each well and incubated for 24 h. The optical density of the resuspended well contents was measured at 595 nm in a 96-well plate reader (Bio-Rad Laboratories). Growth inhibition was defined as the lowest concentration of peptide that reduced growth by more than 90%. MBCs and minimal fungicidal concentrations were evaluated by plating the contents of wells without visible growth onto TSA, SA, or YMA plates and incubating them for 24 to 72 h and defined as the lowest concentration of peptide that prevented any residual colony formation.
Kill-curve studies.
Thirty microliters of 32 μg/ml rGal-6 and sGal-6 and 128 μg/ml sGal-6 (final concentrations, 16 and 64 μg/ml) were mixed with 30 μl of 2.5 × 105 CFU/ml (rGal-6) or 2.5 × 106 CFU/ml (sGal-6) C. perfringens mid-logarithmic phase culture in minimal TSB medium and anaerobically incubated at 37°C. At various time points, a 50-μl bacterial suspension aliquot was taken and diluted 50-fold in TSB medium, of which 100 μl was plated on TSA medium. The number of CFU was counted after overnight incubation at 37°C. As a negative control, the bacterial suspension was incubated with 30 μl of minimal TSB medium.
Transmission electron microscopy.
Mid-logarithmic-phase C. perfringens cells (2 × 108 CFU/ml) were treated with sGal-6 (final concentration, 0, 1.56, 6.25, 12.5, and 25 μg/ml) for 30 min at 37°C under anaerobic conditions. After treatment, bacterial pellets were prefixed in Karnovsky's reagent (2% paraformaldehyde, 2.5% glutaraldehyde, 0.25 mM CaCl2, 0.5 mM MgCl2 in 80 mM sodium cacodylate buffer, pH 7.4) and postfixed in 2% osmium tetroxide buffered in 0.1 M cacodylate (pH 7.4) for 2 h. The samples were block stained with 2% uranyl acetate for 1 h and subsequently dehydrated in acetone (50, 70, 80, 96, and 100%). The cells were immersed in acetone/durcupan resin (1:1) overnight, immersed in pure durcupan resin (Fluka, Buchs, Switzerland) for 2 h, and embedded in durcupan resin at 60°C. Ultrathin sections (thickness, 50 nm), stained with lead citrate (30), were examined in a CM10 electron microscope (Philips, Amsterdam, The Netherlands).
RESULTS
Gal-6 localization in digestive tract tissues.
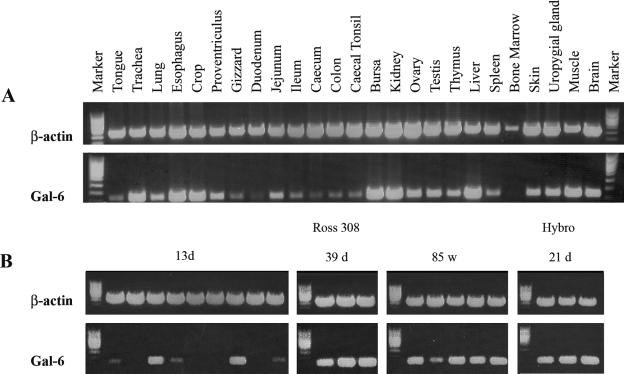
In 6-week-old Ross 308 chicken broilers, the highest Gal-6 mRNA expression was found in the trachea, esophagus, crop, bursa, kidney, liver, and muscle (Fig. 1A). Moderate Gal-6 expression was observed in the uropygial gland, brain, proventriculus, ovary, testis, and skin, whereas low expression levels were observed in the lung, thymus, and spleen and throughout the intestinal tract. Gal-6 mRNA was not detected in bone marrow. Additional investigation of Gal-6 mRNA expression levels in the crop tissue of chickens of different ages and breeds (Fig. 1B) revealed that it was strongly and equally expressed in adult Ross 308 chickens and 3-week-old Hybro chicken broilers. Yet, in crop tissues from 2-week-old Ross 308 chicken broilers, levels of Gal-6 mRNA were low or not detectable in seven of the nine animals investigated.
FIG. 1.
Tissue gene expression of chicken gallinacin-6. (A) cDNA fragments were amplified from the pooled total RNA of three animals per tissue (1 μg) with one-step reverse transcription (RT)-PCR (40 cycles). cDNA fragments of β-actin were amplified (33 cycles) with the same RNA samples as a control. (B) cDNA fragments were amplified from total RNA samples (1 μg) obtained from the crop tissue of individual animals of different breeds (Ross 308, Hybro) and age with one-step RT-PCR (40 cycles); cDNA fragments of β-actin were amplified (33 cycles) with the same RNA samples as a control.
Variability in β-defensin expression levels between animals of the same age and breed could indicate a tissue-specific upregulation and/or inducibility. Therefore the genomic region upstream of the Gal-6 gene was searched for the presence of transcription factor binding sites associated with β-defensin regulation. A putative TSS was predicted 70 bp upstream of the first nucleotide of the ATG start codon, preceded by a TATA box (−29 bp) and a CAAT box at −111 bp (Fig. 2). Further analysis of the Gal-6 upstream region using the JASPAR and Matinspector programs indicated the presence of several putative transcription factor binding sites: nuclear factor kappa beta (NF-κB; −506 bp), activator protein 1 (AP-1; −1,011 bp, −2,700 bp, −2,872 bp), and nuclear factor interleukin-6 (NF-IL-6; −1,108 bp, −1,949 bp, −2,630 bp, −2,777 bp).
FIG. 2.
Nucleotide sequence of the Gal-6 gene promoter region deduced from the published cDNA sequence (accession no. AY621324). Nucleotides are numbered relative to the predicted TSS (arrow). The partial exon 1 sequence is shown in uppercase, and the putative 5′ untranslated region is in italics. The following putative transcription factor binding sites are in boldface type and underlined: TATA, CCAAT box, NF-κB, AP-1, and NF-IL-6.
Clustal-W alignment of Gal-6 with avian and mammalian β-defensins.
An NCBI database pBLAST search using the Gal-6 precursor sequence showed more similarity between Gal-6 and mammalian β-defensins than other avian β-defensins (Fig. 3). Gal-6 was most similar to human β-defensin-1 (43% identity, 51% conserved), primate BD1-like defensins (43 to 45% identity, 54 to 58% conserved), bovine neutrophil β-defensins (33 to 44% identity, 54 to 61% conserved), and sheep β-defensin-2 (41% identity, 52% conserved). However, with the exception of gallinacin-8 (40% identity, 45% conserved), much less similarity was observed between Gal-6 and other avian β-defensins.
FIG. 3.
Alignment of gallinacin-6 prepropeptide sequence with other avian and mammalian β-defensins. Mature peptide sequences are underlined.
A comparison of cleavage sites between propiece and antimicrobial peptide domains of β-defensins with known cleavage sites was used to predict a mature Gal-6 peptide sequence. A putative cleavage site was found between Ala26 and Asp27 (QEDA↓D) which resembled proteolytic cleavage sites in human β-defensin-1 and its primate orthologs (hBD-1, ptBD-1, hcBD-1; GHRS↓D) and bovine neutrophil β-defensin-1 (BNBD-1, QGIS↓D).
Production of synthetic gallinacin-6.
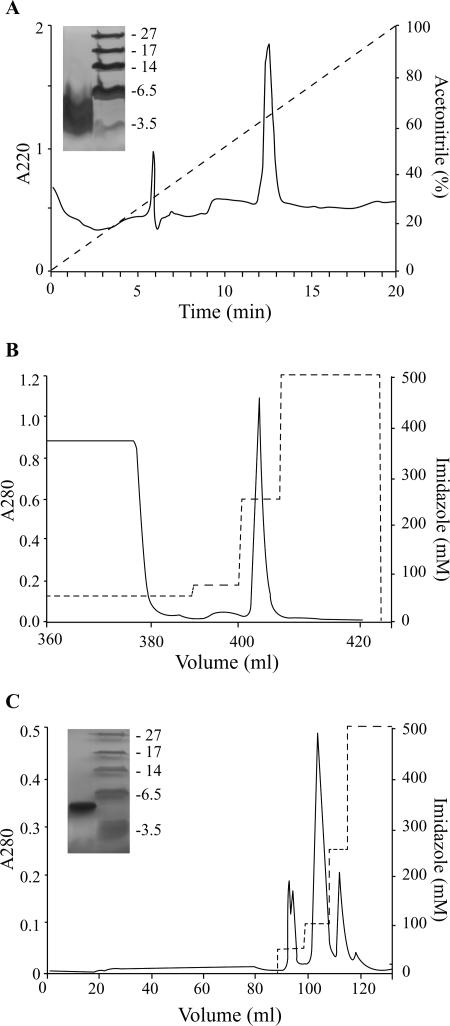
sGal-6, synthesized by Fmoc solid-phase chemistry, was slowly refolded in buffer by air oxidation to obtain the peptide in its thermodynamically most stable form. Final purification of folded peptide by reversed-phase chromatography resulted in elution of a single peak at a 12.46-min retention time (Fig. 4A). Lyophilized sGal-6 was dissolved in water and characterized by SDS-PAGE, amino acid analysis, and mass spectrometry. SDS-PAGE analysis of purified sGal-6 showed a single band migrating at approximately 4.2 kDa (Fig. 4A). Amino acid analysis of the purified mature product was consistent with the expected sGal-6 composition and did not detect significant impurities. The molecular mass of 4,282 ± 4 Da [M+] determined by MALDI-TOF analysis was in accordance with the calculated molecular mass of 4,285 Da for synthetic Gal-6.
FIG. 4.
Purification of synthetic and recombinant Gal-6. (A) Synthetic peptide was loaded on a C8 RP-HPLC column and eluted with a 20-min linear 0 to 100% gradient of acetonitrile in 0.1% trifluoroacetic acid at a 1-ml/min flow rate. Peaks were detected at 220 nm. The purity of synthetic Gal-6 was verified on a Tricine-SDS-PAGE gel. MALDI-TOF analysis showed a monoisotopic mass of 4,282 ± 4 Da, in accordance with the calculated mass. (B) Fusion protein HGH-His8-TEV-rGal-6 was purified from cell supernatant by affinity chromatography. Bound fusion protein was eluted from the HisTrap nickel column with 250 mM imidazole in buffer A (25 mM Tris, 300 mM NaCl, pH 8.2). (C) Dialyzed fusion protein was proteolytically cleaved by TEV protease for 2 h at 30°C and reapplied to the nickel affinity column. The cleaved mature recombinant gallinacin-6 was eluted from the column at 50 mM imidazole in buffer A. The purity of rGal-6 was verified by Tricine-SDS-PAGE. The molecular mass of 4,313 ± 4 Da, determined by MALDI-TOF analysis, was in accordance with the calculated mass.
Production and characterization of recombinant gallinacin-6.
Recombinant mature Gal-6 was expressed in HEK293-EBNA cells as a fusion protein with human growth hormone and subsequently purified by affinity chromatography (Fig. 4B and 4C) as described above. SDS-PAGE (Fig. 4C) and amino acid analysis of the purified mature product did not detect any impurities in the rGal-6 sample. The total yield was typically 125 μg/liter culture of HEK293-EBNA cells. Further characterization of rGal-6 using N-terminal amino acid sequencing showed complete homology of the first 37 amino acids with the expected product. This corresponds to the mature sequence (as indicated in Table 2) with the Asp residue replaced by a glycine and serine residue at the N terminus. These two amino acids are part of the TEV cleavage site and remain part of the Gal-6 N terminus after TEV treatment. MALDI-TOF analysis of the purified rGal-6 peptide resulted in two observed peaks corresponding to the MH+ and MH2+ ions of a peptide with a molecular mass of 4,313 ± 4 Da, which corresponds well with the calculated mass of 4,314 Da. Quantitative analysis of sulfhydryl groups revealed that no cysteine residues were available for complexing with the Ellman's reagent, indicating, in combination with the MALDI-TOF results, that all cysteines were involved in intramolecular disulfide linkages.
TABLE 2.
Amino acid sequences and properties of synthetic and recombinant gallinacin-6
| Peptide | Amino acid sequence | Lengtha | Charge |
|---|---|---|---|
| Gal-6 propeptide | QEDADTLACRQSHGSCSFVACRAPSVDIGTCRGGKLKCCKWAPSS | 45 | +2 |
| sGal-6 | DTLACRQSHGSCSFVACRAPSVDIGTCRGGKLKCCKWAPSS | 41 | +4 |
| rGal-6 | GSTLACRQSHGSCSFVACRAPSVDIGTCRGGKLKCCKWAPSS | 42 | +5 |
Length is measured in amino acids.
Antimicrobial activity of synthetic and recombinant peptide.
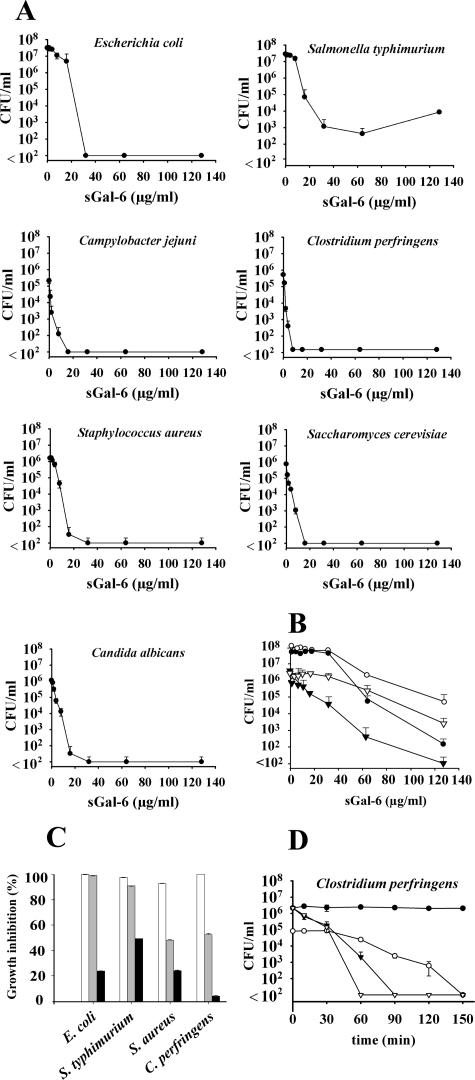
The dose-dependent survival of synthetic Gal-6-treated bacteria and yeasts was tested in colony-counting assays (Fig. 5A). A fast decline in surviving cells was observed for C. jejuni, C. perfringens, S. aureus, C. albicans, and S. cerevisiae, survival of these strains was reduced at 16 μg/ml to below the detection level of 100 cells/ml. A slower decline was observed in the assays performed with E. coli and S. enterica serovar Typhimurium; even at 128 μg/ml sGal-6, S. enterica serovar Typhimurium cells were not completely eradicated. Additionally, broth microdilution assays were performed to compare synthetic and recombinant Gal-6 antimicrobial spectra. Both peptides were active against gram-negative bacteria, gram-positive bacteria, and yeasts (Table 3). Synthetic Gal-6 inhibited C. jejuni (MIC = 64 μg/ml), E. coli (MIC/MBC = 64 μg/ml), C. perfringens (MIC/MBC = 8 μg/ml), and S. pyogenes (MIC/MBC = 64 μg/ml); no inhibition was observed within these assays for S. enterica serovar Typhimurium, P. aeruginosa, S. aureus, and B. cereus. The yeasts C. albicans and S. cerevisiae were inhibited at low concentrations of sGal-6 (MIC = 8 μg/ml); prolonged incubation at the highest concentration tested (128 μg/ml) showed this inhibition to be fungistatic of nature in this assay. Since the recombinant peptide yield was relatively low, broth microdilution assays were performed with 10 μl of 105 CFU/ml instead of the 25 μl of 106 CFU/ml used for synthetic peptide testing. The antimicrobial spectrum observed for recombinant gallinacin-6 was similar to that of synthetic Gal-6, although less potent. Recombinant Gal-6 was active against C. jejuni (MIC = 64 μg/ml; MBC = 128 μg/ml), C. perfringens (MIC = 8 μg/ml), C. albicans (MIC = 32 μg/ml), and S. cerevisiae (MIC = 32 μg/ml). As seen for sGal-6, the recombinant peptide showed bactericidal and fungistatic activity in broth microdilution assays. pH measurements indicated that, in contrast to the colony count assays performed in 1,000-fold-diluted TSB minimal medium, bacterial cultures in 100-fold-diluted TSB minimal media significantly decreased the pH of the assay medium during the 3-h incubation period if the medium was not buffered. In colony count assays, sGal-6 inhibition of E. coli in 10 mM sodium phosphate buffer was investigated at pH 5.5, 6.0, 6.5, and 7.0 (data not shown). E. coli inhibition curves were similar at all pH values, indicating that sGal-6 killing was pH independent in this range (data not shown). Exposure to sGal-6 in the presence of 10 mM sodium phosphate and 1% TSB significantly increased bacterial survival (Fig. 5B). Despite the 3-log unit decline in survival for E. coli and a 4-log unit decline in survival for C. perfringens observed at 64 μg/ml, at 128 μg/ml, only C. perfringens survival was reduced to below the detection limit under these conditions. The presence of low salt (20 mM NaCl) did not affect sGal-6-mediated growth inhibition for E. coli and S. enterica serovar Typhimurium, whereas growth inhibition of S. aureus and C. perfringens decreased to ∼50% (Fig. 5C). In the presence of high salt (150 mM NaCl), growth inhibition was reduced to 49% for S. enterica serovar Typhimurium, 24% for E. coli, and S. aureus and 4% for C. perfringens.
FIG. 5.
Concentration- and time-dependent inhibition of the growth of bacteria and yeasts by synthetic and recombinant gallinacin-6. The data are means ± standard deviations (n = 3). (A) In colony-counting assays, E. coli, S. enterica serovar Typhimurium, C. jejuni, C. perfringens, S. aureus, C. albicans, and S. cerevisiae (∼2.5 × 106 CFU/ml) were incubated for 3 h with various concentrations of synthetic Gal-6. S. cerevisiae cells were counted after 24 h of incubation on agar media at 25°C. All other strains were counted after 24 h of incubation on agar media at 37°C. (B) Additional colony-counting assays were performed with E. coli (•), S. enterica serovar Typhimurium (○), S. aureus (▿), and C. perfringens (▾) in 1% TSB containing 10 mM phosphate buffer (pH 6.5) as described above. (C) Inhibition of E. coli, S. enterica serovar Typhimurium, S. aureus, and C. perfringens in 1% TSB containing 10 mM phosphate buffer, pH 6.5 (white bars), supplemented with 20 mM (gray bars) or 150 mM (black bars) NaCl. (D) The killing kinetics of 16 μg/ml sGal-6 (▾), 64 μg/ml sGal-6 (▿), and 16 μg/ml rGal-6 (○) were determined against logarithmic phase C. perfringens cells in minimal medium (∼2.5 × 106 CFU/ml for sGal-6, ∼2.5 × 105 CFU/ml for rGal-6) in kill-curve studies. During a 150-min incubation time at 37°C under anaerobic conditions, aliquots were removed at various times and incubated overnight at 37°C on TSA plates, and surviving colonies were counted. Bacteria resuspended in minimal medium in the absence of antimicrobial peptide and subjected to the same experimental conditions served as controls (•). All kill-curve studies were performed in duplicate.
TABLE 3.
Antimicrobial activity of synthetic gallinacin-6 by broth microdilution method
| Microorganism | Resulta (μg/ml) for:
|
|||
|---|---|---|---|---|
| sGal-6
|
rGal-6
|
|||
| MIC | MBC | MIC | MBC | |
| Gram-negative | ||||
| Campylobacter jejuni | 64 | >128 | 64 | 128 |
| Salmonella enterica serovar Typhimurium | >128 | >128 | >128 | >128 |
| Escherichia coli | 64 | 64 | >128 | >128 |
| Pseudomonas aeruginosa | >128 | >128 | >128 | >128 |
| Gram-positive | ||||
| Clostridium perfringens | 8 | 8 | 8 | 8 |
| Streptococcus pyogenes | 64 | 64 | NDb | ND |
| Staphylococcus aureus | >128 | >128 | >128 | >128 |
| Bacillus cereus | >128 | >128 | <128 | >128 |
| Fungi | ||||
| Candida albicans | 8 | >128 | 32 | >128 |
| Saccharomyces cerevisiae | 8 | >128 | 32 | >128 |
Determination of MICs and MBC. Final concentrations of 0 to 128 μg/ml peptide were incubated with 106 CFU/ml (sGal-6) or 105 CFU/ml (rGal-6) of each bacterial or fungal strain. All assays were performed in triplicate.
ND, not determined.
Killing kinetics of synthetic and recombinant peptide.
Kill-curve studies were performed with synthetic and recombinant Gal-6 peptide against a logarithmic-phase C. perfringens culture. A 3-log unit decrease in viable C. perfringens cells was observed after 60 min when treated with 16 μg/ml sGal-6 (Fig. 5D). Treatment with 64 μg/ml sGal-6 decreased C. perfringens survival within 60 min to below the detection limit of 100 cell/ml. However, treatment with recombinant Gal-6 resulted only in a 0.5-log unit decline in surviving cells after 60 min of treatment and needed up to 150 min of incubation time to reduce colony counts to practically zero. No effect on C. perfringens growth was detected in the presence of minimal TSB medium (negative control). Thus, again, sGal-6 killing of C. perfringens proved to be more potent than that of rGal-6.
Effects of sGal-6 treatment on the ultrastructure of C. perfringens.
To elucidate the nature of the killing mechanism(s) used by Gal-6, C. perfringens cells were treated for 30 min with 1.56 to 25 μg/ml sGal-6 and were investigated by transmission electron microscopy.
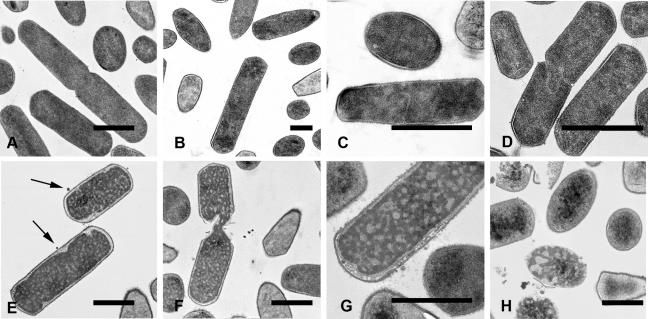
Compared to the control treatment (Fig. 6A), sGal-6 treatment resulted in dose-dependent morphological changes. At 1.56 μg/ml sGal-6, granulation of intracellular material (Fig. 6B) and irregular septa in dividing cells were observed (Fig. 6C). At 6.25 μg/ml, similar observations were made for an increasing fraction of cells (Fig. 6D). Cells treated with 12.5 μg/ml sGal-6 showed other characteristics, such as retracting cytoplasm and membrane leakage (Fig. 6E), whereas lysis at the septa of dividing cells (Fig. 6F) became more prominent. At 25 μg/ml, the majority of cells developed a ghost-like appearance combined with detachment of the cytoplasmic membrane from its peptidoglycan layer and concomitant membrane fragmentation (Fig. 6G) and resulted more often in complete lysis (Fig. 6H). Control (minimal TSB medium only) treated bacteria showed none of these effects.
FIG. 6.
Transmission electron microscopy of C. perfringens cells incubated with synthetic Gal-6. (A) Bacteria incubated in minimal medium for 30 min were undamaged. In contrast, bacteria incubated for 30 min with increasing concentrations of sGal-6 showed dose-dependent changes in ultrastructure. Granulation of intracellular material was already observed at 1.56 μg/ml (B). Irregular septum formation in dividing cells was observed at 1.56 μg/ml (C) and 6.25 μg/ml (D). At 12.5 and 25 μg/ml, cells showed retracting cytoplasm (E), lysis at the septa of dividing cells (F), cytoplasmic membrane degradation (G), and complete cell lysis (H).
DISCUSSION
The predominant Gal-6 expression found in the urogenital tract, liver, and bursa corroborated results from earlier reports (24, 46). However, in contradiction to our findings, these studies showed no significant β-defensin expression in the proximal part of the digestive tract. We found high expression levels of β-defensin gallinacin-6 in the esophagus, crop, and to a lesser extent, in the proventriculus (glandular stomach). These differences could be due to variability in β-defensin expression between individual animals, choice of breed, animal age, and experimental procedures. The similarly high levels of Gal-6 mRNA found in the crop tissues obtained from Hybro and Ross 308 broilers showed that the high Gal-6 expression in crop tissue is not breed dependent. However, the highly variable Gal-6 mRNA levels found in crop tissues of 13-day-old Ross 308 animals does suggest that Gal-6 may be developmentally regulated.
To investigate the possibility of a gallinacin-6 upregulation or inducibility in the upper digestive tract, the genomic sequence flanking the Gal-6 gene was searched for the presence of transcription binding sites. Several β-defensins have been demonstrated to be upregulated by transcription factors involved in inflammation. Indeed, putative transcription binding sites were found for NF-κB, NF-IL-6, and AP-1 in the 5′ region flanking the Gal-6 gene. Inducible β-defensins, such as hBD-2 (2) and bovine tracheal antimicrobial peptide (TAP) (7), have been shown to be upregulated through the NF-κB pathway by a variety of inflammatory mediators. Transcription factors NF-κB and NF-IL-6 have been demonstrated to synergistically participate in activation of numerous innate immune responses (3, 27) and are both markedly conserved in the promoter regions of TAP, hBD-2, and insect defensins attacin and diptericin (7). Additionally, AP-1, which plays a role in both basal and inducible transcription of numerous genes (12), may even be required in cooperation with NF-κB to achieve full gene expression, as demonstrated for monocyte chemoattractant protein 1, tissue factor, and hBD-2 genes (25, 26, 42, 44). Our findings suggest that Gal-6 expression might be induced or upregulated in the chicken digestive tract via NF-κB and AP-1 pathways.
The fact that high β-defensin mRNA levels were found in crop tissue was not unexpected. Chickens practice coprophagy to recover vitamins, amino acids, and other nutrients produced by their hindgut bacteria (28). The crop, an extension of the esophagus, is especially well developed in chickens and other gallinaceous birds, serves to temporarily store food when the stomach is full, and may hold its contents for up to 24 h before it passes into the glandular stomach. Hence, it requires an adequate local innate immune system.
To investigate the antimicrobial properties of Gal-6, synthetic and recombinant Gal-6 peptides were produced (Table 2) and tested against gram-positive bacteria, gram-negative bacteria, and yeasts using colony-counting assays (Fig. 5A) and broth microdilution assays (Table 3). In colony-counting assays, synthetic Gal-6 peptide showed strong bactericidal and fungicidal activity against all investigated strains, including food-borne pathogens Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and Clostridium perfringens (Fig. 5A). However, the magnitude of activity seemed dependent on the assay used, since sGal-6 showed higher activity and a broader antimicrobial spectrum in the colony-counting assay than in the broth microdilution assay (Table 3). The differences between both types of assays can be explained in part by the greater stringency of the broth microdilution assay, i.e., after the killing reaction has been terminated by dilution, surviving microorganisms form single colonies on agar plates but can fully outgrow in liquid media. Additionally, the conditions for resuscitation of sublethally damaged cells seem to be more favorable in the broth microdilution assay than in direct plating on agar media (4, 43). Colony count assays performed with minimal media containing 1:100- instead of 1:1,000-diluted TSB showed only a marginal increase in bacterial growth for the investigated species and a markedly increased bacterial survival (Fig. 5B). It is well described that the microbicidal activity of cationic peptides is influenced by environmental factors (i.e., temperature, pH, ionic strength, cationicity) and microbial growth phase (20, 35, 38). Logarithmic growth phase organisms have been observed to be more susceptible to cationic peptide-mediated killing than organisms in a stationary growth phase, but exceptions have been noted (15, 20), indicating that this phenomenon is target species specific. A 10 mM sodium phosphate buffer at neutral pH is commonly used to study the antimicrobial properties of cationic peptides. Yet several reports suggest that the presence of phosphate buffer could interfere with cationic peptide-mediated inhibition (11, 34), although these effects were attributed to ionic strength.
However, in our experiments, addition of 20 mM NaCl did not affect the bactericidal activity of sGal-6, and even in the presence of 150 mM NaCl, sGal-6 still had bactericidal activity against most bacteria (Fig. 5C). Hence, the increased bacterial survival in these experiments can be explained neither by ionic strength differences nor by an increased nutrient availability.
The production of recombinant Gal-6 involved proteolytic cleavage of the HGH-His8-TEV-rGal-6 fusion protein by TEV protease and resulted in a 42-amino-acid peptide in which the Asp1 residue was replaced by a Gly-Ser N terminus (Table 2). Despite the higher positive charge of the recombinant peptide, thought to be associated with increased antimicrobial activity (6), broth microdilution assays showed that rGal-6 was far less potent than sGal-6 (Table 3). Antcheva et al. (1) made a similar observation for a variant of hBD-2 lacking its N-terminal aspartic acid (Asp4) residue. (−D)hBD-2 was demonstrated to be less structured and had a markedly lower antimicrobial activity than hBD-2. Circular dichroism spectra of (−D)hBD-2 peptide showed little α-helical content in aqueous solution and in the presence of SDS micelles, whereas the hBD-2 spectrum was compatible with the presence of an α-helical stretch, as observed in the crystal structure (19). The slower permeabilization and killing kinetics observed for (−D)hBD-2 toward E. coli ML-35 suggest that the structure of the N-terminal stretch plays a role in mediating interaction with the bacterial membrane.
To elucidate the mechanism(s) involved in gallinacin-6-mediated killing of bacteria, the killing kinetics and morphology of food-borne pathogen C. perfringens cells were examined after treatment with synthetic Gal-6. In support of the broth microdilution assays, kill-curve studies of recombinant and synthetic gallinacin-6 showed a much faster killing mechanism and a greater efficacy for synthetic Gal-6. Similar kinetics have been reported for synthetic tick defensin A; treatment with MICs of defensin A killed Micrococcus luteus cells within 60 min (29). A 1.5, 0.5, and 1 log unit decrease in survival of S. aureus 710A cells within 60 min was observed after treatment with synthetic hBD-2, (MIC), Hylobates concolor β-defensin 3 (4 × MIC), and human β-defensin-3 (8× MIC), respectively, indicating an even slower but effective mechanism (31). Killing and permeabilization kinetics observed for these peptides (29, 31) indicate that cell lysis itself is not the primary mode of β-defensin-mediated bacterial killing. Apparently, a certain level of cell wall permeabilization is required for the β-defensin molecules to reach intracellular targets, possibly affecting DNA replication, RNA, and protein synthesis (23).
Examination of sGal-6-treated C. perfringens by transmission electron microscopy showed dose-dependent morphological effects, as seen for other defensins. A 30-min treatment of 108 CFU/ml C. perfringens cells with sGal-6 concentrations ranging from 1.56 to 25 μg/ml induced dose-dependent changes, such as clumping of intracellular material and irregular septum formation during cell division at lower concentrations. At higher concentrations, most cells showed signs of cytoplasm retraction and detachment of the cytoplasmic membrane from the peptidoglycan layer, sometimes resulting in mesosome-like structures. Prior to complete lysis, often originating at the cell poles or at the septa of dividing cells, many cells developed a ghost-like appearance. Similar observations were described by Lee et al. (22), who observed chromatin condensation in Haemophilus influenza after a 30-min treatment with 10 μg/ml hBD-2. Condensation of plasmid-sized DNA molecules can be induced by low concentrations of chemical agents with a cation valence of +3 or greater (37), including cationic peptides, such as spermine, spermidine, polylysine, and protamine (5, 36). DNA condensation has also been observed by treatment with protein synthesis inhibitors, such as rifampin, chloramphenicol, and puromycin (49). E. coli cells treated for 30 min with 50 μg/ml HE2β1, an antibacterial peptide with similarity to β-defensins, inhibited DNA, RNA, and protein synthesis and caused extensive granulation and cytoplasmic retraction (47). Mesosome-like structures have been observed for bacteria treated with defensins, the artificial peptides Bac2A-NH and CP11CN, and antibiotics, such as trimethoprim and rifampin (13, 38). The phenomenon of an antibacterial mechanism specifically aimed against septum formation and resulting in lysis near the septa of dividing cells has been reported for cationic peptides indolicidin and tick defensin A (14, 29). The morphological changes induced by low concentrations of sGal-6 and the lack of massive complete lysis at the highest concentrations tested show cell lysis not to be the primary mechanism in Gal-6-mediated killing of prokaryotic cells.
In conclusion, to our knowledge, this is the first report of a chicken β-defensin highly expressed in the digestive tract and displaying strong bactericidal activity against food-borne pathogens. The high expression levels found in crop tissue indicate an important role for Gal-6 in chicken innate immunity. The presence of putative transcription factor binding sites involved in β-defensin induction and upregulation make Gal-6 an excellent target to improve chicken digestive tract health and food safety by dietary modulation.
Acknowledgments
We thank Laura van Weeren and Ton Ultee of the Center for Cell Imaging (Department of Biochemistry and Cell Biology, Faculty of Veterinary Medicine, Utrecht University) for their assistance with the transmission electron microscopy images and Wieger Hemrika (Protein expression facility, Academic Biomedical Centre, Utrecht University) for his advice on recombinant peptide production.
This work was supported by a research grant (Adaptation and Resistance Programme) from the Animal Science Group (Wageningen University and Research Centre) and the Faculty of Veterinary Medicine (Utrecht University), The Netherlands.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Antcheva, N., M. Boniotto, I. Zelezetsky, S. Pacor, M. V. Verga Falzacappa, S. Crovella, and A. Tossi. 2004. Effects of positively selected sequence variations in human and Macaca fascicularis β-defensins 2 on antimicrobial activity. Antimicrob. Agents Chemother. 48:685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randell. 2000. CD14-dependent lipopolysaccharide-induced β-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., J. K. Cheshire, S. Akira, T. Kishimoto, and P. Woo. 1993. The role of NF-κB and NF-IL6 transactivating factors in the synergistic activation of human serum amyloid A gene expression by interleukin-1 and interleukin-6. J. Biol. Chem. 268:25624-25631. [PubMed] [Google Scholar]
- 4.Bovill, R. A., and B. M. Mackey. 1997. Resuscitation of ‘non-culturable’ cells from aged cultures of Campylobacter jejuni. Microbiology 143:1575-1581. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, L. R., M. Corzett, and R. Balhorn. 1999. Protamine-induced condensation and decondensation of the same DNA molecule. Science 286:120-123. [DOI] [PubMed] [Google Scholar]
- 6.Dathe, M., and T. Wieprecht. 1999. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1462:71-87. [DOI] [PubMed] [Google Scholar]
- 7.Diamond, G., V. Kaiser, J. Rhodes, J. P. Russell, and C. L. Bevins. 2000. Transcriptional regulation of β-defensin gene expression in tracheal epithelial cells. Infect. Immun. 68:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durocher, Y., S. Perret, and A. Kamen. 15 January 2002, posting date. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed]
- 9.Ellman, G. L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:70-77. [DOI] [PubMed] [Google Scholar]
- 10.Evans, E. W., F. G. Beach, K. M. Moore, M. W. Jackwood, J. R. Glisson, and B. G. Harmon. 1995. Antimicrobial activity of chicken and turkey heterophil peptides CHP1, CHP2, THP1, and THP3. Vet. Microbiol. 47:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernie-King, B. A., D. J. Seilly, and P. J. Lachmann. 30 March 2004, posting date. The interaction of streptococcal inhibitor of complement (SIC) and its proteolytic fragments with the human beta defensins. Immunology doi: 10.1111/j.0019-2805.2004.01837.x. [DOI] [PMC free article] [PubMed]
- 12.Foletta, V. C. 1996. Transcription factor AP-1, and the role of Fra-2. Immunol. Cell Biol. 74:121-133. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, C. L., D. Moyles, T. J. Beveridge, and R. E. W. Hancock. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, C. L., A. Rozek, A. Patrzykat, and R. E. W. Hancock. 2001. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 276:24015-24022. [DOI] [PubMed] [Google Scholar]
- 15.Ganz, T., M. E. Selsted, D. Szklarek, S. S. L. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisow, M. J. 1991. Characterizing recombinant proteins. Bio/Technology 9:921-922, 924. [DOI] [PubMed] [Google Scholar]
- 17.Hidaka, Y., M. Ohno, B. Hemmasi, O. Hill, W.-G. Forssmann, and Y. Shimonishi. 1998. In vitro disulfide-coupled folding of guanylyl cyclase-activating peptide and its precursor protein. Biochemistry 37:8498-8507. [DOI] [PubMed] [Google Scholar]
- 18.Higgs, R., D. J. Lynn, S. Gaines, J. McMahon, J. Tierney, T. James, A. T. Lloyd, G. Mulcahy, and C. O'Farrelly. 2005. The synthetic form of a novel chicken beta-defensin identified in silico is predominantly active against intestinal pathogens. Immunogenetics 57:90-98. [DOI] [PubMed] [Google Scholar]
- 19.Hoover, D. M., K. R. Rajashankar, R. Blumenthal, A. Puri, J. J. Oppenheim, O. Chertov, and J. Lubkowski. 2000. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275:32911-32918. [DOI] [PubMed] [Google Scholar]
- 20.Koo, S.-P., M. R. Yeaman, and A. S. Bayer. 1996. Staphylocidal action of thrombin-induced platelet microbicidal protein is influenced by microenvironment and target cell growth phase. Infect. Immun. 64:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy, D. J., C. E. Dann III, P. Longo, B. Perman, and K. X. Ramyar. 2000. A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr. Purif. 20:500-506. [DOI] [PubMed] [Google Scholar]
- 22.Lee, H.-Y., A. Andalibi, P. Webster, S.-K. Moon, K. Teufert, S.-H. Kang, J.-D. Li, M. Nagura, T. Ganz, and D. J. Lim. 5. May 2004, posting date. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Infect. Dis. http://www.biomedcentral.com/1471-2334/4/12. [DOI] [PMC free article] [PubMed]
- 23.Lehrer, R. I., A. Barton, K. A. Daher, S. S. L. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynn, D. J., R. Higgs, S. Gaines, J. Tierney, T. James, A. T. Lloyd, M. A. Fares, G. Mulcahy, and C. O'Farrelly. 2004. Bioinformatic discovery and initial characterisation of nine novel antimicrobial peptide genes in the chicken. Immunogenetics 56:170-177. [DOI] [PubMed] [Google Scholar]
- 25.Mackman, N., K. Brand, and T. S. Edgington. 1991. Lipopolysaccharide-mediated transcriptional activation of the human tissue factor gene in THP-1 monocytic cells requires both activator protein 1 and nuclear factor κB binding sites. J. Exp. Med. 174:1517-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, T., P. M. Cardarelli, G. C. Parry, K. A. Felts, and R. R. Cobb. 1997. Cytokine induction of monocyte chemoattractant protein-1 gene expression in human endothelial cells depends on the cooperative action of NF-κB and AP-1. Eur. J. Immunol. 27:1091-1097. [DOI] [PubMed] [Google Scholar]
- 27.Matsusaka, T., K. Fujikawa, Y. Nishio, N. Mukaida, K. Matsushima, T. Kishimoto, and S. Akira. 1993. Transcription factors NF-IL6 and NF-κappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 90:10193-10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montrose, M. S., S. M. Shane, and K. S. Harrington. 1985. Role of litter in the transmission of Campylobacter jejuni. Avian Dis. 29:392-399. [PubMed] [Google Scholar]
- 29.Nakajima, Y., J. Ishibashi, F. Yukuhiro, A. Asaoka, D. Taylor, and M. Yamakawa. 2003. Antibacterial activity and mechanism of action of tick defensin against gram-positive bacteria. Biochim. Biophys. Acta 1624:125-130. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahl, H.-G., U. Pag, S. Bonness, S. Wagner, N. Antcheva, and A. Tossi. 2005. Mammalian defensins: structures and mechanism of antibiotic activity. J. Leukoc. Biol. 77:466-475. [DOI] [PubMed] [Google Scholar]
- 32.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 33.Schuster, R. 1988. Determination of amino acids in biological, pharmaceutical, plant and food samples by automated precolumn derivatization and high-performance liquid chromatography. J. Chromatogr. 431:271-284. [DOI] [PubMed] [Google Scholar]
- 34.Selsted, M. E., D. Szklarek, T. Ganz, and R. I. Lehrer. 1985. Activity of rabbit leukocyte peptides against Candida albicans. Infect. Immun. 49:202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selsted, M. E., D. Szklarek, and R. I. Lehrer. 1984. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect. Immun. 45:150-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen, D., and D. M. Crothers. 1986. Condensation of chromatin: role of multivalent cations. Biochemistry 25:1495-1503. [DOI] [PubMed] [Google Scholar]
- 37.Sen, D., and D. M. Crothers. 1986. Influence of DNA-binding drugs on chromatin condensation. Biochemistry 25:1503-1509. [DOI] [PubMed] [Google Scholar]
- 38.Shimoda, M., K. Ohki, Y. Shimamoto, and O. Kohashi. 1995. Morphology of defensin-treated Staphylococcus aureus. Infect. Immun. 63:2886-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townes, C. L., G. Michailidis, C. J. Nile, and J. Hall. 2004. Induction of cationic chicken liver-expressed antimicrobial peptide 2 in response to Salmonella enterica infection. Infect. Immun. 72:6987-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Plas, R. M., L. Gomes, J. A. Marquart, T. Vink, J. C. Meijers, P. G. de Groot, J. J. Sixma, and E. G. Huizinga. 2000. Binding of von Willebrand factor to collagen type III: role of specific amino acids in the collagen binding domain of vWF and effects of neighboring domains. Thromb. Haemost. 84:1005-1011. [PubMed] [Google Scholar]
- 41.van Dijk, A., E. J. A. Veldhuizen, A. J. A. M. van Asten, and H. P. Haagsman. 2005. CMAP27, a novel chicken cathelicidin-like antimicrobial protein. Vet. Immunol. Immunopathol. 106:321-327. [DOI] [PubMed] [Google Scholar]
- 42.Vora, P., A. Youdim, L. S. Thomas, M. Fukata, S. Y. Tesfay, K. Lukasek, K. S. Michelsen, A. Wada, T. Hirayama, M. Arditi, and M. T. Abreu. 2004. β-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J. Immunol. 173:5398-5405. [DOI] [PubMed] [Google Scholar]
- 43.Wai, S. N., Y. Mizunoe, A. Takade, and S. Yoshida. 2000. A comparison of solid and liquid media for resuscitation of starvation- and low-temperature-induced nonculturable cells of Aeromonas hydrophila. Arch. Microbiol. 173:307-310. [DOI] [PubMed] [Google Scholar]
- 44.Wehkamp, K., L. Schwichtenberg, J.-M. Schröder, and J. Harder. 2006. Pseudomonas aeruginosa- and IL-1β-mediated induction of human β-defensin-2 in keratinocytes is controlled by NF-κB and AP-1. J. Investig. Dermatol. 126:121-127. [DOI] [PubMed] [Google Scholar]
- 45.Witte, W. 21 December 2000, posting date. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents doi: 10.1016/S0924-8579(00)00301-0. [DOI] [PubMed]
- 46.Xiao, Y., A. L. Hughes, J. Ando, Y. Matsuda, J.-F. Cheng, D. Skinner-Noble, and G. Zhang. 13. August 2004, posting date. A genome-wide screen identifies a single β-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genomics doi: 10.1186/1471-2164-5-56. [DOI] [PMC free article] [PubMed]
- 47.Yenugu, S., K. G. Hamil, F. S. French, and S. H. Hall. 24 August 2004, posting date. Antimicrobial actions of the human epididymis 2 (HE2) protein isoforms, HE2alpha, HE2beta1 and HE2beta2. Reprod. Biol. Endocrinol. doi: 10.1186/1477-7827-2-61. [DOI] [PMC free article] [PubMed]
- 48.Yu, P.-L., S. D. Choudhury, and K. Ahrens. 2001. Purification and characterization of the antimicrobial peptide, ostricacin. Biotechnol. Lett. 23:207-210. [Google Scholar]
- 49.Zusman, D. R., A. Carbonell, and J. Y. Haga. 1973. Nucleoid condensation and cell division in Escherichia coli MX74T2 ts52 after inhibition of protein synthesis. J. Bacteriol. 115:1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]