Abstract
The salt tolerance of rice (Oryza sativa) correlates with the ability to exclude Na+ from the shoot and to maintain a low cellular Na+/K+ ratio. We have identified a rice plasma membrane Na+/H+ exchanger that, on the basis of genetic and biochemical criteria, is the functional homolog of the Arabidopsis (Arabidopsis thaliana) salt overly sensitive 1 (SOS1) protein. The rice transporter, denoted by OsSOS1, demonstrated a capacity for Na+/H+ exchange in plasma membrane vesicles of yeast (Saccharomyces cerevisiae) cells and reduced their net cellular Na+ content. The Arabidopsis protein kinase complex SOS2/SOS3, which positively controls the activity of AtSOS1, phosphorylated OsSOS1 and stimulated its activity in vivo and in vitro. Moreover, OsSOS1 suppressed the salt sensitivity of a sos1-1 mutant of Arabidopsis. These results represent the first molecular and biochemical characterization of a Na+ efflux protein from monocots. Putative rice homologs of the Arabidopsis protein kinase SOS2 and its Ca2+-dependent activator SOS3 were identified also. OsCIPK24 and OsCBL4 acted coordinately to activate OsSOS1 in yeast cells and they could be exchanged with their Arabidopsis counterpart to form heterologous protein kinase modules that activated both OsSOS1 and AtSOS1 and suppressed the salt sensitivity of sos2 and sos3 mutants of Arabidopsis. These results demonstrate that the SOS salt tolerance pathway operates in cereals and evidences a high degree of structural conservation among the SOS proteins from dicots and monocots.
Rice (Oryza sativa) is one of the most important cereal crops in tropical and temperate regions of the world. Among all common environmental stresses, salinity is a major factor decreasing the yield in rice cultivation in coastal areas and in irrigated farmlands. Problems associated with salinity are water deficit imposed by the greater osmolarity of the soil solution and the cellular damage inflicted by excessive ion accumulation in plant tissues. Comparison of rice subspecies and varieties differing in tolerance to salinity has shown that greater tolerance correlates with the ability to exclude Na+ from the shoot and maintain a low Na+/K+ ratio (Golldack et al., 2003; Lee et al., 2003; Ren et al., 2005). For instance, the salt-sensitive variety IR29 accumulated Na+ in leaves at 5- to 10-fold greater concentrations than the salt-tolerant lines BK or Pokkali (Golldack et al., 2003). In contrast, shoot K+ concentration per se showed no relation to salinity tolerance in japonica spp. and only weak correlation in indica spp. varieties (Golldack et al., 2003; Lee et al., 2003). Because steady accumulation of Na+ is what injures the cells of leaves at moderate salinity levels (Flowers et al., 1991; Munns, 1993), restricting the translocation of Na+ is a mechanism for salt tolerance that plays a major role in rice (Lee et al., 2003; Ren et al., 2005). The gene SKC1/HKT8, responsible for a major quantitative trait locus imparting a high K+/Na+ balance in shoots and salt tolerance, encodes an Na+-selective transporter of the HKT family that regulates long-distance transport of Na+ (Ren et al., 2005). SKC1/HKT8 participates in reabsorption of Na+ at the xylem parenchyma, thereby restricting the buildup of toxic concentrations of Na+ in photosynthetic tissues (Ren et al., 2005). The related rice gene HKT1 is preferentially expressed in root xylem parenchyma and in cells adjacent to phloem vessels in leaves, suggesting that it could also be involved in the regulation of long-distance transport of Na+ (Golldack et al., 2002). The mechanism by which rice roots take up Na+ is uncertain. Anatomical discontinuities in the root endodermis may lead to uncontrolled apoplastic bypass flow of ions and their subsequent discharge into the vascular bundle (Yeo et al., 1987; Yadav et al., 1996). In this process, ion transporters would play a minor role, or none at all, and the natural variability of salt tolerance among cultivars would be determined by genes controlling developmental traits (Koyama et al., 2001). However, the kinetics of Na+ uptake by rice roots is consistent with enzymatic processes driven by ion transporters (Garciadeblas et al., 2003). Although the molecular identities of these transporters remain to be established, the kinetic properties of OsHKT1 in heterologous systems recapitulate those of whole roots (Garciadeblas et al., 2003). In wheat (Triticum aestivum), TaHKT1 is primarily expressed in the root cortex and down-regulation of TaHKT1 by RNAi reduced Na+ uptake and enhanced salt tolerance, indicating that TaHKT1 mediated Na+ uptake (Laurie et al., 2002).
Sodium extrusion at the root-soil interface, as well as some level of Na+ efflux in every other cell type to achieve ion homeostasis, is presumed to be of critical importance for the salt tolerance of glycophytes (Tester and Davenport, 2003). Indeed, efficient efflux of Na+ to the soil solution must function in the roots of several species to minimize net uptake because unidirectional influx of Na+ is rapid and greatly exceeds the rate of accumulation (Tester and Davenport, 2003). In wheat roots, high rates of Na+ efflux were inferred because net uptake was very low relative to unidirectional influx (Davenport et al., 2005). The Na+/H+ antiporter salt overly sensitive (SOS)1 is the only Na+ efflux protein at the plasma membrane of plants characterized so far. Mutants of Arabidopsis (Arabidopsis thaliana) lacking SOS1 are extremely salt sensitive and have combined defects in Na+ extrusion and in the long-distance transport of this ion from root to shoot (Qiu et al., 2002; Shi et al., 2002). SOS1 is primarily expressed at the root tip epidermis and in xylem parenchyma at the xylem-symplast boundary throughout the plant (Shi et al., 2002). At the root-soil interface, SOS1 would act extruding the excess of Na+ ions from root epidermal cells. In addition, analysis of the Na+ root-shoot partition in the sos1 mutant under different saline regimes indicated that SOS1 also participated in the redistribution of Na+ between roots and shoot in a complex manner (Shi et al., 2002). Under moderate saline stress (25 mm NaCl) sos1 mutant plants accumulated less Na+ in their aerial parts than the wild type, indicating that SOS1 functions in loading Na+ into the xylem for controlled delivery to the shoot. By contrast, at high salinity (100 mm NaCl), the roots and aerial parts of sos1 mutant plants accumulated more Na+ than wild-type plants, which could be caused by the breakdown of Na+ exclusion at the root epidermis and to the large electrochemical gradient of Na+ across the xylem-symplast boundary (Shi et al., 2002; Pardo et al., 2006). In addition, it has been suggested that, under severe salinity stress, the difference in Na+ concentration between xylem sap and xylem parenchyma cells could be of greater magnitude than the corresponding pH gradient, which would result in reversal of SOS1 activity (assuming an electroneutral exchange) and retrieval of Na+ from the xylem (Shi et al., 2002; Tester and Davenport, 2003). The activity of the SOS1 exchanger is regulated through protein phosphorylation by the SOS2/SOS3 kinase complex (Qiu et al., 2002; Quintero et al., 2002). SOS2 is a Ser-Thr protein kinase belonging to the SNF1-related kinase (SnRK)3 family (Gong et al., 2004; Kolukisaoglu et al., 2004). SOS3 is a myristoylated Ca2+ sensor belonging to the recoverin-like family of SOS3-like Ca2+ sensor/binding proteins (SCaBPs)/calcineurin B-like (CBL) proteins (Gong et al., 2004; Kolukisaoglu et al., 2004). Upon Ca2+ binding, SOS3 undergoes dimerization and enhances the protein kinase activity of SOS2 (Guo et al., 2001; Sanchez-Barrena et al., 2005). Besides activating SOS2, SOS3 was also shown to recruit SOS2 to the plasma membrane to achieve efficient interaction with SOS1 (Quintero et al., 2002). Mutant plants deficient in either SOS2 or SOS3 share the salt-sensitive phenotype of sos1 plants (Zhu, 2000).
We have begun to characterize Na+ efflux proteins of rice by isolating a SOS1 homolog, which is encoded by a single-copy gene. We show that OsSOS1 functions as a plasma membrane Na+/H+ antiporter in yeast (Saccharomyces cerevisiae) cells and that, like its Arabidopsis counterpart, it is phosphorylated and activated by the SOS2-SOS3 protein kinase complex. Ectopic expression of OsSOS1 suppressed the growth defects of an Arabidopsis sos1 mutant line. We have also identified the homologs of AtSOS2 and AtSOS3 (OsCIPK24 and OsCBL4, respectively), which coordinately regulate the activity of OsSOS1. These results show that the SOS pathway for salt tolerance operates in cereals.
RESULTS
Cloning of OsSOS1 and Sequence Analyses
A homology-based computer search identified a rice 290-bp expressed sequence tag (EST) clone (rice subsp. japonica cv Nipponbare; accession no. C71771) encoding a putative homolog of the Arabidopsis Na+/H+ antiporter SOS1. The EST clone was obtained from the Ministry of Agriculture, Forestry and Fisheries (MAFF) DNA Bank (Japan) and the entire cDNA insert was sequenced. Sequence comparison with the Arabidopsis SOS1 protein indicated that the rice EST clone encoded a full-length SOS1 homolog, although the open reading frame (ORF) was presumably interrupted by two unspliced introns. These conclusions were supported by sequence comparisons that were extended to include additional SOS1 homologs from other plant species. Sequences of SOS1 homologs from the moss Physcomitrella patens and the seagrass Cymodocea nodosa are available from public databases (accession nos. CAD91921 and CAD20320, respectively). Sequences from the Arabidopsis relative Thellungiella halophila and from the facultative halophyte Mesembryanthemum crystallinum were kindly provided by Valery Poroyko and Hans Bohnert (University of Illinois). Multiple sequence alignments of these polypeptides (data not shown) indicated a high degree of amino acid sequence colinearity among SOS1 homologs and evidenced the presence of two intervening nucleotide sequences of 805 and 160 bp in the rice EST clone at positions +750 and +970, respectively (numbering relative to the ATG codon in the spliced rice cDNA). That these intervening sequences were, in fact, unprocessed introns was confirmed by the analysis of the rice genome sequence at the corresponding locus. To obtain a fully spliced cDNA that could be used for functional studies, reverse transcription (RT)-PCR amplification was performed with rice RNA as a template and oligonucleotides that primed the amplification of a fragment spanning the region that contained the two unprocessed introns in the EST clone. The amplified fragment was sequenced to confirm the removal of all introns and the fidelity of the polymerase and was subsequently subcloned into the EST clone replacing the unspliced portion of the cDNA. In this way, full-length cDNA was created that encoded a 1,148-amino acid protein with 57.5% similarity to the Arabidopsis SOS1 transporter (Supplemental Fig. S1). A search of the available rice genomic sequences (subsp. indica and subsp. japonica) indicated that the isolated OsSOS1 cDNA corresponded to locus Os12g44360 on chromosome XII (25,285–35,082 bp), with no additional sequences of significant similarity. Therefore, SOS1 is a single-copy gene in rice.
Sodium Transport Activity of OsSOS1 Is Stimulated by the Arabidopsis SOS2/SOS3 Kinase Complex
Cells of the yeast strain AXT3K (Δena1-4 Δnha1 Δnhx1) lack all major sodium transporters essential for the sodium tolerance of yeast and thus are incapable of growing in Arg-phosphate (AP) medium with Na+ concentrations higher than 70 mm (in 1 mm K+; Quintero et al., 2002). The P-type pumps ENA1 to ENA4 and the (Na+,K+)/H+ antiporter NHA1 localize to the plasma membrane and both mediate Na+ efflux (Wieland et al., 1995; Bañuelos et al., 1998), whereas the Na+/H+ antiporter NHX1 drives Na+ sequestration into endosomal compartments (Darley et al., 2000; Quintero et al., 2000). When OsSOS1 was expressed in AXT3K cells from a multicopy vector (pDR195), halotolerance was partially recovered and cells could grow in AP medium containing up to 100 mm sodium (1 mm K+; Fig. 1A).
Figure 1.
Activation of rice SOS1 by the Arabidopsis SOS2/SOS3 kinase complex. A, AXT3K cells transformed with an empty vector (0) or with the indicated combination of SOS genes (1, OsSOS1; 2, AtSOS2; 3, AtSOS3) were grown overnight in selective synthetic dextrose medium. Five microliters of serial decimal dilutions were spotted onto plates of AP medium with 1 mm KCl and supplemented with 0 (data not shown), 100, or 200 mm NaCl. Plates were incubated at 28°C for 3 d. Plasmids used for expression of the SOS proteins were pDR195 for OsSOS1, pFL2T for AtSOS2, pFL3T for AtSOS3, and pFL32T for coexpression of AtSOS2 and AtSOS3. B, Intracellular Na+ content as determined by atomic emission spectrometry. Cells were grown in AP medium with 1 mm KCl and 30 mm NaCl and collected at OD550 = 0.2 to 0.3. Values are the mean and se of three independent cultures of each combination of SOS genes. Units are nanomols of ions per milligram dry weight of cell samples. [See online article for color version of this figure.]
The Arabidopsis AtSOS1 transporter has been shown to be phosphorylated and activated by the SOS2/SOS3 protein kinase complex (Qiu et al., 2002; Quintero et al., 2002). SOS2 is a Ser-Thr protein kinase whose activity and subcellular localization are dependent on the interaction with the calcium sensor protein SOS3 (Halfter et al., 2000; Quintero et al., 2002). To test whether the rice homolog OsSOS1 could also be activated by the Arabidopsis SOS2/SOS3 kinase complex, Arabidopsis SOS2 and SOS3 proteins were coexpressed in AXT3K yeast along with OsSOS1, and the effect of these combinations on the halotolerance of transformants was determined. As shown in Figure 1A, concurrent expression of the three SOS proteins greatly increased the salt tolerance of the transformed cells and afforded growth in media with up to 400 mm sodium, which is near the maximal salt concentration tolerated by wild-type yeast cells in AP medium (data not shown). This enhanced tolerance was observed only when OsSOS1 was present, negating that SOS2 and SOS3 were unmasking endogenous yeast activity. AtSOS2 alone in the presence of OsSOS1 was capable of enhancing the salt tolerance of yeast to an intermediate level lower than that under coexpression of the three SOS proteins, but still considerably higher than that with OsSOS1 alone. Presumably, this enhanced salt tolerance resulted from the basal activity of SOS2 in the absence of SOS3.
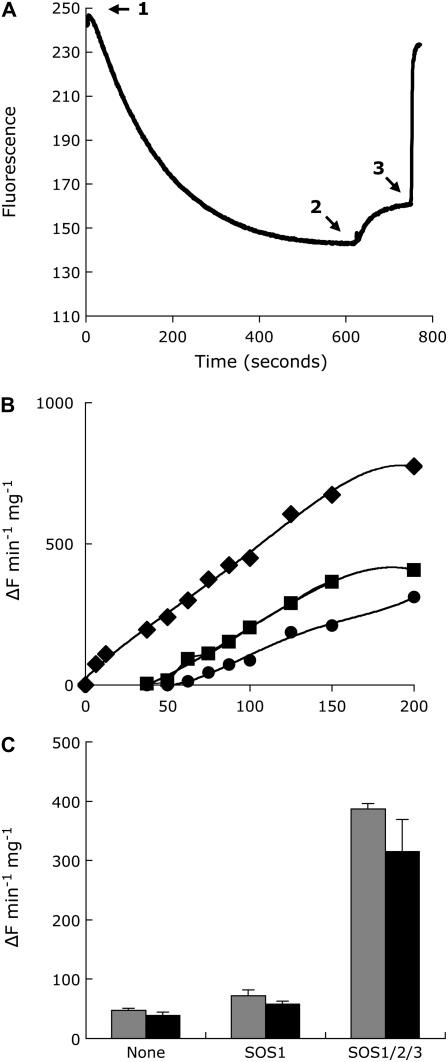
To determine how OsSOS1 conferred greater Na+ tolerance to yeast, we measured the Na+ content of cells expressing various combinations of SOS proteins. Cells expressing OsSOS1 alone and growing in AP with 30 mm NaCl (1 mm KCl) maintained their intracellular sodium levels considerately lower than did control cells transformed with an empty vector (Fig. 1B). The intracellular Na+ content was minimal when the three SOS proteins were coexpressed, well below that of cells expressing OsSOS1 alone or together with SOS2. Thus, the association between the halotolerance and cytoplasmic sodium levels suggests that OsSOS1 functions as a sodium efflux transporter in yeast and that its activity is greatly enhanced by the SOS2/SOS3 kinase complex. To directly demonstrate Na+/H+ antiporter activity of OsSOS1, plasma membrane vesicles were purified by use of aqueous two-phase partitioning from yeast expressing OsSOS1 with and without coexpression of the SOS2/SOS3 kinase complex. Cells were grown on selective AP medium containing 1 mm KCl and transferred to the same medium supplemented with 100 mm NaCl for 1 h to ensure activation of the SOS2/SOS3 kinase complex when present. The purity of vesicle preparations was tested by measuring ATP hydrolysis in the presence of inhibitors of mitochondrial (azide), vacuolar (nitrate), and plasmalemma (vanadate) ATPases. The relative sensitivity of total ATPase activity to these inhibitors demonstrated that vesicle preparations were enriched in plasma membrane (data not shown). Na+/H+ exchange was monitored by the quinacrine fluorescence quenching method. An inside-acid proton gradient (ΔpH) across vesicle membranes was established after the addition of ATP (Fig. 2A). Once ΔpH reached steady state, the addition of sodium salts led to fluorescence recovery (i.e. dissipation of the ΔpH; Fig. 2A). Vesicles isolated from yeast cells expressing OsSOS1 displayed Na+/H+ exchange activity that was greater than background exchange in AXT3K recipient cells over 25 to 200 mm Na2SO4 (Fig. 2B). Maximal Na+/H+ exchange activity, which was detectable at lower Na+ concentrations, was observed in cells coexpressing OsSOS1 and the Arabidopsis SOS2/SOS3 kinase complex, as was expected from activation of the rice Na+ transporter by the SOS2/SOS3 kinase. These differences in Na+/H+ exchange activity are in agreement with the relative tolerance to NaCl and intracellular Na+ content of these transformants (see Fig. 2). To further ensure that ΔpH dissipation was due to Na+/H+ exchange, transport assays were performed with sodium gluconate as a substrate. Relative Na+/H+ exchange rates among vesicle preparations were commensurate at identical Na+ concentrations independently of the salt used (Fig. 2C). Because gluconate is an impermeant anion, unlike sulfate, ΔpH dissipation was concluded to be specific for Na+. Transport assays in the presence of K+ and valinomycin to dissipate the electrical membrane potential produced similar results (data not shown), further indicating that Na+ did not move electrophoretically and that the Na+/H+ exchange mediated by SOS1 was electroneutral. An approximate Km of 29 mm for Na+ was estimated for activated OsSOS1 in the range of 6.5 to 100 mm Na+ after subtracting background transport in control vesicles from the AXT3K strain.
Figure 2.
Na+/H+ antiporter activity of rice SOS1. A, ATP-dependent pH gradient formation in membrane vesicles isolated from yeast cells expressing rice SOS1. An inside-acid ΔpH was formed after the addition of ATP to vesicles (arrow 1). Once fluorescence was stabilized, sodium salts were added to the cuvette (arrow 2) and fluorescence recovery, indicating proton exchange, was monitored for 2 min, after which ΔpH was disrupted by the addition of 25 mm (NH4)2SO4 (arrow 3). Fluorescence is expressed as arbitrary units. B, Na+/H+ exchange as a function of Na2SO4 concentration and the presence of rice OsSOS1, with and without coexpression of the Arabidopsis SOS2/SOS3 kinase complex. Circles, AXT3K cells transformed with empty vector pDR195; squares, AXT3K cells expressing rice OsSOS1 alone; diamonds, AXT3K cells transformed to produce proteins OsSOS1, AtSOS2, and AtSOS3. Na+/H+ exchange activity is given as the proportion of dissipation of the preformed pH gradient per minute and milligrams of membrane protein. C, Specificity of sodium-induced proton exchange. Sodium was added to a final concentration of 75 mm as sulfate (gray bars) or gluconate (black bars) salt. Values are the mean and se of percent fluorescence dissipation of triplicate samples.
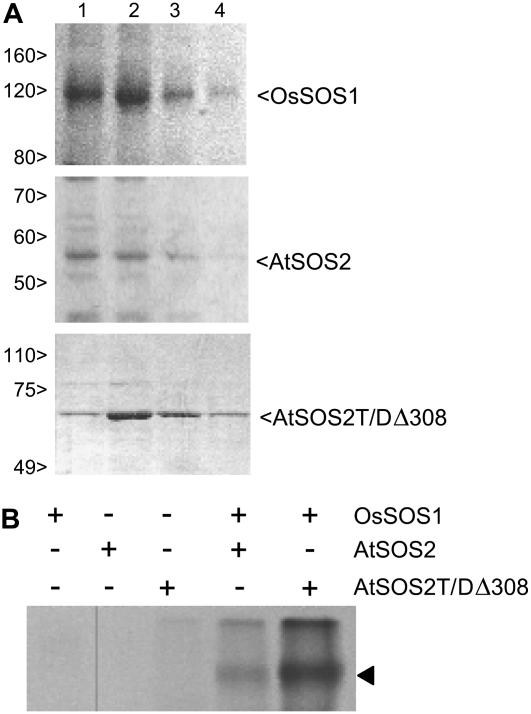
We have previously shown that the Arabidopsis SOS1 protein is a substrate for the SOS2/SOS3 kinase complex (Quintero et al., 2002). Results depicted in Figures 1 and 2 indicate that the activity of the rice SOS1 transporter is also enhanced by the coexpression of the Arabidopsis protein kinase. To confirm that OsSOS1 was phosphorylated by AtSOS2, C-terminal His-tagged OsSOS1 and AtSOS2 were purified from yeast membranes by Ni2+-binding chromatography (Fig. 3A). Also, a mutant form of SOS2 (SOS2T/DΔ308) that is hyperactive and independent of SOS3 in vitro (Guo et al., 2001) was purified as a translational glutathione S-transferase (GST) fusion (Fig. 3A). As shown in Figure 3B, OsSOS1 was weakly phosphorylated by wild-type SOS2 (in the absence of SOS3) compared to the strong phosphorylation catalyzed by the recombinant SOS2T/DΔ308 kinase, thus demonstrating that the rice protein is recognized as a legitimate substrate by the Arabidopsis protein kinase.
Figure 3.
Phosphorylation of rice SOS1 by the Arabidopsis kinase SOS2. A, Recombinant His-tagged OsSOS1, AtSOS2, and GST-fused AtSOS2T/DΔ308 were purified by affinity chromatography. Aliquots (1–4) of the first elution volumes were analyzed for protein purity by SDS-PAGE. Bands corresponding to OsSOS1:His-6x (128 kD), AtSOS2:His-6x (52 kD), and GST:AtSOS2T/DΔ308 (60 kD) are indicated. Standard Mrs are shown on the left. B, Purified proteins were combined as indicated in protein kinase reaction assays. Aliquots of phosphorylation reactions were resolved by SDS-PAGE and exposed to x-ray films. Arrowhead indicates the 128-kD band pertaining to phosphorylated OsSOS1.
Complementation of the Arabidopsis sos1 Mutant
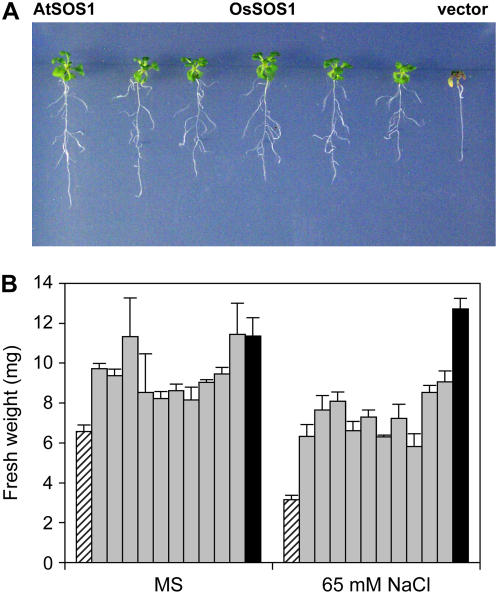
In Arabidopsis, AtSOS1 has been shown to fulfill two important roles pertaining to sodium tolerance, namely, restriction of net Na+ uptake by roots and control of xylem loading for long-distance transport (Shi et al., 2002). To extend the functional comparisons between OsSOS1 and AtSOS1 to whole plants, full-length cDNA encoding the rice transporter was transformed into the Arabidopsis mutant sos1-1, which bears a 14-bp gene deletion that completely abrogates AtSOS1 activity (Shi et al., 2000; Qiu et al., 2002). Transgenic seedlings (T1 generation) were selected on Murashige and Skoog medium with kanamycin and then transferred to Murashige and Skoog supplemented with 50 mm NaCl to test genetic complementation (Fig. 4). Ten of 16 transgenic T1 seedlings tested continued growing in the presence of salt (Fig. 4A) and were successfully transferred to soil for further analyses. In all 10 complemented lines, expression of the OsSOS1 transgene was confirmed by RT-PCR with the use of rice-specific primers, and the sos1-1 genetic background was verified by diagnostic PCR of the 14-bp deletion that defines this mutant allele (data not shown; Shi et al., 2000). Mutant sos1-1 plants showed a moderate growth defect in Murashige and Skoog plates, which was further exacerbated by the imposition of moderate salinity stress (Fig. 4B). Phenotypic suppression of the Arabidopsis sos1-1 mutant by rice SOS1 cDNA was partial because of the greater growth of congenic lines transformed with the Arabidopsis SOS1 cDNA as compared with transgenic lines expressing the rice protein, both in the absence and presence of NaCl (Fig. 4B). These results indicate that the rice SOS1 protein can substitute for the endogenous transporter of Arabidopsis, albeit not completely.
Figure 4.
Complementation of Arabidopsis sos1-1 mutant by rice OsSOS1. A, Six-day-old seedlings grown on Murashige and Skoog agar medium were transferred to Murashige and Skoog medium supplemented with 50 mm NaCl and imaged after 14 d of salt treatment. Left, Transgenic sos1-1 mutant seedling expressing AtSOS1 from the cauliflower mosaic virus 35S promoter. Middle, five independent transgenic lines of sos1-1 mutants expressing the rice OsSOS1 gene under the control of the 35S promoter. Right, sos1-1 mutant seedling transformed with empty vector pBI321. B, Quantitation of seedling growth, expressed as fresh weight after 14 d in Murashige and Skoog medium with and without supplemental 65 mm NaCl. Data are the mean and se of fresh-weight values of three to six individual seedlings from each line. Dashed bars, sos1-1 mutant seedlings transformed with empty vector pBI321; gray bars, sos1-1 mutant seedlings transformed with the rice OsSOS1 gene under the control of the 35S promoter; black bars, sos1-1 mutant seedlings transformed with Arabidopsis SOS1. [See online article for color version of this figure.]
OsSOS1 Transcript Levels in Response to Salt Stress
To learn whether the expression of OsSOS1 is modified under salt stress, a northern-blot membrane was prepared with total RNA purified from roots and shoots of rice plants subjected to 100 mm NaCl salinity stress for 3, 15, and 48 h. Hybridization was performed with a probe corresponding to the C-terminal part of OsSOS1 and radiometric signals were quantified by densitometric scanning of autoradiograms. The relative signal intensities were normalized by reprobing the blots with an 18S rRNA gene probe. Basal OsSOS1 expression levels were detected in roots and shoots of control, nontreated plants (Fig. 5). After 3-h salt treatment, roots showed a slow and transient increase in the level of OsSOS1 transcripts, which reached a maximal 6-fold induction 15 h after the onset of salt stress (Fig. 5). By contrast, the effect of salt stress in shoots was the opposite. After 3-h salt treatment, the level of OsSOS1 transcripts decreased, reaching a 5-fold reduction in mRNA abundance relative to basal levels in control plants (Fig. 5). This decrease was transient and the mRNA abundance slowly recovered, reestablishing near-basal levels after 48 h of salt treatment. These results further suggest a functional role of OsSOS1 in the response of rice plants to salt stress. The changes in mRNA abundance in roots over the first day after transfer to salt were likely related to the osmotic shock rather than to ionic stress, whereas the up-regulation in roots and leaves at later times after turgor recovery (48 h) was the likely result of sodic stress.
Figure 5.
Transcript abundance of OsSOS1 in response to salt stress. Total RNA purified from roots and shoots of rice plants subjected to salt stress with 100 mm NaCl in hydroponic culture medium for 0, 3, 15, and 48 h. Hybridization was performed with a probe prepared from OsSOS1 cDNA. RNA sample loading was normalized by hybridization with a probe derived from radish (Raphanus sativus) 18S rDNA.
Identification of a Rice Kinase Complex That Activates OsSOS1
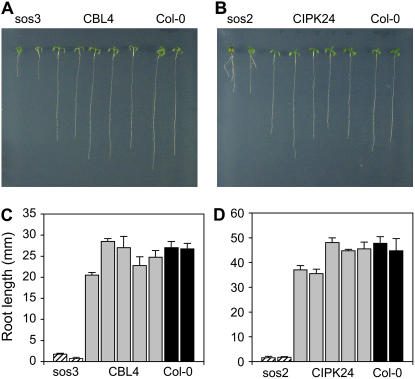
Analysis of the rice genome allowed for the identification of up to 30 CBL-interacting protein kinases (CIPK)/SOS2-like protein kinases (PKS) of the SnRK3 family and 10 CBL/SCaBP-interacting calcium sensors (Kolukisaoglu et al., 2004). On the basis of amino acid sequence comparisons (Thompson et al., 1997), the rice proteins most similar to the Arabidopsis SOS3 protein are OsCBL4 (Os05g45810), OsCBL7 (Os02g18880), and OsCBL8 (Os02g18930), with 66.2%, 67.1%, and 66.7% identity to AtSOS3, respectively (Supplemental Fig. S2). Like AtSOS3, all three rice homologs are predicted to be N-myristoylated proteins, a posttranslational modification that is essential for SOS3 functionality (Ishitani et al., 2000). The remaining OsCBL proteins show significantly lower similarity scores (Hwang et al., 2005). All CBL proteins contain four conserved elongation factor hand motifs for Ca2+ binding, separated by intervening sequences of fixed length. Size and sequence variations among CBL proteins are therefore restricted largely to N- and C-terminal extension from the conserved core (Kolukisaoglu et al., 2004). Because the N-terminal part of AtSOS3 is more similar to that of OsCBL4 than OsCBL7 and OsCBL8 (Supplemental Fig. S2) and OsCBL4 localizes to the plasma membrane as does SOS3 (Hwang et al., 2005), full-length cDNA of OsCBL4 was amplified by RT-PCR with total mRNA isolated from rice cv Nipponbare used as a template as described in “Materials and Methods.” To test its putative functional identity with AtSOS3, OsCBL4 was cloned in the plasmid pDR195 to express the rice protein from the PMA1 gene promoter. The resulting plasmid, pDROsCBL4, was transformed in the yeast strain YP890, expressing AtSOS1 from chromosomal integration, with or without AtSOS2 as the interacting protein kinase. As shown in Figure 6A, OsCBL4 was competent for interaction with AtSOS2 and activation of AtSOS1, as expected if OsCBL4 were a functional homolog of AtSOS3. Moreover, expression of OsCBL4 suppressed completely the salt sensitivity of the Arabidopsis mutant sos3-1 (Fig. 7, A and C). The sos3-1 mutant bears a 9-bp deletion starting at position 759 from the translation start codon (Liu and Zhu, 1998). Five sos3-1 lines (T2 plants) showing genetic complementation were further analyzed, and all tested positive for expression of OsCBL4 on RT-PCR. They also tested positive for the sos3-1 genetic background on diagnostic PCR for the signature 9-bp deletion (data not shown). Together, these data indicate that OsCBL4 can functionally substitute for AtSOS3.
Figure 6.
Functional interactions between Arabidopsis SOS proteins and rice counterparts. Strains YP890 carrying the integration of the PGK1PRO:AtSOS1:CYC1TER cassette (A and C), YP1021 with the analogous integration PMA1PRO:OsSOS1:ADH1TER with the SOS1 cDNA from rice (D), and AXT3K cells transformed with plasmid pSOS1-1 for the expression of AtSOS1 (B) were transformed with plasmids directing the expression of the regulatory proteins SOS2 and SOS3 from Arabidopsis or CIPK24 and CBL4 from rice, as indicated in each case. CIPK24Δ309 bears a C-terminal deletion rendering a constitutive, CBL-independent protein kinase. Yeast cells were grown overnight in selective synthetic dextrose medium. Five microliters of serial decimal dilutions were spotted onto plates of AP medium with 1 mm KCl and 200 mm NaCl. Plates were incubated at 28°C for 3 to 4 d.
Figure 7.
Complementation of Arabidopsis sos2 and sos3 mutants by rice CIPK24 and CBL4. Six-day-old seedlings of mutants sos3-1 (A) and sos2-2 (B) transformed with cDNAs of rice genes CBL4 (A) or CIPK24 (B), respectively, were transferred to Murashige and Skoog medium supplemented with 100 mm (CBL4 and sos3) or 75 mm NaCl (CIPK24 and sos2) and imaged after 14 d of growth. From left and right, two wild-type plants, five complemented lines, and two mutant plants transformed with empty vectors are depicted. The root length of seven individual plants from each of these lines was measured after 14 d in salinized media. C, sos3 mutant and rice CBL4. D, sos2 mutant and rice CIPK24. In both plots, dashed columns represent the root length attained by mutant lines; gray columns are complemented lines; and back columns are wild-type Col-0. [See online article for color version of this figure.]
The Arabidopsis protein kinase SOS2/CIPK24 is most similar to the uncharacterized rice proteins OsCIPK24 (Os06g40370; 68.2% identity) and OsCIPK8 (protein accession no. BAD87720; 62% identity). Because OsCIPK8 is also more similar to Arabidopsis AtCIPK8 than to AtSOS2 (Kolukisaoglu et al., 2004), we focused our initial analysis on OsCIPK24. Full-length cDNA was amplified by RT-PCR with total mRNA isolated from rice cv Nipponbare used as a template as described in “Materials and Methods.” Oligonucleotides for amplification were designed according to the available sequence of a potential full-length cDNA sequence of OsCIPK24 (database entry AK102270) coding for a putative 454-amino acid polypeptide. cDNA was cloned in the yeast vector p414GPD and transformed in the yeast strain YP890 (PGK1:AtSOS1:CYC1), with and without the potentially interacting protein OsCBL4. Unexpectedly, OsCIPK24 failed to activate AtSOS1 in the yeast system (data not shown). To test whether this failure was due to the inability of OsCIPK24 to interact with OsCBL4 or to recognize AtSOS1 as a substrate, a constitutive CBL-independent form of OsCIPK24 was produced by deletion of the conserved C-terminal autoinhibitory domain present in SnRK3 kinases (Albrecht et al., 2001; Guo et al., 2001). On the basis of the known localization of the autoinhibitory domain of AtSOS2 (Guo et al., 2001), a C-terminal truncation of OsCIPK24 was produced by introducing a stop codon at the Leu residue at position 315 (Supplemental Fig. S3). The truncated protein was tested for its ability to activate AtSOS1 or the rice protein OsSOS1, with negative results in both cases (data not shown), which suggests that the OsCIPK24 was not a functional homolog of AtSOS2 or that the protein being produced in yeast was not biologically active. Close inspection of the predicted ORF of the cDNA amplified based on the EST database entry AK102270 revealed that a conceptual translation starting at the downstream ATG codon corresponding to Met in position +7 predicted a shorter protein with greater similarity to the amino-terminal part of AtSOS2 than the one produced by the larger ORF present in the OsCIPK24 cDNA amplified initially (Supplemental Fig. S3). Hence, seven-amino acid shorter versions of OsCIPK24, with or without the C-terminal deletion at the Leu residue at position 309 (numbering relative to the new start codon; denoted OsCIPK24Δ309), were cloned in plasmid p414GPD and tested in yeast cells. As depicted in Figure 6B, the constitutive protein kinase OsCIPK24Δ309 was able to activate AtSOS1 in the absence of OsCBL4, which demonstrates that the polypeptide with the shortened N terminus was biologically active. Moreover, coexpression of the complete OsCIPK24 (i.e. starting at the second Met, but with the intact C-terminal inhibitory domain) interacted productively with OsCBL4 to activate the SOS1 exchangers from both Arabidopsis (AtSOS1) and rice (OsSOS1), but failed to fully activate these exchangers in the absence of OsCBL4 (Fig. 6, C and D). Finally, the OsCIPK24 kinase (short N terminus) was also able to suppress the salt sensitivity of an Arabidopsis sos2-2 mutant completely (Fig. 7, B and D). The sos2-2 allele contains a 2-bp deletion corresponding to nucleotides 1,521 and 1,522 relative to the translational start codon of SOS2 (Liu et al., 2000). Six transgenic lines (T2 plants) showing genetic complementation for salt tolerance tested positive for expression of the OsCIPK24 transgene on RT-PCR and for the signature deletion of the sos2-2 mutant allele on diagnostic PCR (data not shown). Together, these results strongly suggest that OsCIPK24 is the functional rice homolog of Arabidopsis SOS2. They also show that the OsCIPK24-OsCBL4 complex forms a functional module with OsSOS1 to achieve sodium tolerance.
DISCUSSION
Our genetic and biochemical analyses show that a rice protein, OsSOS1, with significant sequence similarity to the SOS1 sodium exchanger from Arabidopsis, is also its functional homolog. First, OsSOS1 suppressed the Na+ sensitivity of a yeast mutant lacking the major Na+ efflux systems at the plasma membrane [the array of ENA1-ENA4 Na+ pumps and the (Na+,K+)/H+ exchanger NHA1] by a mechanism that reduced the net cellular Na+ content (Fig. 1). Furthermore, plasma membrane preparations from yeast transformants expressing OsSOS1 demonstrated greater capacity for Na+/H+ exchange (Fig. 2). Second, the Arabidopsis protein kinase complex SOS2/SOS3, which positively controls the activity of AtSOS1 (Qiu et al., 2002; Quintero et al., 2002), also stimulated the activity of OsSOS1, both in vivo (Fig. 1) and in plasma membrane vesicles (Fig. 2, B and C), and OsSOS1 was recognized as a genuine phosphorylation substrate by AtSOS2 (Fig. 3B). Third, OsSOS1 suppressed the growth defect, both in the absence and presence of salt, of a sos1-1 mutant of Arabidopsis (Fig. 4). Taken together, these results demonstrate that OsSOS1 is a functional homolog of AtSOS1, and they represent the first molecular and biochemical characterization of a Na+ efflux protein from monocots. Rice OsSOS1 catalyzed Na+/H+ exchange in plasma membrane vesicles derived from the yeast strain AXT3K in which genes encoding the Na+ efflux proteins ENA1 to ENA4 and NHA1 had been deleted. Although these proteins account for most of the Na+ efflux in yeast plasma membranes and the cellular tolerance to Na+ (Bañuelos et al., 1998), some capacity for Na+/H+ exchange still remained in plasma membrane vesicles of the strain AXT3K at Na+ concentrations greater than 50 mm (Fig. 2B). Expression of OsSOS1 alone imparted moderate enhancement of Na+/H+ exchange above that background activity. In contrast, coexpression of the rice SOS1 with the activating SOS2/SOS3 complex from Arabidopsis significantly increased the Na+/H+ exchange in vesicles. An approximate Km of 29 mm for Na+ was estimated for activated OsSOS1, which is similar to the Km of 23 mm Na+ that has been estimated for AtSOS1 in plasma membrane vesicles of Arabidopsis plants subjected to salt exposure to induce the activity of AtSOS1 (Qiu et al., 2003). Accordingly, the salt tolerance imparted to yeast cells by OsSOS1 and AtSOS1 was comparable (Fig. 6, A and D).
The rice OsSOS1 protein was phosphorylated and activated by the Arabidopsis SOS2/SOS3 proteins (Figs. 2 and 3), which strongly suggests that the mechanistic details of the biochemical regulation of SOS1 proteins are conserved among these species and that, as a consequence, functional homologs of the SOS2 and SOS3 proteins should exist in rice. On the basis of protein sequence similarity, functional tests in yeast, and genetic complementation of Arabidopsis mutants, we have isolated likely candidates as the functional rice homologs of the SOS2/SOS3 kinase complex. The rice proteins OsCIPK24 and OsCBL4 were able to activate the rice transporter OsSOS1 and they could also be exchanged with their Arabidopsis counterparts to form heterologous protein kinase modules that were fully competent to activate the SOS1 Na+/H+ antiporters from both Arabidopsis and rice in yeast cells (Figs. 1 and 6). Their ability to complement the sos2 and sos3 mutations of Arabidopsis implies that they are able to form heterologous protein kinase complexes in planta also (Fig. 7). We have previously shown that overexpression of SOS2 or SOS3 failed to increase the salt tolerance of complemented mutant lines above wild-type levels (Guo et al., 2004), which is consistent with the data presented in Figure 7 showing that the overexpression of the rice proteins brought the salt tolerance of the Arabidopsis mutants close, but not beyond, that of wild-type plants. These results show a high degree of structural conservation among the SOS proteins from dicots and monocots. Contrary to OsSOS1, which is a single-copy gene in rice, OsCIPK24 and OsCLB4 belong to a gene family. Although the complete genomes of subsp. japonica and subsp. indica are assembled only in part, in both cases the same set of 10 CBLs and 30 CIPKs have been identified in silico (Kolukisaoglu et al., 2004). Hence, it remains a possibility that other rice CIPK and CBL isoforms could also have tested positive in our functional analyses, especially for OsCBL4, for which there were at least two other candidates, OsCBL7 and OsCBL8, with high sequence similarity and predicted N-terminal myristoylation (Kolukisaoglu et al., 2004; Hwang et al., 2005). In rice, the predicted polypeptides of OsCBL1, OsCBL4, OsCBL5, OsCBL7, and OsCBL8 are potential substrates for N-myristoylation, a lipid modification that promotes protein-protein or protein-membrane interactions in eukaryotic cells (Kolukisaoglu et al., 2004). However, only OsCBL4 has been localized to the plasma membrane (Hwang et al., 2005), which is consistent with a role in SOS1 activation (Quintero et al., 2002). Nonetheless, because CIPK-PKS protein kinases can interact with more than one CBL-SCaBP protein (Guo et al., 2001; Kolukisaoglu et al., 2004), whether OsCBL4, OsCBL7, and/or OsCBL8 are functionally redundant needs to be investigated. The phenotypic analysis of the growing number of annotated insertional mutants in rice should enable the unequivocal identification, by genetic criteria, of the isoforms of CBLs and CIPKs that are relevant for salt tolerance.
Root hairs and epidermal cells are primary sites for controlled uptake of inorganic ions that enter the symplastic pathway. In addition, ions move apoplastically across the cortex with the bulk flow of water and solutes until they reach the endodermis barrier that prevents further diffusion into the vasculature and entails the selective uptake of ions into the symplast. In a saline environment, the cell-to-cell pathway imparts greater selectivity and reduced ion uptake than the bulk flow of water and solutes along the root apoplastic pathway. The relative contribution of each pathway to net flow of water and solutes is affected by environmental factors, including salinity (Steudle and Peterson, 1998; De Boer and Volkov, 2003). Typically, under stress conditions, the apoplastic pathway is reduced by a process that involves the lignosuberization of root tissues (Azaizeh and Steudle, 1991; Cruz et al., 1992; Sanchez-Aguayo et al., 2004). Notwithstanding this general model, sodium uptake has been suggested to occur in rice roots primarily via apoplastic bypass flow and leakage into the xylem (Yeo et al., 1987). This apoplastic pathway is presumed to involve discontinuities along the rhizodermal and endodermal barriers (Yeo et al., 1987; Yadav et al., 1996). Consequently, the uptake of sodium in rice in saline conditions was proposed to be controlled by genes affecting root anatomy rather than membrane transport processes (Koyama et al., 2001). However, even if uncontrolled apoplastic flow were the predominant pathway for Na+ entry, the root vascular cylinder, an enzymatic Na+ efflux mechanism, would still be of critical importance to minimize cellular Na+ accumulation in tissues along the plant axis. The plasma membrane Na+/H+ antiporter SOS1 could likely fulfill this role and prevent cellular injury, thus contributing to whole-plant salt tolerance, particularly at moderate salinity levels that do not cause catastrophic physiological failure. Low salt concentration may not, in itself, be damaging to rice (Yeo et al., 1991); it is the progressive increase in internal concentration that leads to cellular damage and growth impairment (Munns, 1993). As long as growth is sustained to keep the concentration of salt in leaf tissues low by dilution, significant damage could be averted because the long-term buildup of salt in leaves is what ultimately leads to injury (Flowers et al., 1991). Although enhanced ion compartmentation in vacuoles may increase the salt tolerance of rice (Ohta et al., 2002; Fukuda et al., 2004), this species is a typical glycophyte that relies primarily on Na+ exclusion for growth in saline environments (Golldack et al., 2003; Lee et al., 2003).
In support of the critical involvement of ion transport processes in Na+ accumulation by rice roots, the ability of salt-tolerant lines to exclude Na+ depends on the K+ concentration in the growth medium. At high to low micromolar K+ concentrations, the salt-tolerant lines Pokkali and BK selectively excluded Na+ while keeping the internal K+ concentration fairly constant. In contrast, at low micromolar K+, these lines accumulated Na+ similar to the salt-sensitive line IR29 (Golldack et al., 2002, 2003). Physiological measurements of ion uptake by rice roots indicate the existence of separate high-affinity uptake pathways for K+ and Na+, which have been suggested to be mediated by HAK-KUP transporters and HKT transporters, respectively (Garciadeblas et al., 2003). The rice isoform OsHKT1 is expressed in root epidermal and cortical cells (Golldack et al., 2002) and its kinetic parameters are similar to those displayed by whole roots (high-affinity Na+ influx that is blocked by K+), whereas the isoform OsHKT4 is expressed in shoots and mediates fast Na+ influx with low affinity (Garciadeblas et al., 2003). Recently, a major quantitative trait locus for shoot K+ content and salt tolerance in rice identified the gene SKC1 encoding an HKT-type protein corresponding to OsHKT8 (Garciadeblas et al., 2003; Ren et al., 2005). SKC1/HKT8 was preferentially expressed in the parenchymal cells surrounding xylem vessels and up-regulated by salt stress (Ren et al., 2005). The SKC1/HKT8 isoform from the relatively salt-tolerant variety Nona Bokra was more active in the facilitation of Na+ transport across the plasma membrane than its counterpart from the salt-sensitive Koshihiraki variety, which suggests that a greater capacity for Na+ retrieval from the xylem by SKC1/HKT8 in Nona Bokra plants was the basis for their salt tolerance. Together, these results are consistent with facilitated Na+ transport in saline conditions, particularly at low K+, and support the concept that ion transport systems and their capacity for Na+:K+ selectivity are of physiological relevance for the salt tolerance of rice. By extension, they also imply that mechanisms restricting net Na+ content, including active efflux at the plasma membrane to counteract ion loading, would be determinants for salt tolerance. A precise assessment of the relative importance of SOS1 in the salt tolerance of rice awaits the availability of knockout mutants or RNAi lines.
MATERIALS AND METHODS
Isolation of Rice cDNAs
An EST clone (accession no. C71771) encoding a putative SOS1 homolog in rice (Oryza sativa) cv Nipponbare was obtained from the MAFF DNA Bank (Japan). The deduced ORF was apparently interrupted by two unprocessed introns of 805 and 160 bp at nucleotide positions +750 and +970, respectively (numbering relative to the ATG codon in the spliced cDNA). To obtain a fully spliced cDNA, RT-PCR was performed with rice RNA as a template and the primers RSOS1A (5′-GCGTCGACAATCCATGGACAATCCCGAGGCGG) and RSOS1B (5′-TGTGATAAAATTGGATCCAATGAATGCC), which annealed at the putative initiation codon (underlined) and next to an internal BamHI site, respectively. The RT-PCR reaction yielded a fragment containing a fragment of the ORF, from nucleotides +1 to +2,432, which was subcloned in the vector pCR-BluntII-TOPO (Invitrogen) and fully sequenced to confirm the continuity of the ORF and the fidelity of the amplification. Finally, the amplified fragment was reintroduced in the original EST clone as a SalI-BamHI fragment, replacing the equivalent portion in the original cDNA clone with unprocessed introns. The sequence of the corrected cDNA has been deposited in GenBank (accession no. AY785147).
A putative full-length cDNA clone (AK101368) corresponding to OsCBL4 (according to the nomenclature of Kolukisaoglu et al., 2004) guided the design of oligonucleotides OsCBL4F (5′-TCGCCATGGGATGCGCGTCGT-3′) and OsCBL4R (5′-TATTTTCAGTCATGGGCTTCT-3′) used for the amplification of the ORF of OsCBL4 by RT-PCR with total mRNA from rice cv Nipponbare as a template. The amplified cDNA was fully sequenced to assess the absence of errors. Likewise, the full-length cDNA clone AK102270 encoding OsCIPK24 was used to design the oligonucleotides OsK24F (5′-GGCGGATGGGAGGGGAGGAGG-3′) and OsK24R (5′-CCAGGCTAGCATGTGGCTGTC-3′) for the amplification of the OsCIPK24 by RT-PCR. Amplification from the second Met at position +7 was done with primer OsK24M2F (5′-CGCGGATCCGCGGATGGCGGCGGGGAGGA-3′). Truncation of the C-terminal part of OsCIPK24 at the Leu residue at position 315/309 was achieved by PCR with an oligonucleotide of sequence (5′-CACCTAAAGAGGGCCACCATC-3′), which introduced a stop codon, and primers OsK24F and OsK24M2F, respectively.
Plasmid Constructs
Plasmid pDROsSOS1 for expression of OsSOS1 in yeast (Saccharomyces cerevisiae) cells under the control of the PMA1 gene promoter was constructed by subcloning the OsSOS1 cDNA as a 3.7-kb SalI-NotI fragment in XhoI-NotI restriction sites of multicopy vector pDR195 (Rentsch et al., 1995). A C-terminal His-tagged version of OsSOS1 for protein purification was created by PCR using the high-fidelity Pfu polymerase (Promega) and the primers 5′-AAATGGCAACACATGAGCTCAGGG-3′ and 5′-TGAGCGGCCGCTCAGTGATGGTGATGGTGATGTCGATCAGCAGCGCT-3′. The OsSOS1:His-6x cDNA was subcloned as a BglII-NotI fragment into pDROsSOS1 to produce plasmid pDROsSOS1H. A His-tagged version of wild-type AtSOS2 was produced by PCR using the primers 5′-AGAAGCTTATGACAAAGAAAA-3′ and 5′-GAGCGGCCGCAAACGTGATTG-3′. The PCR product was digested with HindIII and NotI and subcloned in the yeast expression vector pYESHis, a derivative of pYES2 (Invitrogen) modified to contain the epitope RGSH6x (Venema et al., 2002). The OsCBL4 cDNA was cloned in pDR195 as a XhoI-BamHI fragment to create plasmid pDROsCBL4. All versions of the OsCIPK24 cDNA (full length or bearing N- and C-terminal truncations) were cloned as EcoRI-EcoRI fragments in the yeast expression vector p414GPD (Mumberg et al., 1995). Plasmids pFL2T, pFL3T, and pFL32T used for the expression in yeast of Arabidopsis (Arabidopsis thaliana) SOS2, SOS3, and SOS2/SOS3 proteins, respectively, and plasmid pSOS1-1 for the expression of AtSOS1, have been described elsewhere (Quintero et al., 2002). The plant expression vector pBIOsSOS1 was obtained by two-step cloning of OsSOS1 fragments. First, a 2.3-kb SalI-BamHI fragment was inserted into the XhoI and BamHI sites of pBI321. Next, a 1.3-kb BamHI-BamHI was inserted in the right orientation to reconstitute a full-length OsSOS1 cDNA. Plasmid pBI321 is a derivative of pBI121 in which the XbaI through SacI cloning sites of plasmid pCR2.1 (Invitrogen) were inserted between XbaI and SacI sites of pBI121 (CLONTECH), thus excising the β-glucuronidase coding region of pBI121 and creating additional enzyme restriction sites for cloning. Plasmids pBIOsCIPK24 and pBIOsCBL4 were constructed on the pBI321 backbone for the expression of OsCIPK4 and OsCBL4 in Arabidopsis.
Arabidopsis Transformation and Complementation Test
The constructs pBIAtSOS1 (Shi et al., 2000), pBIOsSOS1, pBIOsCIPK24, pBIOsCBL4, and empty vector pBI321 were introduced into the Agrobacterium GV3101 strain, and the resulting bacterial clones were used to transform sos1-1, sos2-2, and sos3-1 Arabidopsis mutants by vacuum infiltration (Clough and Bent, 1998). The viral 35S gene promoter is leaky in Agrobacterium and the residual expression of SOS1 is deleterious to Agrobacterium in the presence of sodium (J. Martínez-Atienza, unpublished data). To minimize the selection of reorganized constructs, Agrobacterium was grown in Luria-Bertani medium in which 1% KCl substituted for NaCl. Kanamycin-resistant T2 transgenic plants were selected and subjected to complementation tests on Murashige and Skoog agar medium supplemented with NaCl, as indicated for each case. Culture was in an environmentally controlled chamber at 22°C and a 16-h light/8-h dark cycle with photosynthetically active radiation of 30 μmol m−2 s−1.
Rice Plant Growth Conditions
Rice cv Nipponbare seeds were germinated in sterile conditions at 28°C temperature, 100% relative humidity, and kept in darkness for 5 d. Seedlings were then transferred to a hydroponic culture system consisting of boxes containing 8 L of aerated nutrient solution and holding up to 20 plants each. Nutrient medium consisted of 0.09 mm (NH4)2SO4, 0.05 mm KH2PO4, 0.05 mm KNO3, 0.03 mm K2SO4, 0.06 mm Ca(NO3)2, 0.07 mm MgSO4, 0.11 mm Fe-EDTA, 4.6 μm H3BO3, 1.8 μm MnSO4, 0.3 μm ZnSO4, 0.3 μm CuSO4, pH 5 to 5.6 (Miyamoto et al., 2001). The hydroponic containers were placed in a growth chamber set to a light/dark cycle of 16/8 h daily (photosynthetically active radiation 300 μmol m−2 s−1), 25°C to 20°C day/night temperature, and 60% to 40% relative humidity. Nutrient solution was changed every 7 d. After 3 weeks of growth in hydroponic medium, rice plants were subjected to salt stress by adding NaCl to 100 mm final concentration. Samples of roots and shoots were collected at 0, 3, 15, and 48 h after treatment onset and then frozen in liquid nitrogen.
RNA Isolation and Northern Blot
Total RNA was isolated from roots and shoots of salt-treated rice plants and 40 μg from each sample were resolved on 1.25% (w/v) agarose-formaldehyde gel and transferred to Hybond-N nylon membranes (Amersham) according to standard methods (Sambrook et al., 1989). A radioactive (32P) probe was prepared from a 1.7-kb HindIII fragment of the OsSOS1 C terminus by use of a random priming kit (Amersham). Probe hybridization was performed overnight at 65°C in a buffer containing 7% (w/v) SDS, 0.5 m Na2HPO4, 1% (w/v) bovine serum albumin, 1 mm EDTA, pH 7.2. The final wash of filters was at 65°C in 0.1× SSC with 0.1% (w/v) SDS. To estimate the relative abundance of OsSOS1 mRNA in various samples, RNA loading was normalized through hybridization with the EcoRI fragment G of the radish (Raphanus sativus) ribosomal 18S RNA gene (Delcasso-Tremousaygue et al., 1988).
Yeast Strains and Media
Yeast AXT3K strain (Δena1∷HIS3∷ena4, Δnha1∷LEU2, Δnhx1∷KanMX4; Quintero et al., 2002) is a derivative of W303-1B (MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100). Strain YP890 was derived from AXT3K by chromosomal integration of a PGK1:SOS1:CYC1 expression cassette into the 3′ end of the CYC1 gene (Guo et al., 2004). Strain YP1021, carrying a chromosomal integration of a rice OsSOS1 expression cassette (PMA1:OsSOS1:ADH1) was constructed by removing a 1.3-kb ApaI-SnaBI fragment spanning the URA3 marker and the 2Δ DNA replicon from plasmid pDROsSOS1. The resulting plasmid was linealized at the PMA1 promoter with KpnI and transformed into strain AXT3K carrying plasmid pFL32T. Plasmid pFL32T expresses the SOS2-SOS3 kinase complex and integrative transformants were selected on AP medium supplemented with 200 mm NaCl based on the gain of function of SOS1 activity from the PMA1:OsSOS1:ADH1 cassette through its activation by SOS2/SOS3. Subsequently, plasmid pFL32T was segregated out in rich, nonselective medium. Yeast transformation was done by the polyethylene glycol-lithium acetate method (Elble, 1992) and transformants were selected on solid synthetic dextrose drop-out media, except when indicated otherwise. Na+ tolerance tests were performed in the alkali cation-free medium AP (Rodriguez-Navarro and Ramos, 1984) supplemented with 1 mm KCl and with NaCl as indicated for each experiment. For ion content measurements, yeast cells were collected during exponential growth (OD550 = 0.2) in liquid AP medium and their Na+ content was determined by atomic emission spectrometry after acidic extraction (Rodriguez-Navarro and Ramos, 1984).
OsSOS1:His-6x Purification and Phosphorylation Assays
Yeast extracts of cells expressing His-tagged OsSOS1 and AtSOS2 proteins were obtained as described (Serrano, 1988). AtSOS2:His-6x was purified from the soluble fraction by chromatography on a 1-mL column with nickel nitrilotriacetic acid resin (Qiagen) without further processing. To purify OsSOS1:His-6x, membrane proteins were purified, solubilized, and chromatographed on the nickel nitrilotriacetic acid resin as described elsewhere (Quintero et al., 2002). Construction and purification of a GST:SOS2T/DΔ308 translational fusion has been described also (Guo et al., 2001). Purified OsSOS1:His-6x (100 ng) was used as substrate for phosphorylation by the Arabidopsis SOS2 and SOS2T/DΔ308 protein kinases (100 ng) in 50 μL of kinase buffer (20 mm Tris·HCl, pH 8.0, 5 mm MgCl2, 1 mm CaCl2, 1 mm dithiothreitol [DTT]). Reactions were started by adding ATP (0.2 mm with 1 μCi of [γ-32P] ATP), incubated at 30°C for 30 min, and stopped with 15 μL of 4× SDS-PAGE sample buffer. Aliquots were then resolved by SDS-PAGE and the gel exposed to x-ray films.
Purification of Plasma Membrane Vesicles
Plasma membrane was isolated from yeast cells by the aqueous two-phase system (Menendez et al., 1995). Yeast cells were first cultivated in 200 mL AP (1 K) medium containing selected amino acids at 30°C for 1 d with shaking (200 rpm), and then transferred into 1,000 mL fresh medium and cultured at 30°C with shaking (200 rpm) until the culture reached an OD600 of 3 to 4. To ensure activation of the SOS2/SOS3 kinase complex, NaCl was then added to a final concentration of 100 mm and incubation continued for 1 h before harvesting. Cells were resuspended in 5 mm Tris-HCl, pH 7.5, 700 mm sorbitol to an absorbance of 6 at 800 nm. The volume of the suspension was then measured and mixed with one-fourth of the volume with lyticase (Sigma-Aldrich) dissolved in the above medium at 300 units/mL. DTT was then added in to 6.5 mm final concentration, and the mixture incubated at 30°C for 1 h with gentle shaking (100 rpm). After lyticase treatment, the resulting protoplasts were recovered by centrifugation at 3,000g for 5 min, washed with the above medium containing 1 mm DTT, and centrifuged again. The pellet was resuspended in 15 mL ATPase-inducing medium containing 15 mm MES-Tris, pH 6.5, 500 mm sorbitol, 100 mm Glc, and incubated at 30°C for 10 min with gentle shaking (100 rpm). Osmotic lysis was thereafter induced with 30 mL osmotic lysis medium containing 25 mm MES-Tris, pH 6.5, 5 mm EDTA, 1 mm DTT, 0.2% casein hydrolysate, 0.2% bovine serum albumin, and 1 mm phenylmethylsulfonyl fluoride on ice for 5 min, assisted by a glass homogenizer, and then centrifuged to remove cell wall fragments at 300g and 4°C for 3 min. The supernatant was collected and centrifuged at 35,000g and 4°C for 15 min. The pellet was resuspended in a 9-g membrane suspension medium containing 5 mm potassium phosphate, pH 7.8, 330 mm Suc, and 1 mm DTT, and mixed quickly with 27 g of two-phase solution with the final composition of 5.7% dextran T-500 (w/w), 5.7% polyethylene glycol 3,350 (w/w), 5 mm potassium phosphate, pH 7.8, 330 mm Suc, 1 mm EDTA, and 1 mm DTT. The two-phase system was settled on ice for 30 min until phases separated. The upper and lower phases were collected separately, diluted with 10-fold the fraction volume of 15 mm MES-Tris, pH 6.5, 330 mm Suc, 1 mm DTT, and centrifuged at 65,000g and 4°C for 50 min. The membranes were resuspended in the same medium and frozen in liquid nitrogen and stored at −70°C until use.
Na+/H+ Exchange Assays
The formation of ΔpH was established by the activity of the plasma membrane H+-ATPase and Na+/H+ exchange activity was measured as a Na+-induced dissipation of ΔpH with the pH value sensitive fluorescent probe quinacrine in the following reaction mixture (1 mL): 5 μm quinacrine, 50 mm BTP-HCl, pH 7.5, 25 mm BTP-HEPES, pH 7.5, 250 mm mannitol, 4 mm MgSO4, and 50 μg plasma membrane protein. The reaction mixture was placed in a fluorescence spectrophotometer (Hitachi F-2500) and equilibrated in the dark with stirring for 5 min before fluorescence measurement. The assay was initiated with the addition of 3 mm ATP. When ΔpH reached steady state, equal amounts of various concentrations of Na2SO4 or sodium gluconate stock solutions were added to the reaction mixture. To determine initial rates of Na+/H+ exchange, the change of relative fluorescence was measured during the first 30 s after the addition of sodium salts. Specific activity was calculated by dividing the initial rate of fluorescence recovery, expressed as a ratio of the preformed pH gradient, by the mass of plasma membrane protein in the reaction and time (ΔF mg−1 min−1, where ΔF = F30 − F0/Fmax − Fmin). The change of pH value was measured at excitation and emission wavelengths of 430 and 500 nm, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY785147.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic relationships of SOS1 homologs from various plant species.
Supplemental Figure S2. Amino acid sequence alignment of AtSOS3 from Arabidopsis and OsCBL4, OsCBL7, and OsCBL8 from rice.
Supplemental Figure S3. Amino acid sequence alignment of AtSOS2 from Arabidopsis and OsCIPK24 from rice.
Supplementary Material
Acknowledgments
We are grateful to the MAFF DNA Bank (Japan) for biological materials. We thank Alonso Rodríguez-Navarro for his helpful advice.
This work was supported by the Ministerio de Educación y Ciencia (grant no. BIO2003–08501–CO2–01 to J.M.P.), Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria (grant no. CPE03–006–C6–3 to J.M.P.), Junta de Andalucía (grant no. CVI–148 to F.J.Q. and J.M.P.), and by the National Institutes of Health (grant no. R01GM59138 to J.-K.Z.). J.M.-A. was supported by a Formacion Profesorado Universitario FPU fellowship from the Ministerio de Educación y Ciencia.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José M. Pardo (pardo@cica.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azaizeh H, Steudle E (1991) Effects of salinity on water transport of excised maize (Zea mays) roots. Plant Physiol 97 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos MA, Sychrová H, Bleykasten-Grosshans C, Souciet JL, Potier S (1998) The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology 144 2749–2758 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cruz RT, Jordan WR, Drew MC (1992) Structural changes and associated reduction of hydraulic conductance in roots of Sorghum bicolor L. following exposure to water deficit. Plant Physiol 99 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley CP, Wuytswinkel OCM, Woude K, Mager WH, De Boer AH (2000) Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J 351 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in Durum wheat. Plant Physiol 137 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer AH, Volkov V (2003) Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ 26 87–101 [Google Scholar]
- Delcasso-Tremousaygue D, Grellet F, Panabieres F, Ananiev ED, Delseny M (1988) Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur J Biochem 172 767–776 [DOI] [PubMed] [Google Scholar]
- Elble R (1992) A simple and efficient procedure for transformation of yeast. Biotechniques 13 18–20 [PubMed] [Google Scholar]
- Flowers TJ, Hajibagher MA, Yeo AR (1991) Ion accumulation in the cell walls of rice plants growing under saline conditions: evidence for the Oertli hypothesis. Plant Cell Environ 14 319–325 [Google Scholar]
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45 146–159 [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34 788–801 [DOI] [PubMed] [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ (2003) Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol Biol 51 71–81 [DOI] [PubMed] [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Munoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 31 529–542 [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu JK (2004) The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement of its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis. Plant Cell 16 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YS, Bethke PC, Cheong YH, Chang HS, Zhu T, Jones RL (2005) A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol 138 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 1667–167811006339 [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama ML, Levesley A, Koebner RMD, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125 406–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJ, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32 139–149 [DOI] [PubMed] [Google Scholar]
- Lee KS, Choi WY, Ko JC, Kim TS, Gregorio GB (2003) Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216 1043–1046 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97 3703–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945 [DOI] [PubMed] [Google Scholar]
- Menendez A, Larsson C, Ugalde U (1995) Purification of functionally sealed cytoplasmic side-out plasma membrane vesicles from Saccharomyces cerevisiae. Anal Biochem 230 308–314 [DOI] [PubMed] [Google Scholar]
- Miyamoto N, Steudle E, Hirasawa T, Lafitte R (2001) Hydraulic conductivity of rice roots. J Exp Bot 362 1835–1846 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119–122 [DOI] [PubMed] [Google Scholar]
- Munns R (1993) Physiological processes limiting plant growth in saline soils: some dogmas and hypotheses. Plant Cell Environ 16 15–24 [Google Scholar]
- Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, Hayakawa T (2002) Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett 532 279–282 [DOI] [PubMed] [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles on cellular homeostasis and stress tolerance. J Exp Bot 57 1181–1199 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Barkla BJ, Vera-Estrella R, Zhu JK, Schumaker KS (2003) Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol 132 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR, Pardo JM (2000) Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett 471 224–228 [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37 1141–1146 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WBV (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370 264–268 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro A, Ramos J (1984) Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol 159 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanchez-Aguayo I, Rodriguez-Galan JM, García R, Torreblanca J, Pardo JM (2004) Salt stress enhances xylem development and expression of S-adenosyl-methionine synthase in lignifying tissues of tomato plants. Planta 220 278–285 [DOI] [PubMed] [Google Scholar]
- Sanchez-Barrena MJ, Martinez-Ripoll M, Zhu JK, Albert A (2005) The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345 1253–1264 [DOI] [PubMed] [Google Scholar]
- Serrano R (1988) H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol 157 533–544 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steudle E, Peterson CA (1998) How does water get through roots? J Exp Bot 49 775–788 [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K, Quintero FJ, Pardo JM, Donaire JP (2002) The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem 277 2413–2418 [DOI] [PubMed] [Google Scholar]
- Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK (1995) The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J 14 3870–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Flowers TJ, Yeo AR (1996) The involvement of the transpirational flow in sodium uptake by high- and low-sodium transporting lines of rice developed through intravarietal selection. Plant Cell Environ 19 329–336 [Google Scholar]
- Yeo AR, Lee KS, Izard P, Boursier PJ, Flowers TJ (1991) Short and long term effects of salinity on leaf growth in rice (Oryza sativa). J Exp Bot 42 881–889 [Google Scholar]
- Yeo AR, Yeo ME, Flowers TJ (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J Exp Bot 38 1141–1153 [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.