Abstract
Resistance conferred by the Mi-1 gene from Solanum peruvianum is effective and widely used for limiting root-knot nematode (Meloidogyne spp.) yield loss in tomato (Solanum lycopersicum), but the resistance is ineffective at soil temperatures above 28°C. Previously, we mapped the heat-stable resistance gene Mi-9 in Solanum arcanum accession LA2157 to the short arm of chromosome 6, in a genetic interval as Mi-1 and the Cladosporium fulvum resistance gene Cf2. We developed a fine map of the Mi-9 region by resistance and marker screening of an F2 population and derived F3 families from resistant LA2157 × susceptible LA392. Mi-1 intron 1 flanking primers were designed to amplify intron 1 and fingerprint Mi-1 homologs. Using these primers, we identified seven Mi-1 homologs in the mapping parents. Cf-2 and Mi-1 homologs were mapped on chromosome 6 using a subset of the F2. Cf-2 homologs did not segregate with Mi-9 resistance, but three Mi-1 homologs (RH1, RH2, and RH4) from LA2157 and one (SH1) from LA392 colocalized to the Mi-9 region. Reverse transcriptase-polymerase chain reaction analysis indicated that six Mi-1 homologs are expressed in LA2157 roots. We targeted transcripts of Mi-1 homologs for degradation with tobacco (Nicotiana tabacum) rattle virus (TRV)-based virus-induced gene silencing using Agrobacterium infiltration with a TRV-Mi construct. In most LA2157 plants infiltrated with the TRV-Mi construct, Mi-9-meditated heat-stable root-knot nematode resistance was compromised at 32°C, indicating that the heat-stable resistance is mediated by a homolog of Mi-1.
Root-knot nematodes (Meloidogyne spp.) are root endoparasites of numerous crops worldwide and are the most damaging nematode pest in agriculture (Sasser, 1980). Tomato (Solanum lycopersicum, formerly Lycopersicon esculentum; Peralta and Spooner, 2005) is a highly susceptible host of several species of root-knot nematodes and incurs yield losses from root-knot infection in warm temperate to tropical regions as well as in greenhouse and other controlled environment production systems. Yield of susceptible tomato cultivars can be reduced by 50% or more in infested fields (Johnson, 1998). The use of resistant cultivars carrying gene Mi-1 has proved to be highly effective as a nematode management strategy, and resistant cultivars produce normal yields on infested land (Roberts and May, 1986). As a result, this gene has been exploited extensively in the last two decades for modern tomato cultivar development.
Gene Mi-1 was introgressed into tomato from its wild relative Solanum peruvianum (formerly Lycopersicon peruvianum; Peralta and Spooner, 2005) and is the only commercially available source of resistance to root-knot nematodes in this crop. Mi-1 confers resistance to three species of root-knot nematodes: Meloidogyne arenaria, Meloidogyne incognita, and Meloidogyne javanica (Dropkin, 1969a). In addition to resistance to the three root-knot nematode species, Mi-1 also confers resistance to certain biotypes of potato aphid (Macrosiphum euphorbiae; Rossi et al., 1998; Vos et al., 1998) and two biotypes of the whitefly Bemisia tabaci (Nombela et al., 2003). The Mi-1 gene belongs to the class of resistance (R) genes that contains a coiled-coil, nucleotide-binding site (NBS), and Leu-rich repeats (LRRs; Milligan et al., 1998). Although Mi-1-mediated resistance has proven to be highly effective for root-knot nematode control, the resistance is inactive above 28°C soil temperature (Holtzmann, 1965; Dropkin, 1969b). The breakdown of Mi-1-mediated resistance due to high temperature has been reported in both greenhouse and field tomato production systems (Philis and Vakis, 1977; Tzortzakakis and Gowen, 1996; Noling, 2000). In fact, temperature sensitivity appears to be a characteristic of several root-knot nematode R genes, having been described in other crop species, including alfalfa (Medicago sativa; Griffin, 1969), sweet potato (Ipomoea batatas; Jatala and Russell, 1972), and cotton (Gossypium hirsutum; Carter, 1982).
Reproduced primarily by outcrossing, S. peruvianum is comprised of a genetically heterogeneous group of plants referred to as the S. peruvianum complex (Rick, 1979). This wild relative of tomato has proven to be a rich source of disease resistance (Atherton and Rudich, 1986). For example, new sources of root-knot nematode resistance have been identified in accessions of S. peruvianum (Ammati et al., 1985, 1986). Inheritance studies of some of these new resistance traits have revealed the presence of additional genes that segregate independently of Mi-1 (Cap et al., 1993; Veremis and Roberts, 1996b). These genes were distinguished according to resistance phenotype at high temperature or resistance to Mi-1-virulent nematode isolates (Veremis and Roberts, 1996a, 1996b). To date, only two of these novel loci have been mapped: the resistance in S. peruvianum accession 126443 clone 1MH and in accession LA2157 (Yaghoobi et al., 1995; Veremis et al., 1999). S. peruvianum accession 126443 clone 1MH has both heat-stable resistance and resistance to Mi-1-virulent root-knot nematodes (Yaghoobi et al., 1995). Both heat-stable resistance and resistance against virulent nematodes are mediated by single dominant genes. It is not clear, however, whether the heat-stable resistance and the resistance to Mi-1-virulent nematodes are mediated by the same gene or by tightly linked genes (Yaghoobi et al., 1995; Veremis and Roberts, 1996a).
In LA2157, an accession belonging to the ancient Maranon race complex of S. peruvianum from the Maranon drainage area located in northern Peru, the heat-stable root-knot nematode resistance is also mediated by a single dominant gene, Mi-9 (Veremis et al., 1999). Recently, the Maranon races from northern Peru were reclassified, and accession LA2157 was assigned to a new distinct species, Solanum arcanum (Peralta et al., 2005). Mi-9 confers resistance to Mi-1-avirulent isolates of M. arenaria, M. incognita, and M. javanica at 25°C and 32°C but does not confer resistance to Mi-1-virulent nematodes (Ammati et al., 1986; Veremis et al., 1999; Veremis and Roberts, 2000). Unlike most S. arcanum accessions, LA2157 is self compatible. Using a true F2 segregating population, Mi-9 was mapped to chromosome 6 (Veremis et al., 1999). Mi-9 was further mapped to the short arm of chromosome 6 between markers CT119 and C8B, a similar genetic interval as Mi-1 (Ammiraju et al., 2003).
Many R genes are members of gene families that seem to be clustered (Michelmore and Meyers, 1998). In these clusters, arrays of paralogs exist that confer resistance to members of distinct groups of pathogens or to multiple variants of a single pathogen (Kesseli et al., 1994; Bendahmane et al., 1999; van der Vossen et al., 2000). Pseudogenes and members with unknown functions also exist within these clusters. Clusters of R genes could be tightly organized or could be spaced over several megabases (Meyers et al., 1998; Noel et al., 1999). In the Mi-1 locus on the short arm of tomato chromosome 6, Mi-1 and six homologs exist in two distinct clusters about 300 kb apart (Vos et al., 1998; Seah et al., 2004). The cluster containing Mi-1 (also known as Mi-1.2) has two additional members, Mi-1.1 and Mi-1.3, and is located near the centromeric proximal end of the chromosome (Kaloshian et al., 1998; Milligan et al., 1998). Mi-1.3 is a pseudogene, while Mi-1.1 and Mi-1 are both transcribed genes with over 91% sequence identity (Milligan et al., 1998). Of these two genes, only Mi-1 conferred resistance to root-knot nematodes and insects (Milligan et al., 1998; Rossi et al., 1998; Nombela et al., 2003). The short arm of chromosome 6 is characterized by clusters of disease R genes besides Mi-1 and Mi-9. Cladosporium fulvum R genes Cf-2 and Cf-5 (Dixon et al., 1996, 1998), genes Ol-4 and Ol-6 conferring resistance to Oidium neolycopersici (Bai et al., 2005), alfalfa mosaic virus R gene, Am (Parrella et al., 2004), and possibly Ty-1 and Bw-5 conferring resistance to tomato yellow leaf curl virus and Ralstonia solanacearum, respectively (Zamir et al., 1994; Thoquet et al., 1996), also map to this region. Only Cf-2 and Cf-5 have been cloned and encode receptors with N-terminal LRR regions and transmembrane domains, suggesting that R genes with distinct motifs are also clustered (Dixon et al., 1996, 1998).
Because members of distinct classes of R genes could be clustered in the same chromosomal region, identifying which type of R gene confers resistance to a specific pathogen requires extensive fine mapping, and, possibly, transforming more than one R gene type into susceptible genotypes to identify the gene in question. Recent development of virus-induced gene silencing (VIGS) technology allows assessment of the functional role of genes by targeting their transcripts for degradation (Lu et al., 2003; Burch-Smith et al., 2004). This technique also could be used to target members of a gene family by eliminating functional redundancy and correlating their roles to a specific function. Alternatively, this approach could be used to assess whether a novel function is conferred by a member or members of a known gene family. In this article, we describe the use of VIGS to assess whether the heat-stable root-knot nematode R gene Mi-9 is a Mi-1 homolog. We use the word homolog as an inclusive term to refer to orthologs or paralogs. First, we describe a molecular genetic analysis of the Mi-9 region on the short arm of S. arcanum chromosome 6. We then identified Mi-1 homologs in the Mi-9 donor S. arcanum accession LA2157 that cosegregate with the heat-stable nematode resistance. Using VIGS targeted to silence Mi-1 homologs in S. arcanum LA2157, we showed that Mi-9 is likely a homolog of Mi-1.
RESULTS
Identifying Recombinants in the Mi-9 Region
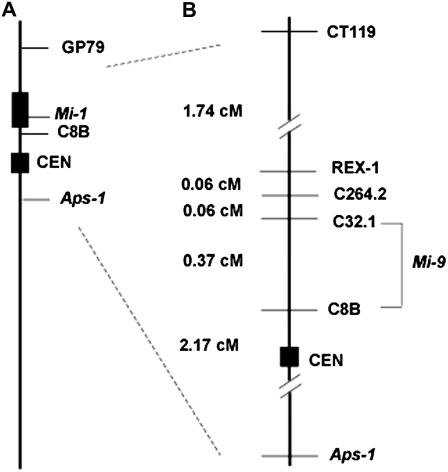
Earlier, Mi-9 was mapped between markers CT119 and C8B, located on the telomeric distal end and centromeric proximal end of the short arm of chromosome 6, respectively (Fig. 1A). To further delimit the Mi-9 location, we screened the F2 population used previously and derived from a cross between the heat-stable root-knot nematode resistance S. arcanum accession LA2157 and the root-knot nematode susceptible accession LA392 (Ammiraju et al., 2003). A total of 589 F2 progeny plants were scored for DNA markers CT119, Rex-1, C8B, and Aps-1 (Supplemental Table S1). Segregation ratios for CT119, Rex-1, and Aps-1 codominant markers were in agreement with the expected 1:2:1 ratios (P = 0.40 for CT119, P = 0.85 for Rex-1, and P = 0.36 for Aps-1).
Figure 1.
Genetic map of Mi-1 and Mi-9 on the short arm of tomato chromosome 6. A, Mi-1 location and flanking markers. Thick bar represents the introgressed region from S. peruvianum in the tomato cv Motelle. B, Mi-9 linkage map on the short arm of chromosome 6 generated by the mapping population S. arcanum accessions LA2157 × LA392. Numbers on the left of the vertical line indicate genetic distances (centimorgans).
Fifty-four recombinants were identified that had recombination between CT119 and Aps-1 (Table I). All F2 recombinants had a single recombination event. The largest group of recombinants, with 30 members, had a recombination event between Aps-1 and C8B (F2 classes 4–7; Table I). Within this group, the recombinant F2 class 5 separated Aps-1 and the heat-stable nematode resistance and localized Mi-9 above Aps-1, confirming the earlier finding. This finding was further supported by F3 segregation analysis of F2 families in class 4 and class 5. Heat-stable nematode resistance in the F3 populations from these families segregated in a 3 resistant:1 susceptible ratio as expected based on χ2 statistics (Supplemental Table S2). Initially, while performing F3 segregation marker analysis, we noticed unexpected segregation in F3 plants that originated from a single F2 fruit. We interpreted the surprising segregation results as the ability of our S. arcanum population to both self fertilize and outcross. To make sure the F3 seeds were from selfed F2 plants, immature flowers on F2 plants were bagged, and seeds from a single fruit were germinated and segregation of plants from the fruit was monitored.
Table I.
Segregation of Mi-9 phenotype and linked markers in F2 population of S. arcanum LA2157 × LA392
Plants were genotyped using PCR and RFLP markers. Genotype designation: (1) homozygous resistant locus, (2) homozygous susceptible, (3) heterozygous, 1/3 resistant allele is dominant, and 2/3 susceptible allele is dominant. Plants were also evaluated for nematode resistance (R) or susceptibility (S) to root-knot nematodes strain VW4 at 32°C.
| Plant | Attributes | No. of Plants | Phenotype | Markers
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CT119 | Cf-2 | Rex-1 | C264.1 | C32.1 | C8B | Aps-1 | ||||
| LA2157 | Parent | – | R | 1 | 1 | 1 | 1/3 | 1 | 1/3 | 1 |
| LA392 | Parent | – | S | 2 | 2 | 2 | 2 | 2/3 | 2 | 2 |
| F2 class 1 | Parental type | 124 | R | 1 | 1 | 1 | nda | nd | 1/3 | 1 |
| F2 class 2 | Parental type | 145 | S | 2 | 2 | 2 | nd | nd | 2 | 2 |
| F2 class 3 | Heterozygote | 264 | R | 3 | 3 | 3 | nd | nd | 1/3 | 3 |
| F2 class 4 | Recombinant | 6 | R | 3 | 3 | 3 | 1/3 | 2/3 | 3b | 1 |
| F2 class 5 | Recombinant | 8 | R | 3 | 3 | 3 | 1/3 | 2/3 | 3b | 2 |
| F2 class 6 | Recombinant | 5 | S | 2 | 2 | 2 | 2 | 2/3 | 2 | 3 |
| F2 class 7 | Recombinant | 11 | R | 1 | 1 | 1 | 1/3 | 1/1 | 1b | 3 |
| F2 class 8 | Recombinant | 5 | R | 3 | 3 | 3 | 1/3 | 2/3 | 1b | 1 |
| F2 class 9 | Recombinant | 1 | R | 2 | 2 | 2 | 2 | 3b | 3b | 3 |
| F2 class 10 | Recombinant | 1 | R | 1 | 1 | 1 | 3b | 3b | 3b | 3 |
| F2 class 11 | Recombinant | 8 | R | 2 | 2 | 3 | 1/3 | 2/3 | 3b | 3 |
| F2 class 12 | Recombinant | 2 | S | 3 | 3 | 2 | 2 | 2/3 | 2 | 2 |
| F2 class 13 | Recombinant | 1c | R | 1 | 3 | 3 | nd | nd | 1/3 | 3 |
| F2 class 14 | Recombinant | 6 | R | 2 | 3 | 3 | nd | nd | 1/3 | 3 |
nd, Not determined.
Allele designation is based on F3 progeny segregation.
Plant died before setting fruits.
Seventeen recombinants (F2 classes 11–14; Table I) were identified between CT119 and Rex-1, and 16 of these recombinants (F2 classes 11, 12, and 14) separated CT119 from Mi-9 and localized Mi-9 to the centromeric proximal end of the arm. Two additional RFLP markers, C264.1 and C32.1, located on the short arm of chromosome 6, were used to further map the recombinants (Table I; Kaloshian et al., 1998). One recombinant from F2 class 10, E27, had recombination between Rex-1 and C264.1 (Table I), and F3 progeny of this family segregated in a 3:1 ratio for heat-stable resistance (Supplemental Table S2), indicating that Mi-9 is located below Rex-1 (e.g. E27-H3, Fig. 2; Supplemental Table S3). One recombinant (F2 class 9; Table I) was also identified between markers C264.1 and C32.1, indicating that Mi-9 is located below C264.1. Five recombinants were identified between C32.1 and C8B (F2 class 8; Table I). The F3 population from these F2 families segregated in a 3 resistant:1 susceptible ratio for heat-stable resistance (Supplemental Table S2). Segregation analysis in four of these families (E42, R26, D52, and R35) indicated recombination between C8B and Mi-9, localizing Mi-9 above C8B in agreement with our earlier finding (Supplemental Tables S2 and S3). The F3 segregation of one of the F2 families (A56) in this class (e.g. A56-E3; Fig. 2) did not separate Mi-9 from C8B and localized Mi-9 below C32.1, further delimiting the location of Mi-9 on the centromeric proximal end of the short arm of chromosome 6. A genetic map of the Mi-9 region was developed by combining the F2 segregation data from this work and our previous results (Fig. 1, A and B).
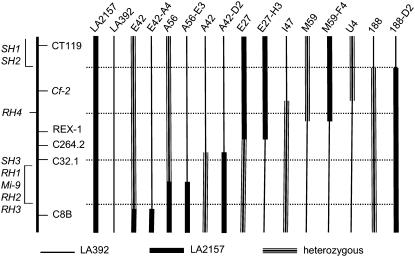
Figure 2.
Genetic map of Mi-9 and key S. arcanum recombinants. A map of the short arm of chromosome 6 is shown on the left. Mi-1 homologs RH1, RH2, RH3, and RH4 are from S. arcanum accession LA2157, while SH1, SH2, and SH3 are from accession LA392. The region in S. arcanum from accession LA2157 is represented by vertical thick bar, from accession LA397 is represented by vertical thin bar, and heterozygous region is represented by vertical multiple line bar. In recombinants, junctions between regions are depicted as midway between flanking markers.
Identification of R Gene Homologs on the Short Arm of Chromosome 6
Members of two distinct classes of R genes are localized to the short arm of chromosome 6 (Ho et al., 1992; Dickinson et al., 1993). These are the Mi-1 gene with coiled coil-NBS-LRR protein motifs and Cf-2 and Cf-5 genes with transmembrane and LRR protein motifs (Dixon et al., 1996, 1998). We developed markers for Cf-2 and Mi-1 and assessed whether Cf-2 or Mi-1 homologs cosegregated with the heat-stable nematode resistance. The Cf-2 primers amplified a fragment of 540 bp in both LA2157 and LA392 (Supplemental Table S1). Restriction with TaqI distinguished the two alleles by cleaving the amplified product from LA392 into two fragments of 380 bp and 160 bp, while the amplification product from LA2157 remained uncut (data not shown).
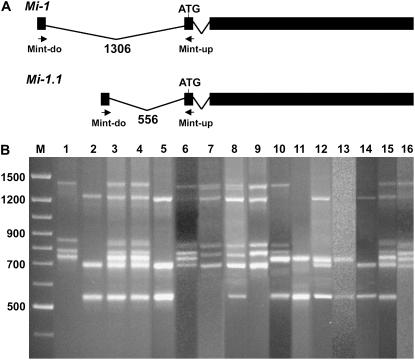
Mi-1 and six homologs are localized to the short arm of tomato chromosome 6 (Seah et al., 2004). To distinguish among Mi-1 homologs, primers Mint-up and Mint-do were designed that annealed to the intron 1 flanking regions (Fig. 3A). Using these primers in PCR with DNA from tomato containing Mi-1, we were able to amplify intron 1 from Mi-1.1 and Mi-1 (Supplemental Fig. S1). The size of the amplified fragments from Mi-1.1 and Mi-1 were 622 and 1,372 bp, respectively. We used intron 1 flanking primers in PCR with DNA from the source of Mi-9, LA2157, and four DNA fragments were amplified (Fig. 3B). We refer to these fragments as RH1, RH2, RH3, and RH4 with sizes 1,372, 844, 786, and 756 bp, respectively. Using the intron 1 primers in PCR with DNA from LA392, the susceptible parent in our genetic cross, three distinct fragments, SH1, SH2, and SH3, with sizes 1,235, 713, and 556, respectively, were amplified (Fig. 3B).
Figure 3.
Mi-1 and homologs in Solanum spp. A, Schematic diagram of Mi-1 and Mi-1.1 genes showing the relative positions of intron 1 flanking primers Mint-do and Mint-up (arrows). Thick horizontal lines represent transcripts and angled lines indicate introns. B, A composite image of Mi-1 homologs amplified using PCR and intron 1 flanking primers and resolved on 2% agarose gels. DNA source in lanes 1 to 16 are from: LA2157, LA392, I47, E42, E42-A4, A56, A56-E3, A42, A42-D2, E27, E27-H3, M59, M59-F4, U4, 188, and 188-D2.
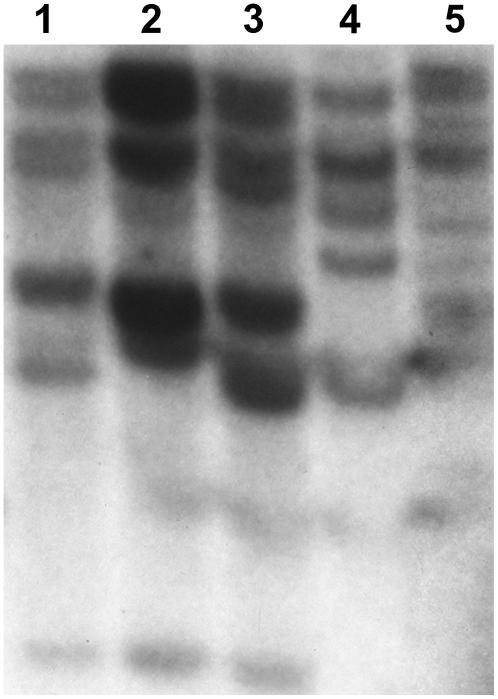
To determine the number of Mi-1 homologs in LA2157, DNA-blot analyses were performed using the NBS fragment from Mi-1 as a probe. The DNA-blot analysis indicated the presence of about six to seven Mi-1 homologs in LA2157 (Fig. 4).
Figure 4.
DNA-blot analysis of NBS fragment of Mi-1. Genomic DNA from tomato cv VFN (Mi-1/Mi-1; lane 1), cv UC82B (mi/mi, lane 2), cv Motelle (Mi-1/Mi-1; lane 3), S. arcanum accessions LA2157 (Mi-9/Mi-9; lane 4), and LA392 (mi/mi; lane 5) were digested with EcoRV, separated on 0.9% agarose gel, blotted onto nylon membrane, and hybridized with radiolabeled probe.
Mapping Cf-2 and Mi-1 Homologs on Chromosome 6
We scored the entire F2 population for Cf-2 alleles and identified 10 recombinants between Cf-2 and Rex-1 (F2 classes 11 and 12; Table I) localizing Cf-2 above Rex-1 to the telomeric distal end of the chromosome. In addition, seven recombinants were identified between Cf-2 and CT119 (F2 classes 13 and 14; Table I) localizing Cf-2 below CT119. Because all members of the recombinant F2 class 11 had heat-stable nematode resistant phenotype, this suggested that Mi-9 is not likely to be a Cf-2 homolog.
The segregation of Mi-1 homologs was determined in over 289 F2 plants that included all recombinant plants. All tested nematode resistant F2 plants with homozygous LA2157 markers located on the short arm of chromosome 6 displayed the four alleles from LA2157 (Supplemental Fig. S1). Moreover, all tested susceptible F2 plants homozygous for LA392 markers in this region displayed the three alleles from LA392 (Supplemental Fig. S1), and all tested heterozygous F2 plants displayed all seven members (data not shown). Taken together, these data suggest that the four Mi-1 homologs from LA2157 and the three homologs from LA392 are localized to the short arm of chromosome 6. This was further confirmed using the recombinants.
The recombinants in F2 classes 4, 5, 8, 11, and 14 displayed all seven Mi-1 homologs (class 4 and 5, data not shown; e.g. E42 and A56 class 8, I47 class 11, 188 class 14; Fig. 3B). All recombinants in F2 class 7 had LA2157 Mi-1 alleles, while all recombinants in the F2 class 6 had LA392 Mi-1 alleles (data not shown). Crossover events among the Mi-1 homologs were identified in members of the F2 classes 8 to 10 and class 12. F3 segregation of four of the F2 class 8 families (E42-A4, R26-D6, D52-B5, and R35-B1; Supplemental Table S3; e.g. E42-A4, Figs. 2 and 3B) indicated that the recombination points are between C32.1 and C8B and that all seven Mi-1 alleles are located above C8B (Fig. 2). Segregation of the F3 population (e.g. A56-E3) of one member of this class, A56, not only supported this finding but also separated Mi-1 homolog RH4 from the three other homologs, RH1, RH2, and RH3, from LA2157 (Figs. 2 and 3B). The location of Mi-1 homologs RH4 above C32.1 was further confirmed by segregation of homologs in A42 and E27-H3 (Figs. 2 and 3B). Mi-1 homolog profiles in M59 and U4 further localized RH4 between Cf-2 and Rex-1 (Figs. 2 and 3B).
The recombination mapping also uncovered the crossover points among Mi-1 homologs from LA392. Segregation of Mi-1 homolog SH3 in E27, E27-H3, and A42-D2 indicates that SH3 is located below C32.1 (Figs. 2 and 3B), while Mi-1 homolog profiles from M59-F4 and 188-D2 indicated that SH1 and SH2 are located above Cf-2 (Figs. 2 and 3B).
Sequence Relationships of Mi-1 Homologs
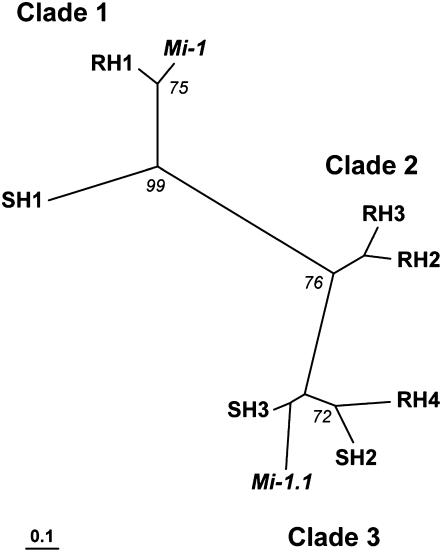
To identify the relationship among the Mi-1 homologs in LA2157 and LA392, the seven amplified intron fragments were sequenced (Supplemental Fig. S2). Fragments representing Mi-1.1 and Mi-1 intron 1 amplified from cv Motelle were also sequenced. Phylogenetic analysis grouped the intron sequences to three distinct clades (Fig. 5). Introns RH1 and SH1 grouped with intron 1 from Mi-1, while RH4, SH2, and SH3 grouped with intron 1 from Mi-1.1 (Fig. 5). A third clade was denoted by RH2 and RH3, and both members had higher sequence identity to Mi-1.1 than Mi-1. Sequence analysis indicated over 99% and 78% sequence identity between Mi-1 intron 1 and RH1 and SH1, respectively. Sequence identity between Mi-1.1 and Mi-1 intron 1 was only 32%. In contrast, intron 1 sequence identity between Mi-1.1 and RH4, SH2, and SH3 ranged between 63% and 77% (Fig. 5). The striking difference among all the intron 1 sequences is the presence of indels scattered along intron 1. One major deletion of about 740 bp differentiates between Mi-1 and the members in clade 2 and clade 3 (Fig. 5; Supplemental Fig. S2).
Figure 5.
An unrooted parsimony tree showing the differentiation of the three groups of Mi-1 intron 1 homologs. The phylogenetic analysis includes Mi-1.1, Mi-1, and homologs from S. arcanum accessions LA2157 (resistance parent homologs, RH) and LA392 (susceptible parent homologs, SH), namely RH1 (EF028059), RH2 (EF028056), RH3 (EF028057), RH4 (EF028058), SH1 (EF028060), SH2 (EF028061), and SH3 (EF028062). Numbers at nodes represent bootstrap values. Scale bar indicates number of substitution per site.
VIGS in S. arcanum LA2157
Mi-1 homologs cosegregated with the Mi-9 heat-stable resistance, which suggested that Mi-9 could be a homolog of Mi-1. To determine whether Mi-9 is a homolog of Mi-1, we targeted transcripts of Mi-1 homologs for degradation using tobacco (Nicotiana tabacum) rattle virus (TRV)-based VIGS. TRV is a bipartite virus (TRV RNA1 [TRV1] and TRV RNA2 [TRV2]) and has been used effectively as a VIGS vector to silence genes in roots from Solanaceae (Ryu et al., 2004; Valentine et al., 2004). To examine whether TRV could be used efficiently to silence genes in S. arcanum, we infiltrated accession LA2157 with an Agrobacterium culture containing TRV1 and TRV2 carrying a fragment of tomato phytoene desaturase gene (TRV-PDS; Liu et al., 2002). Because temperature has an effect on the efficiency of VIGS in tomato (Ekengren et al., 2003), we tested TRV-VIGS at 19°C and 24°C. All LA2157 plants infiltrated with TRV-PDS and maintained at 19°C showed photo-bleaching symptoms (Supplemental Fig. S3A), while only 22% of the infiltrated plants maintained at 24°C showed the symptoms. Silencing within a plant was also more efficient when plants were maintained at 19°C compared to 24°C (Supplemental Fig. S3B).
Silencing of Mi-1 Homologs in Accession LA2157 Using TRV-VIGS
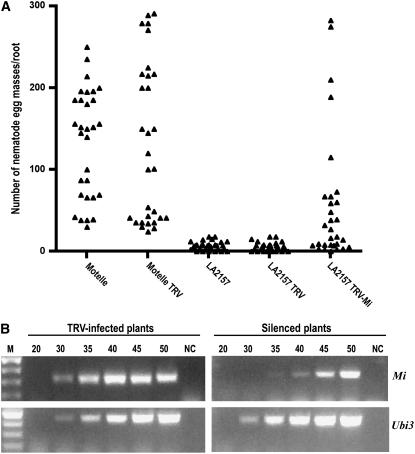
To target Mi-1 homologs in VIGS, a TRV-Mi clone was used (Li et al., 2006). Seedlings of LA2157 and tomato cv Motelle agroinfiltrated with empty vector TRV, TRV-Mi construct, or buffer controls were assayed for heat-stable root-knot nematode resistance at 32°C. Tomato cv Motelle was used as a positive control for Mi-1-mediated nematode resistance breakdown at high temperature. Eight weeks after nematode inoculation, egg masses developed on roots of tomato cv Motelle infiltrated with buffer, indicating success in breaking the Mi-1-mediated resistance (Fig. 6A). In addition, similar numbers of egg masses were developed on Motelle treated with buffer or agroinfiltrated with an empty TRV vector, indicating that Agrobacterium culture and/or TRV did not interfere with nematode infection. All LA2157 plants infiltrated either with buffer or TRV developed low numbers of egg masses, indicating that Mi-9-mediated heat-stable resistance is functional in these plants and that Agrobacterium and/or TRV did not hinder Mi-9-mediated resistance. In contrast, a subset of LA2157 plants agroinfiltrated with the Mi-1 VIGS construct developed large numbers of egg masses, indicating attenuation of Mi-9-mediated heat-stable resistance.
Figure 6.
TRV-Mi VIGS attenuated Mi-9-mediated heat-stable root-knot nematode resistance in S. arcanum accession LA2157. A, Root-knot nematode infection on roots of buffer-infiltrated controls or agroinfiltrated with pTRV1 plus pTRV2 empty vector (TRV) or with pTRV1 plus pTRV2-Mi (TRV-Mi). Triangles represent number of egg masses per root system. Two independent experiments were performed. In each experiment, 15 plants per treatment were used. Data from both experiments are presented. B, Effect of TRV-Mi VIGS on Mi-1 transcript levels in silenced and control roots. Ethidium bromide stained 1.5% agarose gels with RT-PCR products. cDNA was synthesized from total RNA isolated from roots of plants agroinfiltrated with TRV or TRV-Mi. Samples from TRV-Mi infiltrated plants were from roots portions supporting nematode reproduction. Amplification of the tomato ubiquitin Ubi3 gene was used as an internal control for equal cDNA use from control and silenced plants. PCR cycles are as indicated on the top of the sections. Lane M indicates DNA marker, and NC indicates negative control where RNA was used as template in the absence of reverse transcriptase.
Root sections of LA2157 agroinfiltrated with the Mi-1 VIGS construct and harboring egg masses were collected for RNA extraction and used in qualitative evaluation of the relative abundance of Mi-1 transcripts. RNAs from five different roots were subjected to reverse transcription (RT)-PCR analysis using Mi-1 primers (Li et al., 2006). All indicated reduction in Mi-1 transcript levels compared to TRV only agroinfiltrated plants (Fig. 6B). Sequence information of RT-PCR product from uninfected LA2157 root RNAs indicated that six Hmr1 (for homologs of Meloidogyne resistance gene Mi-1) transcripts were amplified with the Mi-1 primers (Supplemental Fig. S4).
DISCUSSION
In this article, we report that the heat-stable root-knot nematode resistance gene Mi-9 is a homolog of Mi-1. Our strategy was based on a combination of candidate gene approach and functional analysis without cloning. The short arm of tomato chromosome 6 is a rich source of disease R genes, and two distinct R gene groups have been cloned from this portion of the chromosome. These are Cf-2 and Cf-5 with LRR and transmembrane domains and Mi-1 with NBS-LRR domains (Dixon et al., 1996, 1998; Kaloshian et al., 1998; Milligan et al., 1998). We considered the possibility that either a homolog of Cf or Mi-1 could confer the heat-stable nematode resistance. Fine mapping ruled out the possibility of a Cf homolog in this role. Because Mi-1 homologs are clustered in two distinct groups, an easily distinguishable feature among Mi-1 homologs was necessary to further map and identify the source of resistance. Only members of the cluster at the centromeric proximal end that included Mi-1, Mi-1.1, and Mi-1.3 have been cloned and sequenced (Milligan et al., 1998). The feature that clearly distinguished between Mi-1 and Mi-1.1 was the size of the first intron. Furthermore, our work indicated that the size of intron 1 also allowed the distinction of several Mi-1 homologs in S. arcanum accessions LA2157 and LA392. However, the PCR approach to amplify intron 1 did not detect all Mi-1 homologs in either accession. DNA-blot analysis indicated the presence of five to seven Mi-1 homologs in these accessions, and RT-PCR indicates six transcribed Mi-1 homologs in LA2157 (Fig. 4; Supplemental Fig. S4). Nevertheless, the PCR approach allowed the distinction among seven Mi-1 homologs and indicated that the intron 1 flanking sequences are conserved in four and three homologs from accessions LA2157 and LA392, respectively.
Our data indicated that three Mi-1 homologs cosegregate with the heat-stable resistance in LA2157. However, it is not clear which one of the three members confers the heat-stable resistance and whether any of these members is a pseudogene. Although six distinct Mi-1 homolog transcripts were identified in LA2157, a subset of these transcripts could be of pseudogene origin. Several Mi-1 homologs are pseudogenes and are expressed in root-knot nematode susceptible and resistant tomato (S. Seah and V. M. Williamson, personal communication). In the S. peruvianum introgressed region, within the Mi locus at the centromeric proximal region, Mi-1 and two homologs (Mi-1.1 and Mi-1.3) are present. Mi-1 and Mi-1.1 have intron 1, but Mi-1.3 does not have any detectable introns (Milligan et al., 1998). Although the primers used to amplify intron 1 will not identify members without introns, all three Mi-1 homologs from LA2157 (RH1, RH2, and RH3) located in the centromeric proximal cluster have intron 1. However, phylogenetic analysis of the intron 1 sequences of Mi-1.1, Mi-1, and homologs from LA2157 and LA392 did not distinguish between members of the two clusters, indicating that extensive sequence exchange among homologs in these clusters has occurred.
S. peruvianum sensu lato is a heterogeneous species complex, and the northern races of this species were considered the ancestral progenitor of the Solanum complex (Rick, 1986; Spooner et al., 2005). Both parental accessions LA2157 and LA392 are from the northern races of S. peruvianum recently renamed as S. arcanum (Peralta et al., 2005). As demonstrated in this work, considerable polymorphism exists between these two accessions. However, our recombination analysis indicates that the marker order is consistent among these two S. arcanum accessions and the introgressed region in tomato from S. peruvianum (Kaloshian et al., 1998). S. peruvianum s. str. PI128657 is the accession that Mi-1 was introduced from and was collected from the southern region of Peru (Veremis and Roberts, 2000). Comparing markers in the Mi region between S. lycopersicum and the S. peruvianum introgressed region, an inversion encompassing Rex-1 and C264.2 has been demonstrated (Seah et al., 2004). The presence of this inversion partly explains the lack of recombination in this region in crosses between root-knot nematode susceptible tomato and resistant tomato that contains S. peruvianum-introgressed region (Kaloshian et al., 1998; Seah et al., 2004). Although the suppression of recombination is partly attributed to the presence of the S. peruvianum-introgressed genome, the suppression is more pronounced in the region spanning the inversion (Liharska et al., 1996; Kaloshian et al., 1998; Seah et al., 2004). While several recombination events were identified in close proximity to the Mi locus, no recombination between C264.2 and Rex-1 was previously reported (Kaloshian et al., 1998). Earlier, the order of these two markers was determined by a physical map using yeast artificial chromosomes and cosmid subclones (Kaloshian et al., 1998).
In this work, we identified a recombination event between these two markers that localized Rex-1 above C264.2 on the telomeric end of the short arm of chromosome 6, in agreement with the physical map of the Mi-1 introgressed region. Because the Rex-1 and C264.2 positions are the same in S. peruvianum accession PI128657 and in S. arcanum accessions LA2157 and LA392, it is therefore likely that the inversion happened in S. lycopersicum after divergence from S. peruvianum sensu lato. Because the previous mapping information was from only one representative of each S. lycopersicum and S. peruvianum genomes, it was difficult to determine in which species the inversion happened (Seah et al., 2004).
Our recombination data mapped CT119 above Cf-2 in S. arcanum. In Solanum pimpinellifolium, the source of Cf-2, CT119 is localized below Cf-2 (Dixon et al., 1995). Cf-2 is a member of a gene family, and different numbers of homologs are reported from S. lycopersicum, which lacks Cf-2, compared to S. pimpinellifolium. In addition, the region spanning CT119 and Cf-2 is about 30 kb in S. lycopersicum (Dixon et al., 1995, 1996). In our population, we identified seven recombinants between CT119 and Cf-2, suggesting that the member amplified in the S. arcanum in our experiments is different than the members in S. pimpinellifolium, and/or this region in S. arcanum encompasses a recombination hot spot.
We also demonstrated in this work that TRV can be used as a functional tool for VIGS in S. arcanum. TRV-VIGS is highly efficient in S. arcanum LA2157 where 100% of the plants infiltrated with TRV-PDS showed photo-bleaching symptoms. Along with S. arcanum, we also tested the efficiency of TRV-VIGS using TRV-PDS in S. peruvianum and S. pimpinellifolium and found high efficiency of silencing in both species (K. Bhattarai and I. Kaloshian, unpublished data). Although TRV-VIGS is highly efficient in LA2157, silencing is not uniform within a plant and within a single leaflet, as visualized by PDS silencing (Supplemental Fig. S3A). The patchy pattern of silencing of TRV-VIGS has also been observed in above-ground parts of tomato (Liu et al., 2002; Ekengren et al., 2003). In TRV-Mi infiltrated roots, root-knot nematode egg masses were clustered in a limited number of patches on the root system, suggesting that the patchy nature of TRV-VIGS in below-ground parts (roots) mirrors the above-ground parts (shoots).
The observed variation in number of egg masses on roots of LA2157 is partly due to the variable efficiency of TRV-Mi silencing and efficiency of TRV-VIGS in roots (Valentine et al., 2004). Comparing TRV-Mi silencing in tomato leaves and roots using aphid and nematode assays, respectively, suggests that silencing in roots is less efficient than in leaves (I. Kaloshian, unpublished data). The ability of nematodes to infect and develop is based on interaction of the nematode with a few distinct root cells where feeding is initiated. If Mi-9 is silenced in these cells, then the nematode is able to initiate successful parasitism and complete its life cycle. However, the absence of Mi-9 silencing in even a few cells at the site of nematode feeding will inhibit parasitism. Variation in numbers of egg masses also could be due to high temperature breaking the Mi-1-mediated resistance. Variable numbers of egg masses were also observed in the Motelle infiltrated with buffer treatment, indicating that temperature may have played a role in nematode infectivity.
Combining a fine-mapping strategy, candidate gene approach with VIGS allowed us to determine the likely nature of Mi-9 without cloning. The development of wide host range virus vectors such as TRV will facilitate the adoption of this approach to a wide variety of plant species and accelerate the identification of the nature of the R genes prior to cloning. This approach will be especially valuable in regions where R genes have been mapped and where clusters of R genes with distinct motifs reside. Because only 23 nucleotides identity is needed between the insert sequence in the VIGS vector and the targeted gene, VIGS will assist in quickly identifying the sequence motif of the R gene in question and result in targeting a limited number of candidate genes for stable transformation (Thomas et al., 2001). As plant genomes and EST databases are completed, TRV-VIGS could be used to target a specific member of a gene family by designing gene-specific VIGS constructs.
Heat instability of R genes is a feature of a number of root-knot nematode R genes from distinct plant families (Griffin, 1969; Jatala and Russell, 1972; Carter, 1982). Because Mi-9 is a homolog of Mi-1, cloning Mi-9 will assist in determining the nature of heat stability.
MATERIALS AND METHODS
Plants Material and Growth Conditions
Two accessions of Solanum arcanum (formerly Lycopersicon peruvianum), LA2157 and LA392, and three tomato (Solanum lycopersicum; formerly Lycopersicon esculentum) cultivars Motelle (Mi-1/Mi-1), VFN (Mi-1/Mi-1), and UC82B (mi/mi), were used in this study. Seeds were sown in seedling trays filled with organic planting mix (Sun Gro Horticulture). Two to 4 weeks after germination, seedlings were either used directly or transplanted into larger containers. To assist in uniform germination, seedling trays were maintained in a greenhouse in an enclosed structure with misters. Plants were maintained in a greenhouse at 22°C to 26°C unless otherwise stated. After germination, seedlings were supplemented with Osmocote (17-6-10; Sierra Chemical) and fertilized biweekly with tomato MiracleGro (18-18-21; Stern's MiracleGro Products).
Nematode Cultures
Cultures of Mi-1-avirulent Meloidogyne incognita isolate VW4 and Project 77 were maintained on tomato cv UC82B in a greenhouse. Eggs were extracted from infected roots by processing in 0.52% (v/v) NaOCl in a Waring blender for 2 min at high speed (Hussey and Barker, 1973). Eggs and root debris were collected on a 43-μm pore diameter sieve. Infective stage juveniles (J2) were obtained by hatching the eggs, as described in Martinez de Ilarduya et al. (2001). J2 were collected every 48 h and used immediately or stored at room temperature for an additional 48 h with aeration.
Nematode Screens
Heat-stable resistance screens were carried out in growth chambers with 16-h-light and 8-h-dark photoperiod and 700 μmol m−2 s−1 light intensity. Five to 6-week-old plants in 10-cm pots filled with steam-sterilized loamy sand were used in these assays. Plants were moved to the growth chamber set at 25°C. The temperature in the chamber was raised gradually over a 2-d period to 32°C. Plants were maintained at 32°C for 2 to 3 d before inoculation. Plants were inoculated with 3,000 J2 and maintained at 32°C for 4 weeks. Plants were then moved to a greenhouse and maintained at 22°C to 26°C for three additional weeks. For F3 screens, 12 to 22 F3 individuals were used per F2 family. Eight weeks after inoculation, nematode reproduction was evaluated by staining roots in 0.001% (w/v) erioglaucine (Sigma-Aldrich). Plants were classified as resistant if the individual root system had less than 20 egg masses, or susceptible if the individual root system had 25 or more egg masses. Susceptible tomato cultivar UC82B was included as control for nematode infectivity, and Mi-1-containing cultivars VFN or Motelle were included as controls for breakdown of Mi-1-mediated resistance.
DNA Isolation and DNA-Blot Analysis
For PCR analyses, DNA was isolated from leaflets using Wizard Genomic DNA purification kit (Promega) according to manufacturer's recommendation or as described in Ammiraju et al. (2003). For DNA-blot analysis, DNA was isolated, restricted with EcoRV restriction enzyme, and DNA blots prepared according to Kaloshian et al. (1998). A 300-bp subclone of Mi-1 containing the NBS region (clone 3-3) was used as probe. The random-primed 32P-labeled probe was prepared from insert amplified from plasmid DNA using PCR. Hybridization was carried out overnight in 50% (v/v) formamide at 42°C, and final washes were done in 0.5× SSC, 0.1% (w/v) SDS at 48°C.
PCR-Based Markers
Aps-1, Rex-1, CT119, and C8B PCR-based markers and conditions used for these markers were described previously (Dixon et al., 1995; Kaloshian et al., 1998; Ammiraju et al., 2003).
Primers were developed to amplify Cf-2.1 (Supplemental Table S1). To distinguish among Mi-1 homologs, we made use of a unique feature that distinguishes between the functional Mi-1 gene and its homolog Mi-1.1. Both Mi-1 and Mi-1.1 have two introns. While the size of intron 2 is similar in both genes, the size of intron 1 is variable, 556 bp in Mi-1.1 and 1,306 bp in Mi-1 (Milligan et al., 1998). The sequences flanking intron 1 in both Mi-1.1 and Mi-1 are identical. To amplify intron 1, primers were designed that annealed to the intron-flanking regions. The primers are Mint-do, 5′-TTCTCTAGCTAAACTTCAGCC-3′ and Mint-up, 5′-TTTTCGTTTTTCCATGATTCTAC-3′. Mint-do primer annealed to nucleotides 1,280 to 1,300 of Mi-1 (GenBank accession no. U65668) and 1,045 to 1,065 of Mi-1.1 (GenBank accession no. U65667), and Mint-up primer annealed to nucleotides 2,629 to 2,651 of Mi-1 and 1,644 to 1,666 of Mi-1.1. PCR conditions were as described for Rex-1, except 3 mm MgCl2 was used. Products were resolved on 1.5% to 2% (w/v) agarose gels.
Sequence Analysis
Nucleotide alignments were performed using ClustalX (Thompson et al., 1997) and edited manually using GENEDOC (http://www.psc.edu/biomed/genedoc). For intron 1 data, intron/exon boundaries were determined based on Mi-1 cDNA sequence information, and intron sequences were used to generate phylogenetic trees using parsimony and maximum likelihood methods in PAUP 4.0 (Sinauer Associates). The HKY + G likelihood model for the intron 1 data matrix was chosen using Winmodeltest (Posada and Crandall, 1998). The parsimony tree was obtained by using a heuristic search implemented in PAUP. Bootstrap value for each node was calculated from 3,000 replicates. Maximum likelihood analysis was similarly conducted with heuristic search using the model defined for the data matrix.
RNA Isolation and RT-PCR
Total RNA was extracted using Trizol (Invitrogen) according to manufacturer's recommendation. Five micrograms of total RNA was treated with 1 unit RNase-free DNase I (Promega), and cDNAs were synthesized as described in Li et al. (2006). In PCR, Mi-1 transcripts were amplified using primers VIGS-F, 5′-CTTGCGTCTACTGACTCTTTCC-3′ and C2S4, 5′-CTAAGAGGAATCTCATCACAGG-3′, and tomato ubiquitin Ubi3 gene transcripts were amplified as an internal control for equal cDNA use from control and silenced plants, as described in Li et al. (2006). To confirm lack of genomic DNA contamination, 200 ng of DNase-treated RNAs were also used as template. High fidelity Platinum Taq DNA polymerase (Invitrogen) was used. The amplified products were analyzed on 1.5% (w/v) agarose gels stained with ethidium bromide, gel purified, and ligated into a TOPO TA cloning vector (Invitrogen). Two independent RT-PCR reactions were performed and cloned. At least 15 recombinant plasmids were sequenced from each cloning.
Virus Constructs Used in VIGS
The TRV vector used in these experiments was previously described (Liu et al., 2002). The TRV-PDS construct was a gift from Dr. Dinesh-Kumar (Yale University). The TRV-Mi construct was engineered by cloning a 300-bp Mi-1 cDNA fragment spanning the carboxy terminal end of the Mi-1 gene and amplified using primers C1/2Do and C2S4 into pTRV2, as described in Li et al. (2006). The resulting plasmid was transformed into Agrobacterium tumefaciens strain GV3101.
Agrobacterium-Mediated Virus Infection
One-milliliter cultures of A. tumefaciens strain GV3101 containing each of the constructs derived from pTRV2, empty vector control, and pTRV1 were grown overnight in Luria-Bertani medium containing 50 μg mL−1 kanamycin and 12.5 μg mL−1 rifampacin at 28°C. Each overnight culture was used to inoculate 50-mL cultures of Luria-Bertani medium containing the same antibiotics, 10 mm MES, and 20 μm acetosyringone. The cultures were grown overnight at 28°C. Agrobacterium cultures were pelleted, resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone), and adjusted to an OD600 value of 0.8. Bacteria were incubated at room temperature for 3 h before use. An equal volume of pTRV1 Agrobacterium culture was mixed with pTRV2-PDS or pTRV2-Mi cultures before infiltration.
The abaxial side of leaflets of 4-week-old seedlings was infiltrated with A. tumefaciens cells (agroinfiltration) using a 3-mL needleless syringe. Seedlings were maintained at either 19°C or 24°C in growth chambers. Ten days after infiltration, seedlings used in nematode resistance assays were transplanted into plastic cups (10 cm diameter, 17 cm deep) filled with sand. Plants were maintained at 19°C for 10 additional days before nematode inoculation.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF028056 to EF028062.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distinct Mi-1 homologs cosegregate with resistant and susceptible phenotypes in F2 population of S. arcanum accessions LA2157 × LA392.
Supplemental Figure S2. Alignment of Mi intron 1 amplified using Mint-up and Mint-do primers from S. arcanum accessions LA2157 and LA392.
Supplemental Figure S3. TRV-mediated silencing in S. arcanum accession LA2157.
Supplemental Figure S4. Alignment of Hmr1 (for homologs of Meloidogyne resistance gene Mi-1) sequences.
Supplemental Table S1. Genetic analysis of F3 progeny.
Supplemental Table S2. Characteristics of PCR markers and respective primers.
Supplemental Table S3. Segregation of Mi-9 phenotype, Mi-1 homologs, and linked markers in parents and key recombinants.
Supplementary Material
Acknowledgments
We thank Drs. Paul Deley and Cheryl Hayashi for help with phylogenetic analysis, Scott Edwards for help with figures, and Qi Li, Daniela Noyes, Amal Khoury, Debrina Johnson, and Yaya Mansour for technical assistance. We are also grateful to Dr. Valerie M. Williamson, University of California, Davis, for sharing unpublished data.
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 98–35300–6350 to I.K. and P.R.) and by the University of California Agricultural Experiment Station (grants to I.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Isgouhi Kaloshian (isgouhi.kaloshian@ucr.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ammati M, Thomason IJ, McKinney HE (1986) Retention of resistance to Meliodogyne incognita in Lycopersicon genotypes at high soil temperature. J Nematol 18 491–495 [PMC free article] [PubMed] [Google Scholar]
- Ammati M, Thomason IJ, Roberts PA (1985) Screening Lycopersicon spp. for new genes imparting resistance to root-knot nematodes (Meloidogyne spp.). Plant Dis 69 112–115 [Google Scholar]
- Ammiraju JS, Veremis JC, Huang X, Roberts PA, Kaloshian I (2003) The heat-stable root-knot nematode resistance gene Mi-9 from Lycopersicon peruvianum is localized on the short arm of chromosome 6. Theor Appl Genet 106 478–484 [DOI] [PubMed] [Google Scholar]
- Atherton JG, Rudich J (1986) The Tomato Crop. A Scientific Basis for Improvement. Chapman and Hall, New York
- Bai YL, van der Hulst R, Bonnema G, Marcel BC, Meijer-Dekens F, Niks RE, Lindhout P (2005) Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol Plant Microbe Interact 18 354–362 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39 734–746 [DOI] [PubMed] [Google Scholar]
- Cap GB, Roberts PA, Thomason IJ (1993) Inheritance of heat-stable resistance to Meloidogyne incognita in L. peruvianum and its relationship to the Mi gene. Theor Appl Genet 85 777–783 [DOI] [PubMed] [Google Scholar]
- Carter WW (1982) Influence of soil temperature on Meloidogyne incognita on resistant and susceptible cotton. J Nematol 14 343–346 [PMC free article] [PubMed] [Google Scholar]
- Dickinson MJ, Jones DA, Jones JDG (1993) Close linkage between the Cf-2/Cf-5 and Mi resistance loci in tomato. Mol Plant Microbe Interact 6 341–347 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 105 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Hatzixanthis K, Ganal MW, Tanksley SD, Jones JDG (1995) High-resolution mapping of the physical location of the tomato Cf-2 gene. Mol Plant Microbe Interact 8 200–206 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrisom K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84 451–459 [DOI] [PubMed] [Google Scholar]
- Dropkin VH (1969. a) Cellular responses of plants to nematode infections. Annu Rev Phytopathol 7 101–122 [Google Scholar]
- Dropkin VH (1969. b) The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: reversal by temperature. Phytopathology 59 1632–1637 [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36 905–917 [DOI] [PubMed] [Google Scholar]
- Griffin GD (1969) Effects of temperature on Meloidogyne hapla in alfalfa. Phytopathology 59 599–609 [Google Scholar]
- Ho J-Y, Weide R, Ma HM, Wordragen MF, Lambert KN, Koornneef M, Zabel P, Williamson VM (1992) The root-knot nematode resistance gene (Mi) in tomato: construction of a molecular linkage map and identification of dominant cDNA markers in resistant genotypes. Plant J 2 971–982 [PubMed] [Google Scholar]
- Holtzmann OV (1965) Effects of soil temperature on resistance of tomato to root-knot nematode (Meloidogyne incognita). Phytopathology 55 990–992 [Google Scholar]
- Hussey R, Barker KR (1973) A comparison of methods of collecting inocula of Meloidogyne species including a new technique. Plant Dis Rep 57 1025–1028 [Google Scholar]
- Jatala P, Russell CC (1972) Nature of sweet potato resistance to Meloidogyne incognita and the effects of temperature on parasitism. J Nematol 4 1–7 [PMC free article] [PubMed] [Google Scholar]
- Johnson AW (1998) Vegetable crops. In G Pederson, G Windham, K Barker, eds, Plant-Nematode Interactions. Agronomy Society of America, Madison, WI, pp 595–635
- Kaloshian I, Yaghoobi J, Liharska T, Hontelez J, Hanson D, Hogan P, Jesse T, Wijbrandi J, Simons G, Vos P, et al (1998) Genetic and physical localization of the root-knot nematode resistance locus Mi in tomato. Mol Gen Genet 257 376–385 [DOI] [PubMed] [Google Scholar]
- Kesseli RV, Paran I, Michelmore RW (1994) Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RFLP and RAPD markers. Genetics 136 1435–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xie Q-G, Smith-Becker J, Navarre D, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling pathways. Mol Plant Microbe Interact 19 655–664 [DOI] [PubMed] [Google Scholar]
- Liharska TB, Koornneef M, van Wordragen M, van Kammen A, Zabel P (1996) Tomato chromosome 6: effect of alien chromosomal segments on recombinant frequencies. Genome 39 485–491 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC (2003) Virus-induced gene silencing in plants. Methods 30 296–303 [DOI] [PubMed] [Google Scholar]
- Martinez de Ilarduya O, Moore AE, Kaloshian I (2001) The tomato Rme1 locus is required for Mi-1-mediated resistance to root-knot nematodes and the potato aphid. Plant J 27 417–425 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO, Zhang Z, Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and birth-and-death process. Genome Res 8 1113–1130 [DOI] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root-knot nematode resistance gene Mi from tomato is a member of leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2112 [PMC free article] [PubMed] [Google Scholar]
- Noling JW (2000) Effects of continuous culture of resistant tomato cultivars on Meloidogyne incognita soil population density and pathogenicity. J Nematol 32 452 [Google Scholar]
- Nombela G, Williamson VM, Muñiz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16 645–649 [DOI] [PubMed] [Google Scholar]
- Parrella G, Moretti A, Gognalons P, Lesage ML, Marchoux G, Gebre-Selassie K, Caranta C (2004) The Am gene controlling resistance to Alfalfa mosaic virus in tomato is located in the cluster of dominant resistance genes on chromosome 6. Phytopathology 94 345–350 [DOI] [PubMed] [Google Scholar]
- Peralta IE, Knapp SK, Spooner DM (2005) New species of wild tomatoes (Solanum section Lycopersicon: Solanaceae) from Northern Peru. Syst Bot 30 424–434 [Google Scholar]
- Peralta IE, Spooner DM (2005) Morphological characterization and relationships of wild tomatoes (Solanum L. section Lycopersicon). Monogr Syst Bot Missouri Bot Gard 104 227–257 [Google Scholar]
- Philis J, Vakis N (1977) Resistance of tomato varieties to the root-knot nematodes Meloidogyne javanica in Cyprus. Nematol Mediterr 5 39–44 [Google Scholar]
- Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14 817–818 [DOI] [PubMed] [Google Scholar]
- Rick CM (1979) Biosystematic studies in Lycopersicon and closely related species of Solanum. In JG Hawkes, RN Lester, AD Skelding, eds, The Biology and Taxonomy of the Solanaceae. Academic Press, London, pp 667–677
- Rick CM (1986) Reproductive isolation in the Lycopersicon peruvianum complex. In WG D'Arcy, ed, Solanaceae: Biology and Systematics. Colombia University Press, New York, pp 477–495
- Roberts PA, May DM (1986) Meloidogyne incognita resistance characteristics in tomato genotypes developed for processing. J Nematol 18 353–359 [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J 40 322–331 [DOI] [PubMed] [Google Scholar]
- Sasser JN (1980) Root-knot nematodes: a global menace to crop production. Plant Dis 64 36–41 [Google Scholar]
- Seah S, Yaghoobi J, Rossi M, Gleason CA, Williamson VM (2004) The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor Appl Genet 108 1635–1642 [DOI] [PubMed] [Google Scholar]
- Spooner DM, Peralta IE, Knapp S (2005) Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes (Solanum L. section Lycopersicon (Mill.) Wettst.). Taxon 54 43–61 [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J 25 417–425 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Olivier J, Sperisen C, Rogowsky P, Prior P, Anais G, Mangin B, Bazin B, Nazer R, Grimsley N (1996) Polygenic resistance of tomato plants to bacterial wilt in the French West Indies. Mol Plant Microbe Interact 9 837–842 [Google Scholar]
- Tzortzakakis EG, Gowen SR (1996) Occurrence of resistance breaking pathotypes of Meloidogyne javanica on tomatoes in Crete, Greece. Fundam Appl Nematol 19 283–288 [Google Scholar]
- Valentine T, Shaw J, Blok VC, Phillips MS, Oparka KJ, Lacomme C (2004) Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol 136 3999–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen EA, van der Voort JN, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe DC, Bakker J, Stiekema WJ, Klein-Lankhorst RM (2000) Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J 23 567–576 [DOI] [PubMed] [Google Scholar]
- Veremis JC, Roberts PA (1996. a) Identification of resistance to Meloidogyne javanica in the Lycopersicon peruvianum complex. Theor Appl Genet 93 894–901 [DOI] [PubMed] [Google Scholar]
- Veremis JC, Roberts PA (1996. b) Relationships between Meloidogyne incognita resistance genes in Lycopersicon peruvianum differentiated by heat sensitivity and nematode virulence. Theor Appl Genet 93 950–959 [DOI] [PubMed] [Google Scholar]
- Veremis JC, Roberts PA (2000) Diversity of heat-stable genotype specific resistance of Meloidogyne in Maranon races of Lycopersicon peruvianum complex. Euphytica 111 9–16 [Google Scholar]
- Veremis JC, van Heusden AW, Roberts PA (1999) Mapping a novel heat-stable resistance to Meloidogyne in Lycopersicon peruvianum. Theor Appl Genet 98 274–280 [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, et al (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol 16 1365–1369 [DOI] [PubMed] [Google Scholar]
- Yaghoobi J, Kaloshian I, Wen Y, Williamson VM (1995) Mapping a new nematode resistance locus in Lycopersicon peruvianum. Theor Appl Genet 91 457–464 [DOI] [PubMed] [Google Scholar]
- Zamir D, Eksteinmichelson I, Zakay Y, Navot N, Zeidan M, Sarfatti M, Eshed Y, Harel E, Pleban T, Vanoss H, et al (1994) Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor Appl Genet 88 141–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.