Abstract
A detailed model of the G2 DNA damage checkpoint (G2DDC) system is presented that includes complex regulatory networks of the mitotic kinase Cdc2, phosphatase Cdc25, Wee1 kinase, and damage signal transduction pathways involving Chk1 and p53. Assumptions on the kinetic equations of the G2DDC are made, and computer simulations are carried out to demonstrate how the various subsystems operate to delay or arrest cell cycle progression. The detailed model could be used to explain various experiments relevant to G2DDC reported recently, including the nuclear export of 14–3–3-bound Cdc25, the down-regulation of cyclin B1 expression by p53, the effect of Chk1 and p53 on Cdc25 levels, and Wee1 degradation. It also is shown that, under certain conditions, p53 is necessary to sustain a G2 arrest.
It is very important to study cell cycle checkpoints because they are windows into the inner workings of the complex process of cell division. Checkpoints can be classified as either intrinsic or extrinsic depending on whether or not they are considered as part of the cell cycle engine (1, 2). The current view of the checkpoint that arrests or delays the cell cycle at G2 as a result of DNA damage (henceforth called the G2 DNA damage checkpoint or G2DDC) is that it is an extrinsic mechanism by which damage signal transduction pathways impinge on the cell cycle machinery. I will discuss how the G2DDC perturbs this machinery where it is vulnerable and analyze the kinetics of the checkpoint.
Models of the G2DDC have been proposed previously (3–5), but they are schematic at best and lack kinetic details. In this paper, I present a detailed molecular mechanism of the G2DDC system and carry out computer simulations based on certain assumptions on the kinetics of the component processes. To simplify my analysis, I first break down the complex G2DDC network into subsystems, understand the intrinsic dynamics of each subsystem, and show how these subsytems interact to elicit a checkpoint response. I will show that my detailed model could explain recent experimental results such as those of Bunz et al. (6), Innocente et al. (7), Lopez-Girona et al. (8), and Toyoshima et al. (9).
The reader is warned that several steps of the detailed model presented here must be regarded as tentative for now because of insufficient or lack of evidence that these steps actually occur in vivo. However, there are good reasons why I think it is not unreasonable to attempt a quantitative study of the G2DDC at this time. In the mathematical field of reaction network analysis, some promising results show that just knowing the structure of the network is sufficient to deduce certain possible behavior of a network independent of the values of the rate parameters; see for example the work of Feinberg and Horn (10). In addition, the type of dynamics a system exhibits usually depends on ratios of rate parameters instead of their absolute values; therefore, it is not necessary that exact parameter values be known to understand the types of behavior that a system can exhibit. As an example, the reader is referred to the pioneering work on cell cycle modeling by Novak and Tyson (11). Important dynamical information can be inferred from the structure of a given network and, because many of the G2DDC pathways have been identified or postulated, a quantitative kinetic study of this important checkpoint system could lead to a better perspective and understanding of this complex experimental system.
The G2DDC and Its Subsystems
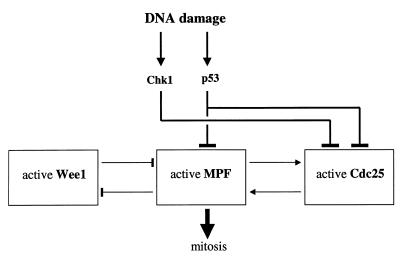
An overview of the G2DDC system is given in Fig. 1, which shows that the basic checkpoint targets are the cyclin-dependent kinase (CDK) Cdc2 and its primary regulators, Cdc25 and Wee1. For its activation, Cdc2 requires binding with cyclin B and the active Cdc2/cyclin B complex often is referred to as MPF (for maturation-promoting factor). I subdivide the G2DDC system into the following subsystems: (i) MPF subsystem, (ii) Cdc25 subsystem, (iii) Wee1 subsytem, and (iv) DNA damage signal transduction pathways. I will discuss the details of these subsytem and show how they are coupled to each other.
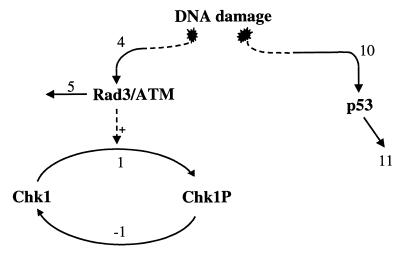
Figure 1.
Schematic diagram of the G2DDC system showing the subsystems involving Wee1, MPF, Cdc25, and signal transduction pathways.
The DNA damage signal transduction pathways summarized in Fig. 1 involve the Chk1 kinase and p53. The Chk1 pathway inhibits the activity of Cdc25 whereas p53 could down-regulate MPF activity [via the induction of the CDK inhibitor p21 (6) or the repression of cyclin B1 transcription (7)] and down-regulate Cdc25 activity via 14–3–3 proteins as will be discussed in more detail below (12).
What is the logic behind the architecture of the G2DDC shown in Fig. 1? I propose a general answer at this point: an antagonistic interaction between MPF and Wee1 exists (Wee1 inhibits MPF activity by tyrosine phosphorylation while MPF phosphorylates Wee1, leading to the latter’s degradation) and Wee1 dominates at first and prevents entry into mitosis; once Cdc25 accumulates inside the nucleus, MPF activity increases rapidly (because of a positive feedback loop between Cdc25 and MPF) and Wee1 eventually loses its battle with MPF, thereby allowing the cell to undergo mitosis. I now will describe the details of the implementation of this picture of the G2-M cell cycle transition and the G2DDC system.
The MPF Subsystem
A certain threshold of MPF activity must be reached for a cell to initiate mitosis; thus, a G2-M checkpoint necessarily involves, directly or indirectly, the regulation of MPF. Excellent reviews on MPF regulation are already available (11, 13), and I give only a summary here. Activation of Cdc2 requires binding with cyclin B and phosphorylation on Thr-161 by a CDK-activating kinase. Cdc2 is inactive when its Tyr-15 and Thr-14 residues are phosphorylated by Wee1/Myt1 kinases, and the phosphatase Cdc25 is required to remove the phosphate groups from these residues and activate Cdc2. I refer the reader to the paper of Novak and Tyson (11) for a more detailed discussion of MPF regulation. I consider the network given in Fig. 2, which, although simplified, is sufficient to represent the transition between the inactive and active forms of MPF.
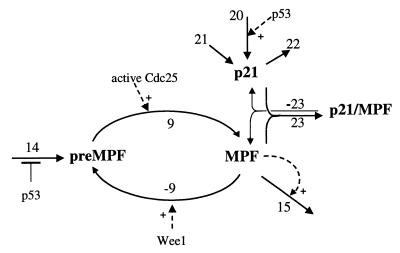
Figure 2.
The MPF subsystem. The reaction numbers correspond to the rate expressions in Table 1.
In Fig. 2, preMPF refers to the inactive Cdc2/cyclin B complexes that are phosphorylated on Tyr-15 and/or Thr-14, and MPF represents the active Cdc2/cyclin B. Activation of MPF from preMPF is represented by a single process (reaction 9) whose rate expression, v9, reflects the contributions of the active forms of Cdc25. The rate expression v9 is given in Table 1; there the factor {[aCdc25]+[aCdc25Ps216]} represents the total active Cdc25 (the different forms of Cdc25 will be discussed later). The active forms aCdc25 and aCdc25Ps216 are assumed to have the same activity (i.e., same value of k9 in Table 1). The second term in v9 represents Cdc25-independent pathways of MPF activation (e.g., see Zheng and Ruderman, ref. 26). The deactivation of MPF (reaction −9 in Fig. 2) is carried out by Wee1/Myt1 kinases and is given the rate expression v−9 in Table 1.
Table 1.
Rate expressions
| v1 = k1[Chk1][Rad3]v−1 = k−1[Chk1P] |
| v2 = k2[Chk1P][aCdc25] + kctak1[aCdc25] |
| v−2 = k−2[aCdc25Ps216] |
| v2′ = k2′[Chk1P][iCdc25] + kctak1′[iCdc25] |
| v3′ = k3′[iCdc25Ps216][14–3–3] |
| v−3′ = k−3′[iCdc25Ps216/14-3-3] |
| v4 = k4v5 = k5[Rad3]v6 = k6[p53] |
| v7 = k7[MPF][iCdc25] + kPlk1[iCdc25]v−7 = k−7[aCdc25] |
| v8 = k8[MPF][Wee1] + k8′[Wee1] v−8 = k−8[Wee1P] |
| v9 = k9{[aCdc25] + [aCdc25Ps216]}[preMPF] + k9′[preMPF] |
| v−9 = k−9[MPF][Wee1] v10 = k10 v11 = k11[p53] |
| v12 = k12[14–3–3] v13 = k13 v14 = k14/(1 + k14′[p53]) |
| v15 = k15[MPF][MPF] v16 = k16 v17 = k17[Wee1P] |
| v18 = k18[MPF][iCdc25Ps216] + kPlk1′[iCdc25Ps216] |
| v−18 = k−18[aCdc25Ps216] v20 = k20[p53] |
| v21 = k21 v22 = k22[p21] |
| v23 = k23[MPF][p21] v−23 = k−23[p21/MPF] |
Reaction 14 is a gross representation of the transcriptional and translational processes for cyclin B, as well as dimerization with Cdc2 and its phosphorylation by CDK-activating kinase, which are involved in forming the inactive preMPF. Furthermore, the translocation of MPF into the nucleus before the onset of mitosis is included in reaction 14. It is well known (11) that the activity of MPF fluctuates in cycling cells; MPF activity is low through most of interphase and increases significantly before mitosis, and then drops when mitosis begins. In the present work, I am not concerned with the oscillations of MPF activity; instead, we focus our attention on the G2-M transition. I make an assumption that reaction 14 is negatively regulated by p53 in accordance with the recent report of Innocente et al. (7) (see the denominator of v14 in Table 1). Note that v14 is assumed to be directly proportional to the rate of cyclin B synthesis because the level of total Cdc2 is fairly constant throughout the cell cycle. Cyclin B degradation is represented by reaction 15 with rate given by v15 in Table 1. The kinetics in v15 accounts for the fact that MPF stimulates its own degradation (11). One also could include the export of cyclin B from the nucleus, which also has been shown to affect the operation of the G2DDC (9).
The CDK inhibitor p21 can interact directly with MPF as represented by reactions 23 and −23. Bunz et al. (6) implicate p21 as a requirement to sustain G2 arrest after DNA damage. Bates et al. (14) also showed that p21 does contribute to a delay in G2 but suggest that the mechanism could be indirect because of the inefficient association of p21 and cyclin B1. In Fig. 2, I assume that p21 forms a complex with active MPF; it also may be possible that p21 complexes with preMPF (15) because of the observation that p21 could block the cell cycle early in G2 or late S-phase before the activation of MPF (14). In my simulations, I will assume inhibitory levels of p21 despite the knowledge that stoichiometric levels of p21 do not abolish MPF kinase activity whereas higher levels of p21 do (16, 17). There are p53-dependent and p53-independent expressions of p21 that are represented by the rates v20 and v21, respectively (see Fig. 2 and Table 1). The rate of degradation of p21 is given by v22, and the rates of reversible association of p21 with MPF are given by v23 and v-23 in Table 1.
The kinetic equations for the MPF subsystem are given in Table 2. To understand the intrinsic dynamics of the MPF subsystem, I integrated the differential equations involving the protein levels [preMPF], [MPF], [p21], and [p21/MPF]. Note that v9 has the factor {[aCdc25]+[aCdc25Ps216]}, which corresponds to the total activity of Cdc25 and, because of the positive feedback loop between MPF and Cdc25, is proportional to active [MPF]. Thus, to simulate the interaction of the MPF subsystem with the Cdc25 subsystem, I replace total active Cdc25 with MPF. Similarly, [Wee1] in the expression of v-9 is replaced with {1/(1+[MPF])} to indicate the inhibitory effect of MPF on Wee1 and vice versa.
Table 2.
Kinetic equations
| Signal transduction subsystem |
| d[Chk1P]/dt = v1 − v−1 |
| d[Rad3]/dt = v4 − v5 |
| d[p53]/dt = v10 − v11 |
| MPF subsystem |
| d[preMPF]/dt = v14 + v−9 − v9 |
| d[MPF]/dt = v9 + v−23 − (v−9 + v23 + v15) |
| d[p21]/dt = v20 + v21 + v−23 − (v22 + v23) |
| d[p21/MPF]/dt = v23 − v−23 |
| Cdc25 subsystem |
| d[iCdc25]/dt = v−7 + v−3′ − (v7 + v2′) |
| d[iCdc25Ps216]/dt = v2′ + v−18 − (v18 + v3′) |
| d[iCdc25Ps216/14–3–3]/dt = v3′ − v−3′ |
| d[aCdc25]/dt = v7 + v−2 − (v−7 + v2) |
| d[aCdc25Ps216]/dt = v2 + v18 − (v−2 + v−18) |
| d[14-3-3]/dt = v6 + v13 + v−3′ − (v3′ + v12) |
| Wee1 subsystem |
| d[Wee1]/dt = v16 + v−8 − v8 |
| d[Wee1P]/dt = v8 − (v−8 + v17) |
The rate expressions vis are given in Table 1.
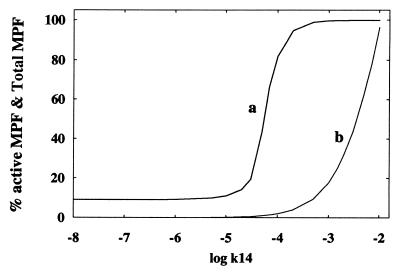
Fig. 3 is a plot of the percentage of active MPF (over total MPF measured at steady state) versus k14 (which is proportional to the rate of formation of preMPF). Also shown in this figure is the increase of [total MPF] with increasing k14. Observe in Fig. 3 that the increase in activity of MPF does not parallel the increase in total MPF. The figure shows a switching threshold value of [total MPF] where a marked increase in the activity of MPF occurs. Note that when log k14 has the value of about −3, practically all of MPF is active.
Figure 3.
Percentage of active MPF (curve a) and total MPF (curve b) versus log k14, the rate of expression of preMPF. Protein levels are measured at steady state and are dimensionless. Parameter values (dimensionless): k9 = k-9 = k-9′ =1.0, k9′ = k20[p53] = k22 =0.1, k15 =10−6, k21 = 0.01, k23 = k-23 =0.0. All integrations carried out in this paper use the routine lsode (35).
The protein p53 affects the MPF subsystem through v20 (corresponding to the expression of p21) and through the negative regulation of cyclin B expression as represented in v14. The effect of these p53 pathways on MPF will be shown later.
The Cdc25 Subsystem
Recent reports on the G2DDC points to Cdc25 as an important link with the cell cycle (3, 5). Important details of Cdc25 regulation are summarized in Fig. 4. The inactive form of Cdc25 (symbolized by iCdc25) is hypophosphorylated and active Cdc25 (symbolized by aCdc25) is obtained after further phosphorylation by the kinase Plk1 (18) and by MPF itself (13, 19). Thus, reaction 7 is given the expression v7 in Table 1. I will assume that [Plk1] is constant and is subsumed in the parameter kPlk1.
Figure 4.
The Cdc25 subsystem. The reaction numbers correspond to the rate expressions in Table 1.
When Cdc25 is phosphorylated on Ser-216, it was shown that 14–3–3 proteins bind with Cdc25 (3, 5, 20–22). Ser-216 phosphorylation of Cdc25 has been shown to be carried out by kinases CTAK1 (23) and Chk1 (22, 24) in vitro. [Recently, Matsuoka, Huang, and Elledge (25) identified Chk2, which, in vitro, could phosphorylate Cdc25 on Ser-216; I will not include this information in my present model.] Although I will assume it in my modeling of the G2DDC, it has not been demonstrated in vivo that Cdc25 phosphorylation on Ser-216 actually occurs in response to DNA damage. The rate expressions for Ser-216 phosphorylation of iCdc25 and aCdc25 are given by v2 and v2′ in Table 1. It is assumed that the Ser-216-phosphorylated inactive Cdc25 (symbolized by iCdc25Ps216) still can be activated by MPF and Plk1 as represented by reaction 18 with rate v18 in Table 1.
In accordance with the experiments of Kumagai et al. (20), I will assume that 14–3–3 proteins can bind only to the Ser-216-phosphorylated inactive form of Cdc25; the rate of this binding is given by v3′ in Table 1. I also assume that iCdc25Ps16/14–3–3 is dephosphorylated at Ser-216 after which 14–3–3 and iCdc25 are separated (4); this process has the rate v-3′ given in Table 1. The rate expressions for the dephosphorylation processes −7, −2, and −18 are given in Table 1.
The last member of this subsystem are the 14–3–3 proteins. According to Hermeking et al. (12), 14–3–3σ are strongly induced after γ-irradiation and by other DNA damaging agents, and the induction depends on p53; the rate of this process is given by v6 in Table 1. Lastly, in this subsystem, v13 is the rate of p53-independent expression of 14–3–3 proteins and v12 is their rate of degradation (see Table 1). I have not included in the present subsystem analysis the effect of 14–3–3 proteins on the intracellular localization of Cdc25 (4, 8) but will do so later in the simulation of the whole checkpoint system.
It has been reported that total Cdc25 levels do not vary significantly during the cell cycle (27). This statement is not true for the nuclear levels of Cdc25 (8) but for the present subsytem analysis I initially assume that the sum of all different species of Cdc25 is constant. The DNA damage signal through the Chk1 pathway affects v2 and v2′, and the p53 pathway affects v6 (see Fig. 4). Note that v7 and v18 are proportional to [MPF], which, in the present analysis of this isolated subsystem, is replaced by the total active Cdc25, i.e., {[aCdc25]+[aCdc25Ps216]}, to account for the positive feedback loop between Cdc25 and MPF.
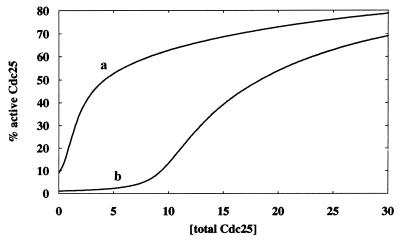
To understand the intrinsic kinetics of the Cdc25 subsystem, the differential equations involving [iCdc25], [iCdc25Ps216], [iCdc25Ps216/14–3–3], [aCdc25], [aCdc25Ps216], and [14–3–3] found in Table 2 are integrated. Fig. 5 shows a plot of the percentage of active Cdc25 (i.e., {[aCdc25] + [aCdc25Ps216]}) versus [total Cdc25]. The effect of two different strengths of the DNA damage signal influencing rates v2 and v2′ via Chk1 are shown in Fig. 5 where curve b corresponds to a 100-fold increase in the signal strength over that of curve a. Fig. 5 demonstrates that the percentage of active Cdc25 depends on [total Cdc25] and the strength of the damage signal. For example, when [total Cdc25] = 5 (see Fig. 5), a 100-fold increase in the damage signal decreases the activity from 50% to 3%. Increased [total Cdc25] renders the system less sensitive to the damage signal.
Figure 5.
Percentage of active Cdc25 (measured at steady state) versus total Cdc25. Curve b corresponds to a 100-fold increase in the DNA damage signal strength over that of curve a. Parameter values (dimensionless) for curve a: k2([Chk1P]+[CTAK1]) = kPlk1 = k12 = kPlk1′ = 0.1, k-2 = k-3′ = k-7 = k-18 = k6[p53] = 0.01, k2′([Chk1P]+[CTAK1]) = k3′ = k7 = k13=k18=1.0. Parameter values for curve b are exactly the same as those of curve a except k2([Chk1P]+[CTAK1]) = 10 and k2′([Chk1P]+[CTAK1]) = 100.
The Wee1 Subsystem
This subsystem is shown in Fig. 6. Observed levels of Wee1 proteins (13) do not allow us to assume that the total Wee1 is constant and therefore I introduce reactions 16 (Wee1 expression) and 17 (Wee1 degradation) with rates v16 and v17, respectively (see Table 1). It has been observed that Wee1 is transiently hyperphosphorylated during M-phase and then degraded (see ref. 13 and references therein). In a recent paper, Michael and Newport (28) showed that Wee1 was degraded in a Cdc34-dependent manner (in Xenopus egg extracts); furthermore, those authors concluded that Wee1 degradation is required for entry into mitosis and that this degradation is inhibited when DNA replication is blocked.
Figure 6.
The Wee1 subsystem. The reaction numbers correspond to the rate expressions in Table 1.
The regulation of the activity of Wee1 by MPF (13) is represented by the first term in the rate v8 in Table 1. Although Chk1 has been shown to phosphorylate Wee1 in vitro (29), this phosphorylation does not alter Wee1 activity toward Cdc2; hence, Chk1 is not assumed to affect process 8. The second term in the expression for v8 accounts for MPF-independent phosphorylation of Wee1. The dephosphorylation of Wee1P (hyperphosphorylated form of Wee1) is assumed to occur with the rate v-8 in Table 1.
The kinetic equations of the Wee1 subsystem are given in Table 2. Because the DNA damage signal via either Chk1 or p53 pathway does not affect v8 or v-8, an analysis of the effect of the DNA damage signal on the Wee1 subsystem separate from the MPF or Cdc25 subsystems is not necessary. Indeed, my previous mathematical analysis of the coupling between the MPF and Wee1 subsystems (31) has suggested that Wee1 may not be an essential target of Chk1.
The DNA Damage Signal Transduction Subsystems
G2 DNA damage signal transduction includes the Rad3/Chk1 and the p53 pathways (see Fig. 7). These pathways impinge on the subsystems described above. The Rad3/Chk1 pathway is still poorly understood, and other proteins that could be involved are still being identified (4). In humans, Rad3 homologs are the ataxia telan-giectasia mutated (ATM) and ATM-related kinases. Recent results suggest that the Rad proteins are activated as a result of DNA damage and act upstream of Chk1. Chk1 is found to be phosphorylated as a result of the damage signal (22, 24, 32).
Figure 7.
DNA damage signal transduction pathways. The reaction numbers correspond to the rate expressions in Table 1.
I will assume in my model that Rad3/ATM protein is activated at some fixed rate v4 because of DNA damage. This protein is assumed degraded at a rate v5 (see Table 1). It also is assumed that a fixed level of total Chk1 exists. Recent work of Matsuoka, Huang, and Elledge (25) also has pointed to another protein kinase, Chk2, which also phosphorylates Cdc25; for simplicity, I will not include Chk2 in my present model.
The other important member of the damage signal transduction is p53. It is well known that G1 arrest involves p53 as a major player (33). Recent work of Hermeking et al. (12) showed that the induction of 14–3–3 proteins is mediated by p53; and recent experiments of Bunz et al. (6) have shown that, indeed, p53 is also important in G2 arrest of the cell cycle. I will assume that because of DNA damage, p53 is activated at the rate v10 (see Table 1) and degraded or inactivated at the rate v11. Recent results (30, 34) suggest that ATM also could contribute to the phosphorylation of p53 in vivo; thus, the Rad3/Chk1 and p53 pathways could be coupled but, for simplicity, my computer simulations assume otherwise.
Kinetics of the Checkpoint System
The entire set of differential equations representing the kinetics of the coupled subsystems is given in Table 2. In the integration and computer simulations of the whole G2DDC system, and as far as understanding the qualitative dynamical behavior of the system, I have assumed that the effects of Plk1 and CTAK1 can be ignored. To simulate the intracellular distribution of Cdc25, I have included a rate vex for the nuclear export of 14–3–3-bound Cdc25 (see ref. 8), and a rate vin for the rate of nuclear import of iCdc25 (see Fig. 8 for expressions of vex and vin).
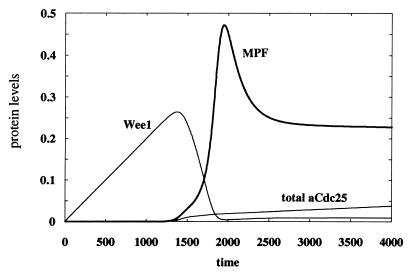
Figure 8.
Active MPF, total active Cdc25, and Wee1 as a function of time. No DNA damage is assumed here, i.e., k4 = k10 = 0. Nuclear export of iCdc25Ps216/14–3–3 has the rate vex = kex[iCdc25Ps216/14–3–3] with kex = 1, and nuclear import of iCdc25 has the rate vin = 10−5. Each of the following parameters has the value 1.0: k1, k5, k7, k18, k9, k11, k13, k-8, k-9, k-23, k14′; each of the following is equal to 0.1: k8, k12, k22, k23, k20; each of the following is equal to 0.01: k6, k17, k2, k2′, k-2, k-7, k-18, k21; the following are set to zero: kPlk1, kPlk1′, kctak1, kctak1′, k8′, k9′, k-3′; k-1 = 10; k14 = 5 × 10−4; k15 = 10−2; k16 = 2 × 10−4; k3′ = 100; [totalChk1] = 1. Initial conditions: [Chk1P] = [iCdc25] = [aCdc25] = [preMPF] = 10−6; [MPF] = 10−8; [iCdc25Ps216] = 2 × 10−5; [Wee1] = 10−3; [iCdc25Ps216/14–3–3] = 0.03; [14–3–3] = 2; all other species are zero. All parameters and variables are dimensionless.
Fig. 8 gives the temporal variations of MPF, Wee1, and total active Cdc25 in the absence of DNA damage. As foreshadowed by the schematic diagram given in Fig. 1, Wee1 decreases sharply as soon as MPF starts increasing autocatalytically. Increasing MPF activity leads to increased phosphorylation and degradation of Wee1 (reactions 8 and 17 in Fig. 6). This result agrees well with Michael and Newport (28) who suggested that Wee1 degradation is a prerequisite for entry into mitosis. Another interesting feature of the kinetics shown in Fig. 8 is the identical switch-on time for the activities of MPF and Cdc25, which is caused by the positive feedback loop between these two proteins (and is reminiscent of the phenomenon of transcritical bifurcation explained in ref. 31).
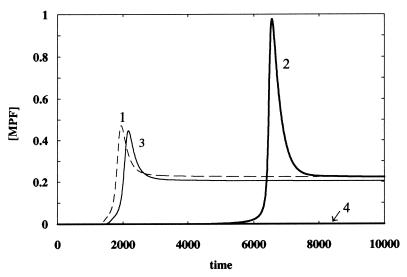
To simulate the DNA damage signal transduction, I investigated the individual as well as combined effects of the Rad3/Chk1 pathway and the p53 pathway on the activation of MPF. Activation of p53 is assumed to increase the rates of reactions 20 and 6 (see Figs. 2 and 4, respectively) and decrease the rate of process 14 (see Fig. 2). The values of the parameters k20, k6 and k14′ associated with the p53 pathway were assigned arbitrarily (see Fig. 8 for values) because no experimental measurements are available. Fig. 9 shows the activity of MPF when there is no DNA damage (curve 1), when only the Rad3/Chk1 pathway is operational (curve 2), when only the p53 pathway is operational (curve 3), and when both pathways are operational (curve 4). The simulations in Fig. 9 were performed to demonstrate a possible explanation of the experiments reported by Bunz et al. (6) regarding the necessity of p53 in sustaining G2 cell cycle arrest after DNA damage. With the parameter values chosen for Fig. 9, the DNA damage signal through the Rad3/Chk1 pathway generates a substantial delay in the switch-on time for MPF (curve 2). However, this delay leads to a doubling of the MPF peak activity (compared with that of curve 1), which then could allow the cell to overcome the G2 delay. Although the choice of parameters for the p53 signal transduction pathway results only in a minor disturbance on MPF activity (curve 3), coupling between the p53 pathway and the Rad3/Chk1 pathway leads to the abrupt disappearance of MPF activity and hence a sustained G2 arrest (curve 4).
Figure 9.
Effect of the DNA damage signal transduction pathways on MPF. Curve 1: no DNA damage, k4 = k10 = 0; curve 2: Chk1 pathway is on, p53 pathways are off, k4 = 0.2, k10 = 0; curve 3: p53 pathways are on, Chk1 pathway is off, k4 = 0, k10 = 0.2; curve 4: both Chk1 and p53 pathways are on, k4 = k10 =0.2. Other parameters and initial conditions are the same as in Fig. 8.
Conclusions
I have presented a detailed model of the G2DDC system based on known or postulated pathways reported in the current literature. A major difficulty in a kinetic study of this complex system is that practically no data on the rate parameters are available, which would render such a quantitative modeling task seemingly pointless. Nevertheless, an important question that could be decided by my detailed G2DDC model is whether or not there exists a set of kinetic parameters that could generate all the well-established experimental behavior of the system. If no such set of parameters exists, then the model must be modified by adding other necessary pathways and/or deleting questionable ones. This is really an essential objective that was carried out in my initial study of the detailed model presented here.
I have shown (Fig. 8) that, because of the mutual negative regulation of each other’s activities, Wee1’s degradation is necessary for MPF’s activation and subsequent entry into mitosis, in accordance with the report of Michael and Newport (28). I also have demonstrated (Fig. 9) that, under certain parameter values, a G2 arrest after DNA damage cannot be sustained by the Rad3/Chk1 pathway without the help of the p53 pathway, in agreement with the experiments of Bunz et al. (6); however, this may only be a special case because my detailed model suggests several points where the checkpoint signal transduction pathways could impinge on the intrinsic G2-M cell cycle machinery and the efficacy of these pathways in generating a G2 arrest could vary according to cell types and other conditions. For example, the Rad3/Chk1 pathway alone can shut off MPF activity by increasing the value of the parameter k4 in Fig. 9. As shown in Figs. 2 and 4, points where the DNA damage signal would influence the cell cycle include reactions 2, 2′, 6, 20, and 14.
Despite the complexity of the different subsystems of the G2DDC, understanding the switching behavior of this checkpoint system is facilitated by analyzing the consequences of the positive coupling between the phosphorylation-dephosphorylation (PD) cycles of MPF and Cdc25. In an earlier analysis of the G2DDC (31), I emphasized the importance of the existence of a transcritical bifurcation point inherent in the coupling between these PD cycles; this bifurcation point gives a threshold value above which both activities of MPF and Cdc25 are switched on at the same time, as seen in Fig. 8. According to ref. 31, the transcritical bifurcation point is given by a critical value of the product p = [total MPF] × [total Cdc25] where values inside the square brackets are total concentrations. The critical value of p is a function of the kinetic parameters involved in the PD cycles of MPF and Cdc25. If either [total MPF] or [total Cdc25] decreases so that their product p is less than the critical value, then both activities of MPF and Cdc25 are turned off. Thus, the nuclear export of 14–3–3-bound Cdc25 could lead to a subcritical value of p in the nucleus and turns MPF activity off, in agreement with Lopez-Girona et al. (8). The observation of Toyoshima et al. (9) on the nuclear export of cyclin B1 and its role in the G2DDC is also readily explained by the resulting decrease in total MPF, which would lower p below its critical value. Lastly, the recent report of Innocente et al. (7) on the negative regulation of cyclin B1 expression by p53 (leading to a decrease in total MPF in the nucleus), which induces a G2 arrest, also could be explained in terms of a resulting subcritical value of the product p. It would be very interesting if such a critical value of p can be experimentally demonstrated to exist.
Acknowledgments
I thank Paul Graves for stimulating discussions and helpful comments. This work is supported by a grant from the Natural Sciences and Engineering Research Council of Canada and a Laurentian University Research Fund grant.
ABBREVIATIONS
- CDK
cyclin-dependent kinase
- MPF
maturation-promoting factor
- G2DDC
G2 DNA damage checkpoint
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Murray A, Hunt T. The Cell Cycle. New York: Freeman; 1993. [Google Scholar]
- 3.Nurse P. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 4.Russell P. Trends Biochem Sci. 1998;23:399–402. doi: 10.1016/s0968-0004(98)01291-2. [DOI] [PubMed] [Google Scholar]
- 5.Weinert T. Science. 1997;277:1450–1451. doi: 10.1126/science.277.5331.1450. [DOI] [PubMed] [Google Scholar]
- 6.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 7.Innocente S A, Abrahamson J L A, Cogswell J P, Lee J M. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nature (London) 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg M, Horn F J M. Chem Eng Sci. 1974;29:775. [Google Scholar]
- 11.Novak B, Tyson J J. J Cell Sci. 1993;106:1153–1168. doi: 10.1242/jcs.106.4.1153. [DOI] [PubMed] [Google Scholar]
- 12.Hermeking H, Lengauer C, Polyak K, He T-C, Zhang L, Thiagalingam S, Kinzler K W, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 13.Poon R Y C, Chau M S, Yamashita K, Hunter T. Cancer Res. 1997;57:5168–5178. [PubMed] [Google Scholar]
- 14.Bates S, Ryan K M, Phillips A C, Vousden K H. Oncogene. 1998;17:1691–1703. doi: 10.1038/sj.onc.1202104. [DOI] [PubMed] [Google Scholar]
- 15.Cai K, Dynlacht B D. Proc Natl Acad Sci USA. 1998;95:12254–12259. doi: 10.1073/pnas.95.21.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pines J. Biochim Biophys Acta. 1997;1332:M39–M42. doi: 10.1016/s0304-419x(96)00042-x. [DOI] [PubMed] [Google Scholar]
- 17.Harper J W. Cancer Surv. 1997;29:91–107. [PubMed] [Google Scholar]
- 18.Kumagai A, Dunphy W G. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 19.Izumi T, Maller J L. Mol Biol Cell. 1993;4:1337–1350. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai A, Yakowec P S, Dunphy W G. Mol Biol Cell. 1998;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng C-Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 23.Peng C-Y, Graves P R, Ogg S, Thoma R S, Byrnes M J, Wu Z, Stephenson M T, Piwnica-Worms H. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 24.Furnari B, Rhind N, Russell P. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka S, Huang M, Elledge S J. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 26.Zheng X-F, Ruderman J V. Cell. 1993;75:155–164. [PubMed] [Google Scholar]
- 27.Girard F, Strausfeld U, Cavadore J-C, Russell P, Fernandez A, Lamb N J C. J Cell Biol. 1992;118:785–794. doi: 10.1083/jcb.118.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael W M, Newport J. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell M J, Raleigh J M, Verkade H M, Nurse P. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 31.Aguda B D. Oncogene. 1999;18:2846–2851. doi: 10.1038/sj.onc.1202462. [DOI] [PubMed] [Google Scholar]
- 32.Walworth N C, Bernards R. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 33.Carr A M. Science. 1996;271:314–315. doi: 10.1126/science.271.5247.314. [DOI] [PubMed] [Google Scholar]
- 34.Canman C E, Lim D-S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 35.Hindmarsch A C. ACM Signum Newslett. 1980;15:10. [Google Scholar]