Abstract
An intriguing new paradigm in plant biology is that systemically mobile mRNAs play a role in coordinating development. In this process, specific mRNAs are loaded into the phloem transport stream for translocation to distant tissues, where they may impact on developmental processes. However, despite its potential significance for plant growth regulation, mRNA trafficking remains poorly understood and challenging to study. Here, we show that phloem-mobile mRNAs can also traffic between widely divergent species from a host to the plant parasite lespedeza dodder (Cuscuta pentagona Engelm.). Reverse transcription-polymerase chain reaction and microarray analysis were used to detect specific tomato (Lycopersicon esculentum Mill.) transcripts in dodder grown on tomato that were not present in control dodder grown on other host species. Foreign transcripts included LeGAI, which has previously been shown to be translocated in the phloem, as well as nine other transcripts not reported to be mobile. Dodders are parasitic plants that obtain resources by drawing from the phloem of a host plant and have joint plasmodesmata with host cortical cells. Although viruses are known to move between dodder and its hosts, translocation of endogenous plant mRNA has not been reported. These results point to a potentially new level of interspecies communication, and raise questions about the ability of parasites to recognize, use, and respond to transcripts acquired from their hosts.
Dodders are obligate stem parasitic plants that have close physical linkages with their hosts. Because of their limited photosynthetic ability and dependence upon the host plant for water and nutrients, a dodder seedling must form connections with a host within several days after germination. Once established on the host, the dodder root system senesces and the mature vegetative plant consists entirely of a yellow-orange stem that twines around host stems and leaves. At points of contact with the host, the coiled dodder stem produces a haustorium, a specialized organ that penetrates host tissues and forms connections with host vascular tissues (Kuijt, 1983). Dodders form direct connections with host xylem and, although the precise nature of phloem connections is still unresolved, evidence also supports direct phloem connections. For example, the phloem-specific dye carboxyfluorescein readily moves from the host into dodder (Birschwilks et al., 2006). Also, transgenic tobacco (Nicotiana tabacum) plants expressing green fluorescent protein in companion cells transmit protein to dodder (Haupt et al., 2001), suggesting that direct transfer of macromolecules may occur. A well-known aspect of host-dodder connections is the transmission of viruses, and a single dodder plant simultaneously parasitizing two hosts may transmit plant viruses from one host to the other (Bennet, 1940, 1944; Johnson, 1941; Hosford, 1967). In addition to direct vascular connections, dodder appears to have cytoplasmic continuity with its host through plasmodesmata. Vaughn (2003) has reported that searching hyphae cells of the dodder haustorium have plasmodesmata that span walls of adjacent dodder hyphae and host cortex cells.
Plasmodesmata are important regulators of cell-to-cell and long-distance movement of macromolecules in plants. Normally, plasmodesmata channels are narrow and allow passive movement of small molecules, whereas macromolecules, such as proteins and nucleic acids, are excluded. However, certain proteins interact with plasmodesmata to increase the size exclusion limit and allow larger molecules to pass through the channel. One category of such proteins are the viral movement proteins (MPs), which facilitate systemic viral spread through plants and have played an important role in shaping our understanding of plasmodesmata function (Carrington et al., 1996). Plants have endogenous counterparts to MPs, termed non-cell-autonomous proteins (Lucas et al., 2001), that mediate the movement of endogenous mRNA from cell to cell. An example of this is the transcription factor KNOTTED1 (KN1), which mediates local cell-to-cell trafficking of the kn1 sense mRNA through plasmodesmata (Lucas et al., 1995; Kim et al., 2001, 2002). Plasmodesmata are also gatekeepers for loading and unloading of materials between phloem sieve elements and companion cells and so play a key role in regulating long-distance transport (Oparka and Turgeon, 1999).
Growing evidence indicates that mRNA is trafficked over long distances in plants through the phloem and that this process plays an important role in regulating plant development. This has come from studies grafting together closely related species or wild-type scions grafted onto mutant or transgenic stocks of the same species. For example, CmPP16, a protein from pumpkin (Cucurbita maxima Duch.), has properties similar to viral MPs, mediates the transport of RNA into phloem sieve elements, and is trafficked into cucumber (Cucumis sativus) scions grafted onto pumpkin stocks (Xoconostle-Cazares et al., 1999). Ruiz-Medrano et al. (1999) cloned several additional mRNAs from phloem sap of pumpkin and demonstrated their phloem localization and mobility into grafted scions. The functional significance of translated messages has been demonstrated in several cases, such as by the movement of a transcript that affects leaf morphology, producing the phenotype Mouse ears (Me) in tomato (Kim et al., 2001). In this study, wild-type tomato scions grafted onto Me root stocks developed leaf morphology typical of the Me mutation. Another study used tobacco root stocks in which nitrate reductase or nitrite reductase expression was suppressed through gene silencing to demonstrate mobility of the silencing mRNA into grafted wild-type scions (Palauqui et al., 1997). Recently, transcripts of GIBBERILLIC ACID-INSENSITIVE (GAI) were shown to be phloem mobile in three plant species, and transgenic tomato and Arabidopsis (Arabidopsis thaliana L. Heyhn.) expressing mutants showed graft-transmissible alterations in the phenotype of developing leaves (Haywood et al., 2005).
It has been estimated that more than 1,000 different mobile transcripts are present in phloem (Lucas and Lee, 2004), but the identities of these mRNAs and their targeting mechanisms remain largely unresolved. Studies of mRNA trafficking are hampered by the challenge of positively identifying the original source of transcripts and have relied on heterografting experiments, such as those described above. Here, we report the detection of tomato transcripts in dodder grown on tomato, including LeGAI. We have also detected three pumpkin transcripts previously characterized as phloem mobile in dodder parasitizing pumpkin. These findings indicate that host-mobile transcripts move into dodder, raising questions about the role of such mobile transcripts in host-parasite communication and suggesting that dodder may provide new insights into the phenomenon of mRNA trafficking.
RESULTS
Translocation of Pumpkin Phloem-Mobile mRNAs into Dodder
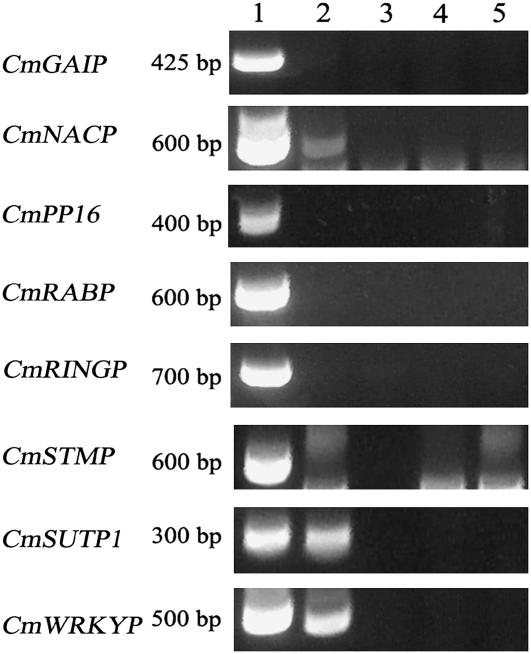
An initial survey of host transcripts in dodder was conducted using pumpkin because it has been a model species for mRNA trafficking and contains several characterized phloem-mobile transcripts (Ruiz-Medrano et al., 1999). The movement of eight of these transcripts from pumpkin to dodder was evaluated. Reverse transcription (RT)-PCR of RNA from dodder grown on pumpkin produced products for three genes, CmNACP, CmSUTP1, and CmWRKYP, which were the same size as products from similar reactions using pumpkin RNA (Fig. 1). Sequencing confirmed that the RT-PCR products from dodder grown on pumpkin were identical to those from pumpkin. The RT-PCR products corresponding to CmNACP, CmSUPT1, and CmWRKYP were also detected in RNA from the pumpkin host, but not from tobacco, dodder grown on tobacco, or the no-template negative control. The remaining pumpkin-mobile transcripts (CmGAIP, CmPP16, CmRABP, CmRINGP, and CmSTMP) were not detected in RNA from control tissues, but were present in the host tissue as evidenced by RT-PCR products from pumpkin RNA.
Figure 1.
Detection of pumpkin-mobile transcripts in dodder using RT-PCR. Tissues assayed included total RNA from lane 1, pumpkin; lane 2, dodder grown on pumpkin; lane 3, tobacco; lane 4, dodder grown on tobacco; and lane 5, no-template negative control.
Amplification of CmNACP, CmSUPT1, and CmWRKYP was consistent among replicates of dodder generated during a single experimental run. However, repetition of the experiment using plants grown at different times of the year and slightly different harvest times yielded variable results. Moreover, a lack of characterized pumpkin sequences beyond those cited here hindered efforts to identify other mobile or nonmobile pumpkin transcripts. Because of these difficulties, a new host species was sought for further research. Although Arabidopsis offered the largest array of genomic resources and supported growth of lespedeza dodder (Cuscuta pentagon Engelm.), it was recalcitrant to establishment of the dodder. Ultimately, tomato was used because it was readily parasitized by dodder and has available genomic resources, such as expressed sequence tag sequences and microarrays.
Microarray Analysis Identifies Tomato Transcripts in Dodder
To identify potential tomato transcripts in dodder using a non-PCR-based method, microarray analysis was performed on RNA from dodder and hosts. Total RNA was extracted from the tomato host and from dodder grown on tomato, Arabidopsis, tobacco, or pumpkin. The host tomato RNA was included to verify that any transcripts detected in the parasite were in fact expressed in the host. Dodder samples grown on tobacco, Arabidopsis, and pumpkin served as controls for dodder genes that may cross-hybridize with tomato array probes, with three different host species used to minimize any host-specific effects on dodder gene expression. Samples were analyzed using Affymetrix GeneChip Tomato Arrays (9,200 transcripts) and transcripts scored for the presence or absence in each sample. Considering that host transcripts present in dodder would be at low levels and diluted with dodder transcripts, a P value of 0.06 in at least two of three biological replicates was used as the threshold for scoring a transcript as being present.
RNA from tomato leaves gave by far the highest number of positives (7,689), reflecting an overall high level of species specificity for the microarray (Table I). A much smaller number of positives (135) were detected in all samples and likely represent cross-hybridization between the array probes and highly conserved plant genes. A small number of transcripts (32) were detected in dodder from tomato but not in tomato leaves, and although it is possible that these derived from other tomato tissues and accumulated in dodder, they may also be due to cross-hybridization. Our main hypothesis was that if tomato transcripts are translocated into dodder, then dodder parasitizing tomato should have more transcripts hybridizing to the tomato microarray than the dodder parasitizing other hosts. This was the case with 474 transcripts detected in dodder grown on tomato as compared to 146 transcripts in dodder grown on other hosts. These 474 transcripts in the dodder parasitizing tomato thus represent the pool of putatively mobile transcripts.
Table I.
Microarray detection of tomato transcripts from tomato, dodder grown on tomato, and dodder grown on other hosts
+ and −, Transcripts present or absent, respectively, in at least two of the three biological replicates.
| Tomato | Dodder on Tomato | Dodder on Other Hosts | No. of Genes | Interpretation |
|---|---|---|---|---|
| + | − | − | 7,689 | Nonmobile tomato-specific transcripts |
| + | + | − | 474 | Mobile tomato transcripts |
| + | + | + | 135 | Conserved, cross-hybridizing transcripts |
| + | − | + | 146 | Cross-hybridization |
| − | + | − | 32 | Mobile tomato or cross-hybridizing transcripts |
| − | − | + | 0 | Cross-hybridization |
| − | + | + | 4 | Conserved, cross-hybridizing transcripts |
| − | − | − | 1,560 | Nonexpressed tomato-specific genes |
RT-PCR Analysis Detects Tomato Transcripts in Dodder Grown on Tomato
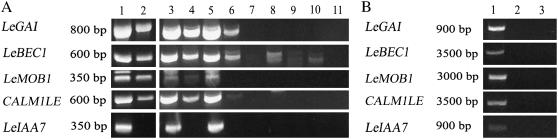
To further investigate genes identified by the microarray experiment as producing mobile transcripts, several putatively mobile transcripts were selected for analysis by RT-PCR. These included beclin 1 (LeBEC1), a protein of unknown function (LeMOB1), an auxin-regulated gene (LeIAA7; Nebenführ et al., 2000), and LeGAI, which was previously reported to produce a mobile transcript in tomato (Haywood et al., 2005) and was detected in one microarray replicate. A putatively nonmobile transcript, calmodulin 1 (CALM1LE), was also included in the analysis.
RT-PCR amplified LeGAI from all three replicates of dodder grown on tomato and the tomato host (Fig. 2A). RT-PCR products were sequenced and those from dodder were found to be identical to those from tomato. Detection of LeGAI in dodder is in agreement with reports that this transcript is phloem mobile (Haywood et al., 2005). Identical reactions using RNA from control tissues (tobacco, pumpkin, and dodder grown on tobacco and pumpkin) did not amplify a product.
Figure 2.
Detection of tomato transcripts in dodder by RT-PCR. A, Total RNA was assayed from the following tissues. Lane 1, Tomato replicate 1; lane 2, dodder grown on tomato replicate 1; lane 3, tomato replicate 2; lane 4, dodder grown on tomato replicate 2; lane 5, tomato replicate 3; lane 6, dodder grown on tomato replicate 3; lane 7, tobacco; lane 8, dodder grown on tobacco; lane 9, pumpkin; lane 10, dodder grown on pumpkin; lane 11, no-template negative control. B, Dodder genomic DNA was assayed for homologs to tomato genes using PCR. Samples assayed were lane 1, pumpkin genomic DNA; lane 2, dodder genomic DNA; and lane 3, no-template negative control.
RT-PCR products corresponding to LeBEC1 were amplified from all replicates of tomato and dodder grown on tomato, and sequencing again confirmed that dodder RNA contained sequences identical to tomato. Products were also amplified from control dodders and pumpkin, but were not the same size or sequence as authentic tomato LeBEC1 and likely represent a conserved gene among these plants.
A band corresponding to LeMOB1 was amplified in two biological replicates of dodder on tomato and all replicates of tomato, but not in any other dodders or control plants. Sequencing showed that LeMOB1 detected in dodder grown on tomato was identical to tomato LeMOB1. A notable aspect of LeMOB1 is that a second band of slightly higher Mr was also amplified from tomato RNA. Sequencing of this band showed that it was an immature RNA of the lower band that included a 58-bp intron. However, only the lower band was detected in dodder grown on tomato, suggesting that mRNA must be fully processed before becoming systemically mobile. Insight into the functional significance of this finding must await further characterization of LeMOB1.
Whereas microarray analysis correctly predicted the presence of LeBEC1 and LeMOB1 in dodder on tomato, the approach was apparently not reliable for CALM1LE and LeIAA7. The putatively nonmobile gene, CALM1LE, was detected in all biological replicates of dodder grown on tomato and in the tomato host. Sequencing showed that these products were identical to tomato, which again suggests trafficking of the CALM1LE transcript. Conversely, a putatively mobile gene, LeIAA7, was not amplified from any samples of dodder grown on tomato, although it was clearly detected in the tomato host. The absence of an RT-PCR product suggests that LeIAA7 does not traffic into dodder. These results highlight the differences between microarrays and RT-PCR in this type of study. Whereas the former is valuable for generating candidate mobile transcripts, the latter is more sensitive in detecting transcripts in dodder (CALM1LE) and has higher sequence specificity to avoid false positives (LeIAA7).
The only alternative explanations for the detection of tomato mRNA in the parasite would be the presence of tomato homologs in dodder or cross-contamination of samples. To address the first possibility, PCR of genomic DNA from tomato and dodder was performed to confirm that the primers used in RT-PCR analysis did not amplify dodder homologs (Fig. 2B). No bands were generated from dodder in these reactions, demonstrating that the primers were specific for tomato. Potential cross-contamination of tissues was avoided by using dodder tissues harvested at least 4 cm from the point of attachment to the host. Furthermore, Mr differences between genomic PCR and RT-PCR products reveal the presence of introns in all the genes studied and confirm that RT-PCR products cannot be attributed to genomic DNA contamination of RNA extracts. The apparently nonmobile LeIAA7 also serves as a control in that the lack of RT-PCR products from dodder RNA indicates that there was no contamination by tomato RNA (Fig. 2A).
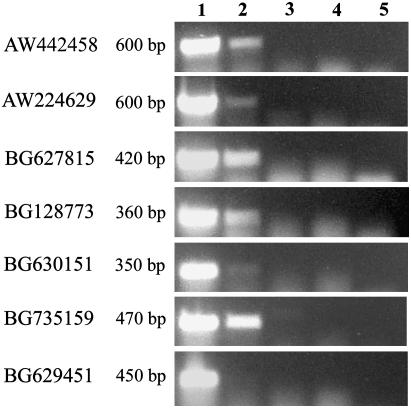
To further assess the value of microarrays as a predictor of mRNA mobility into dodder, we performed RT-PCR for 23 additional genes that were selected randomly from the sets of predicted mobile and nonmobile transcripts. RNA was analyzed from tomato (positive control), dodder grown on tomato, and dodder grown on either tobacco or pumpkin (negative controls). Five of the 24 primer sets tested were considered to be inconclusive because they nonspecifically amplified dodder genes homologous to the intended tomato gene. For the primer sets that gave unambiguous results, six of 14 genes that were predicted by microarray to be mobile produced a band in both the tomato and dodder grown on tomato, but not the control dodders (Fig. 3). Sequencing of the products confirmed that tomato and dodder grown on tomato products were identical. In comparison, four genes from the predicted nonmobile set amplified a product only from tomato RNA, exemplified by BG629451 shown in Figure 3. This supports the value of the microarray as a tool for discovering mobile host genes in dodder.
Figure 3.
Additional tomato transcripts showing evidence of mobility into dodder. Total RNA was assayed from lane 1, tomato; lane 2, dodder grown on tomato; lane 3, dodder grown on tobacco; lane 4, dodder grown on pumpkin; and lane 5, no-template negative control.
Of these six genes showing evidence for transcript mobility, two encode calmodulin-like proteins (AW224629 and BG735159). Others encode a shaggy protein kinase (AW442458), a LIM domain protein (BG627815), a 60S ribosomal protein (BG128773), and a xyloglucan endotransglycosylase/hydrolase 16 protein (BG630151). The nonmobile transcript shown (BG629451) encodes PGR5, a protein involved in electron flow through PSI.
DISCUSSION
We have detected evidence for the movement of transcripts from two host species into dodder. Three pumpkin transcripts previously documented to be phloem mobile were detected in dodder grown on pumpkin (Fig. 1). Ten tomato transcripts, including the phloem-mobile transcript LeGAI, were detected in dodder growing on tomato (Figs. 2 and 3). Together, this suggests that a symplastic continuum exists between host and parasite that involves a previously unreported aspect of macromolecular exchange.
Detection of tomato and pumpkin transcripts in dodder has implications for understanding connections between host and parasite, specifically with respect to the nature of phloem continuity. Ultrastructural studies have indicated that dodder-searching hyphae contacting host sieve elements differentiate into transfer cells, but do not make open connections to the host phloem (Dörr, 1990). Thus, it has been proposed that content of the host phloem must cross host and parasite membranes in an apoplastic transfer process (Christensen et al., 2003). However, Birschwilks et al. (2006) argue that their observed rates of radiolabeled metabolite, dye, and virus translocation from host to dodder can only be explained by symplastic transport. Indeed, any movement of viruses is difficult to explain without direct cytoplasmic continuity, and reports of green fluorescent protein (Haupt et al., 2001) and phytoplasma (Kaminska and Korbin, 1999) movement from host to parasite only support this position. Apart from possible phloem-phloem contact, symplastic connections between the two species are formed by plasmodesmata that span the walls between host cortex cells and developing dodder hyphae (Vaughn, 2003). These connections appear to be transitory and may be occluded as haustoria age, but a dodder plant constantly makes new haustorial connections with its hosts, so it is reasonable that the parasite could have continuous access to host cytoplasm through plasmodesmata connections. Although we cannot distinguish between the movement of host mRNAs through direct phloem-phloem connections or cortex plasmodesmata, the detection of known phloem-mobile host transcripts from hosts in dodder would support direct movement from host phloem.
Considering the transmission of viruses by dodder, it may not be surprising that endogenous mRNAs are also mobile because they use a similar mechanism to traverse plasmodesmata. However, it is interesting that not all phloem-mobile pumpkin transcripts were detected in dodder. GAI was assayed from both pumpkin and tomato, but only found to be mobile from tomato. For pumpkin, only three of eight phloem-associated transcripts were detected in the dodder. Whereas it is possible that such discrepancies may be due to limits of mRNA detection, there may also be a physiological explanation. Foster et al. (2002) described a regulatory surveillance field at the shoot apex that controls the movement of phloem-mobile mRNA and viruses into the apex, with the hypothesized function of protecting the germline from viral infection. A similar regulatory mechanism may exist at the interface of host and parasite to control the movement of host phloem-mobile transcripts. This raises the question of which organism—host or parasite—regulates movement between the species.
The trafficking of host-mobile transcripts into dodder suggests that this parasite may be useful for delineating the mechanisms and functions of mobile RNA within plants. For most species, one of the major challenges associated with studying phloem-mobile RNA is simply assaying phloem content. Heterografting has been an important technique for demonstrating movement of mRNA, but is subject to limitations of graft compatibility and reestablishment of symplastic continuity between stock and scion. The use of dodder may reduce this problem because the parasite haustorium integrates with host tissue to form what has been compared to an extraordinarily successful vegetative graft (Kuijt, 1983). Furthermore, dodder has a broad host range, which is not restricted by typical grafting compatibilities and would allow dodder to be used as a common scion to study the phloem-mobile RNA of diverse plant species. This is illustrated in this study in which dodder was used to detect transcripts from two different host species, tomato and pumpkin.
The genetic distance between the parasite and its host made it possible to perform a survey of phloem- mobile transcripts using microarrays. The 474 putative positives identified by this method (Table I) suggest that this approach has utility as a screening tool, but that validation by RT-PCR is required. In our limited sampling, RT-PCR data agreed with microarray data nearly one-half of the time and the microarray facilitated identification of a new set of mobile transcripts. It should be noted that our microarray analysis was aimed at detecting full-length transcripts so the small RNA population of the phloem (Yoo et al., 2004) was not assayed. It is estimated that the total number of mobile transcripts in the phloem is at least 100 (Ruiz-Medrano et al., 1999) and may exceed 1,000 (Lucas and Lee, 2004), and our detection of 474 tomato transcripts in dodder is consistent with this range.
An important aspect of the host-parasite interaction is the effect of host growth stage and environment on the mobile mRNAs being expressed. It is expected that specific sets of mobile mRNAs will be expressed at various times during development, and Yoo et al. (2004) showed that content of small RNA species in pumpkin phloem sap differed significantly in summer- and winter-grown plants. All of our experiments used mature host plants that were able to support masses of dodder for generating ample tissue for RNA extraction, but it will be of interest to examine dodder parasitizing hosts spanning a range of developmental stages.
Phloem-mobile mRNAs have been suggested to be involved in regulation of gene expression or signal transduction and together play a role in coordinating physiological processes throughout the plant (Ruiz-Medrano et al., 1999; Lucas and Lee, 2004; Haywood et al., 2005). It is therefore interesting that most of the genes with transcripts translocated into dodder have roles in mediating plant response to the environment. GAI influences GA3 sensitivity and is one of the DELLA genes that integrate responses to salt stress in Arabidopsis (Achard et al., 2006). LeBEC1 encodes a beclin 1 protein, which in tobacco acts as a negative regulator of autophagy and senescence during defense response (Liu et al., 2005). Silencing this gene resulted in uncontrolled spread of hypersensitive-response-related programmed cell death, both at the site of initiation and in distal tissues, suggesting a systemic influence. The tomato calmodulin protein CALM1LE is involved in signal transduction of defense genes in response to wounding (Bergey and Ryan, 1999). Identification of two additional calmodulin-like protein transcripts from this relatively small sampling suggests that transcript mobility may be a common feature of this group. Other proteins with mobile transcripts, such as the shaggy protein kinase and LIM domain protein, also have roles in plant growth and development (Schmeichel and Beckerle, 1997; Charrier et al., 2002). The significance of transcript mobility for a xyloglucan endotransglycosylase/hydrolase protein, a 60S ribosomal protein, and the unknown protein encoded by LeMOB1 is not clear, but all will add to the handful of genes already known to have mobile transcripts. Analysis of the remaining candidates identified by microarray will provide a broader base for understanding commonalities between trafficked mRNAs.
We have described a unique situation in which the RNA of two different species coexists in dodder, which raises several questions. For example, does dodder use host transcripts as a way to eavesdrop on host development and thus regulate its own development? Alternatively, does dodder possess a mechanism that restricts or degrades foreign transcripts? In any case, dodder must have evolved mechanisms for managing this situation and understanding these mechanisms will provide insight into dodder biology and mRNA trafficking.
MATERIALS AND METHODS
Plant Growth
Pumpkin (Curcurbita maxima) and Blue Hubbard winter squash (Heirloom Seeds), tobacco (Nicotiana tabacum), and tomato (Lycopersicon esculentum) plants were grown in Metro Mix 360 potting medium (Griffin Greenhouse and Nursery Supplies) under greenhouse conditions. Arabidopsis (Arabidopsis thaliana) was similarly grown in Metro Mix 360 potting media, but in a growth room at 20°C and 8-h light. Lespedeza dodder (Cuscuta pentagona Engelm.) seeds were scarified by soaking in concentrated sulfuric acid for 1 h, rinsed thoroughly in water, and planted around the base of 2-week-old tomato seedlings, 4-week-old Arabidopsis plants, and 7-week-old tobacco plants. In some cases, dodder seedlings did not establish readily on pumpkin, so dodder stems were trained from tomato to 4-week-old pumpkin plants and dodder connections between tomato and pumpkin were severed 2 weeks after the formation of new haustorial connections with pumpkin.
Dodder stems at least 4 cm from the host plant were harvested after 9 to 12 weeks of growth. To obtain a sufficient quantity of dodder tissue, many stems were harvested, representing multiple individuals for each harvest time. Host leaf tissue from parasitized plants was harvested simultaneously. All tissues were immediately frozen in liquid nitrogen and stored at −80°C.
DNA and RNA Extraction
DNA was extracted from host and dodder tissues using a DNeasy plant mini kit (Qiagen) according to the manufacturer's recommendations. RNA (1–1.5 g) was extracted from host and dodder tissues by grinding in liquid nitrogen and extracted using TRIzol reagent (Invitrogen). RNA was precipitated with 100% isopropanol followed by a second precipitation with 100% ethanol and 3 m sodium acetate (pH 5.2). The RNA pellet was dissolved in diethyl pyrocarbonate-treated water and treated with DNase using a DNA-free kit (Ambion).
PCR and RT-PCR Reactions
PCR for gene identification and cloning were performed using Taq PCR master mix kit (Qiagen). Primers for pumpkin CmRBCS, CmIMPORTINα, CmGAIP, CmNACP, CmPP16, CmRABP, CmRINGP, CmSTMP, CmSUTP1, and CmWRKYP genes were as described by Ruiz-Medrano et al. (1999). The PCR reaction using pumpkin primers consisted of an initial melting step at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min followed by a final extension of 8 min at 72°C. RT-PCR reactions were performed using SuperScript one-step RT-PCR with Platinum Taq system (Invitrogen). RT-PCR reactions using the above pumpkin primers were performed with an initial RT step at 50°C for 30 min using oligo(dT) primers, followed by the above PCR program at 55 cycles. Tomato-specific primers used for PCR and RT-PCR analysis are listed in Table II. PCR reactions using tomato primers were performed with an initial melting step at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, the corresponding annealing temperature for 30 s, and 72°C for 1 min followed by a final extension of 8 min at 72°C. RT-PCR reactions using tomato primers were performed with a preliminary RT step at 50°C for 30 min using oligo(dT) primers, followed by the above PCR program at 40 cycles.
Table II.
Tomato genes and specific primers characterized by RT-PCR
| Gene Name | GenBank Accession No. | Forward Primer | Reverse Primer |
|---|---|---|---|
| LeGAI | AY269087 | ttctcacaaaatcatcgaacaagta | ataacaatcgatgaagctcaaaaac |
| LeBEC1 | AW223712 | aatgcatgagggtgctgtct | aactttggtcggaaatgctg |
| LeIAA7 | AF022018.1 | gaacatgttgtctcaaaaaggga | tgaggacatccatcagtttcc |
| LeMOB1 | BE449715 | cgcacgccttctaaaagttc | aagagaggcacacaagtcagaa |
| CALM1LE | M67472 | aggagaaaaatggcagagca | agccgctttaggccactaat |
| AW442458 | ttactatcgtgcgcctgaactt | aagatgtcaaacagtttgtgcca | |
| AW224629 | aggcaagaatgtggaatatcga | aggcaagaatgtggaatatcga | |
| BG627815 | cactggacagcaaaaccgtgttatatt | gcctctactgtaagcatcaccatatt | |
| BG128773 | gcctgctgtcattgttcgtc | aaactgaaacattgcgaaaattct | |
| BG630151 | atgtaaatctttttatacaatgcaagactg | cgcccttcgtagcatcc | |
| BG735159 | atggtgtacttccagttttaaagattaag | cttttagcctattcgacaaggatg | |
| BG629451 | ctagactacaaagcacatatatatatgtttgaa | gaggaaagtgcaaccattaatga |
PCR Product Sequencing
PCR and RT-PCR products were separated by agarose gel electrophoresis and then extracted from the gel using a QIAquick gel extraction kit (Qiagen). Purified products were sequenced (Virginia Bioinformatics Institute), and results were compared against the National Center for Biotechnology Information (NCBI) database to confirm the identity of the products. Host sequences were aligned with dodder sequences using the ClustalW method in Lasergene MegAlign software (DNAStar) to determine the percent identity between the sequences.
Microarray Analysis
Microarray analysis was performed on three biological replicates of dodder grown on tomato, three biological replicates of tomato, two replicates of dodder grown on pumpkin, and one sample each of dodder grown on Arabidopsis and dodder grown on tobacco. Arrays used were the Tomato GeneChip Arrays (Affymetrix). Processing of the chips was performed at the Virginia Bioinformatics Institute. Linear amplification of total RNA was performed using the Ovation biotin RNA amplification and labeling system (NuGEN). The 0.06 P value determined by the Affymetrix analysis program was used as a cutoff for calling genes as present or absent.
Microarray data from this article have been deposited with the EMBL/GenBank data libraries under accession number GSE6736.
Acknowledgments
We thank Tom Lanini for supplying seeds of lespedeza dodder, Verlyn Stromberg for technical assistance, and Brenda Winkel for critical reading of the manuscript.
This work was supported by the Jeffress Memorial Trust (grant no. J–790) and the U.S. Department of Agriculture (Hatch project no. 135657).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James H. Westwood (westwood@vt.edu).
Open Access articles can be viewed online without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94 [DOI] [PubMed] [Google Scholar]
- Bennet CW (1940) Acquisition and transmission of viruses by dodder (Cuscuta subinclusa). Phytopathology 30 2 [Google Scholar]
- Bennet CW (1944) Studies of dodder transmission of plant viruses. Phytopathology 34 905–932 [Google Scholar]
- Bergey DR, Ryan CA (1999) Wound- and systemin-inducible calmodulin gene expression in tomato leaves. Plant Mol Biol 40 815–823 [DOI] [PubMed] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S (2006) Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot 57 911–921 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Kasschau KD, Mahajan SK, Schaad MC (1996) Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8 1669–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen NM, Dörr I, Hansen M, van der Kooij TAW, Schulz A (2003) Development of Cuscuta species on a partially incompatible host: induction of xylem transfer cells. Protoplasma 220 131–142 [DOI] [PubMed] [Google Scholar]
- Dörr I (1990) Sieve elements in haustoria of parasitic angiosperms. In H-D Behnke, R-D Sjölund, eds, Sieve Elements: Comparative Structure, Induction and Development. Springer, Berlin, pp 239–253
- Foster TM, Lough TJ, Emerson SJ, Lee RH, Bowman JL, Forster RLS, Lucas WJ (2002) A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 14 1497–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Oparka KJ, Sauer N, Neumann S (2001) Macromolecular trafficking between Nicotiana tabacum and the holoparasite Cuscuta reflexa. J Exp Bot 52 173–177 [PubMed] [Google Scholar]
- Haywood V, Yu T-S, Huang N-C, Lucas WJ (2005) Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J 42 49–68 [DOI] [PubMed] [Google Scholar]
- Hosford RMJ (1967) Transmission of plant viruses by dodder. Bot Rev 42 387–406 [Google Scholar]
- Johnson F (1941) Transmission of plant viruses by dodder. Phytopathology 31 649–656 [Google Scholar]
- Kaminska M, Korbin M (1999) Graft and dodder transmission of phytoplasma affecting lily to experimental hosts. Acta Physiol Plant 21 21–26 [Google Scholar]
- Kim JY, Yuan Z, Cillia M, Khalfan-Jagani Z, Jackson D (2002) Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci USA 99 4103–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Canio W, Kessler S, Sinha N (2001) Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289 [DOI] [PubMed] [Google Scholar]
- Kuijt J (1983) Tissue compatibility and the haustoria of parasitic angiosperms. In R Moore, ed, Vegetative Compatibility Responses in Plants. Baylor University, Waco, TX, pp 1–12
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Biao D, Hake S (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270 1980–1983 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Lee J-Y (2004) Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol 5 712–726 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Yoo B-C, Kragler F (2001) RNA as a long-distance information macromolecule in plants. Nat Rev Mol Cell Biol 2 849–857 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, White TJ, Lomax TL (2000) The diageotropica mutation alters auxin induction of a subset of the Aux/IAA gene family in tomato. Plant Mol Biol 44 73–84 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R (1999) Sieve elements and companion cells—traffic control centers of the phloem. Plant Cell 11 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ (1999) Phloem and long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126 4405–4419 [DOI] [PubMed] [Google Scholar]
- Schmeichel K, Beckerle M (1997) Molecular dissection of a LIM domain. Mol Biol Cell 8 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC (2003) Dodder hyphae invade the host: a structural and immunocytochemical characterization. Protoplasma 220 189–200 [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B, Xiang Y, Ruiz-Medrano R, Wang H-L, Monzer J, Yoo B-C, McFarland KC, Franceschi VR, Lucas WJ (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98 [DOI] [PubMed] [Google Scholar]
- Yoo B-C, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]