Abstract
Heterologous expression of the Arabidopsis (Arabidopsis thaliana) IPMS1 (At1g18500) and IPMS2 (At1g74040) cDNAs in Escherichia coli yields isopropylmalate synthases (IPMSs; EC 2.3.3.13). These enzymes catalyze the first dedicated step in leucine (Leu) biosynthesis, an aldol-type condensation of acetyl-coenzyme A (CoA) and 2-oxoisovalerate yielding isopropylmalate. Most biochemical properties of IPMS1 and IPMS2 are similar: broad pH optimum around pH 8.5, Mg2+ as cofactor, feedback inhibition by Leu, Km for 2-oxoisovalerate of approximately 300 μm, and a Vmax of approximately 2 × 103 μmol min−1 g−1. However, IPMS1 and IPMS2 differ in their Km for acetyl-CoA (45 μm and 16 μm, respectively) and apparent quaternary structure (dimer and tetramer, respectively). A knockout insertion mutant for IPMS1 showed an increase in valine content but no changes in Leu content; two insertion mutants for IPMS2 did not show any changes in soluble amino acid content. Apparently, in planta each gene can adequately compensate for the absence of the other, consistent with available microarray and reverse transcription-polymerase chain reaction data that show that both genes are expressed in all organs at all developmental stages. Both encoded proteins accept 2-oxo acid substrates in vitro ranging in length from glyoxylate to 2-oxohexanoate, and catalyze at a low rate the condensation of acetyl-CoA and 4-methylthio-2-oxobutyrate, i.e. a reaction involved in glucosinolate chain elongation normally catalyzed by methylthioalkylmalate synthases. The evolutionary relationship between IPMS and methylthioalkylmalate synthase enzymes is discussed in view of their amino acid sequence identity (60%) and overlap in substrate specificity.
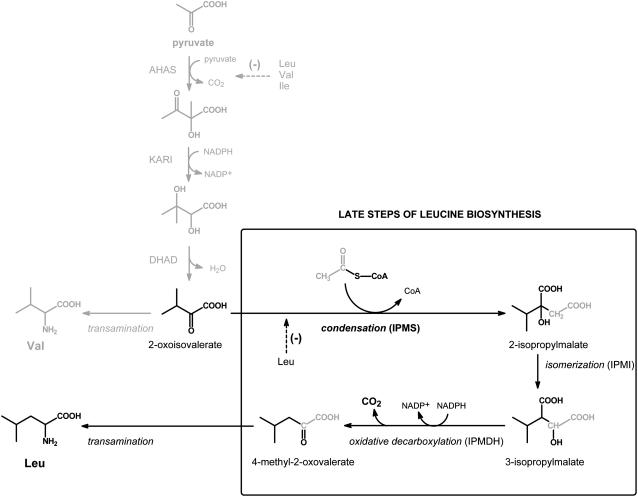
Isopropylmalate synthase (IPMS; EC 2.3.3.13) catalyzes the first dedicated step in Leu biosynthesis, an aldol-type condensation between acetyl-CoA and 2-oxoisovalerate yielding 2-isopropylmalate (Fig. 1). The absence of IPMS and other enzymes of branched-chain amino acid biosynthesis (Leu, Ile, and Val) in monogastric animals has been an important stimulus for the development of herbicides that specifically inhibit the synthesis of branched-chain amino acids in plants with minimal toxicity to animals. Nonetheless, in contrast to Val and Ile, the biosynthesis of Leu in plants is largely unexplored. The available evidence indicates that plants use the pathway depicted in Figure 1, which is the same one found in bacteria and yeast (Singh and Shaner, 1995; Singh, 1999; Coruzzi and Last, 2000).
Figure 1.
The biosynthesis of Leu and Val from pyruvate. The action of acetohydroxyacid synthase (AHAS), ketoacid reductoismerase (KARI), and dihydroxyacid dehydratase (DHAD) yields 2-oxoisovalerate that is either transaminated to Val or subjected to additional reactions specific for Leu biosynthesis. The dedicated step in Leu biosynthesis is the aldol-type condensation between 2-oxoisovalerate and acetyl-CoA that results in formation of 2-isopropylmalate. Isomerization and oxidative decarboxylation by isopropylmalate isomerase (IPMI) and isopropylmalate dehydrogenase (IPMDH) yield 4-methyl-2-oxovalerate that is transaminated to Leu. The enzymes that catalyze the reactions from pyruvate to 2-oxoisovalerate are also involved in biosynthesis of Ile, using 4-oxobutyrate (product of Thr dehydratase) as an initial substrate, but for simplicity have not been depicted. AHAS and IPMS are subject to feedback inhibition as shown with dashed lines.
IPMS activities of plants have been described from crude extracts of maize (Zea mays) embryos (Oaks, 1965), thylakoid fractions of spinach (Spinacia oleracea) chloroplasts (Hagelstein and Schultz, 1993), and soluble chloroplast-enriched preparations of nasturtium (Tropaeolum majus), Diplotaxis tenuifolia, Eruca sativa, and Arabidopsis (Arabidopsis thaliana), all members of the Brassicaceae (Falk et al., 2004). The next step in Leu biosynthesis is isomerization of the IPMS product 2-isopropylmalate to 3-isopropylmalate (Fig. 1), but an isopropylmalate isomerase has until now not been described from any plant source. More is known about the penultimate step in Leu biosynthesis since Wittenbach et al. (1994) partially purified an isopropylmalate dehydrogenase from pea (Pisum sativum) and showed the inhibition of this enzyme by an herbicide. In addition, a cDNA clone encoding an enzyme that dehydrogenates and decarboxylates 3-isopropylmalate to 4-methyl-2-oxovalerate was isolated from both oil seed rape (Brassica napus; Ellerstrom et al., 1992) and potato (Solanum tuberosum; Jackson et al., 1993). Transamination of the resulting 2-oxo acid yields the final product Leu (Singh, 1999).
A gene unambiguously encoding IPMS activity has not yet been identified from any plant source, despite the considerable attention devoted to four genes of Arabidopsis (Columbia [Col]-0) that show similarity to IPMS sequences of other organisms (e.g. Kroymann et al., 2001; Junk and Mourad, 2002; Field et al., 2004; Textor et al., 2004). A principal reason for this attention is the potential role of these genes in glucosinolate biosynthesis. Two of these IPMS-like sequences (At5g23010 and Atg5g23020) are located at the GS-Elong locus on chromosome V, which regulates the side-chain length of the aliphatic glucosinolates in Arabidopsis (Campos de Quiros et al., 2000; Kroymann et al., 2001, 2003).
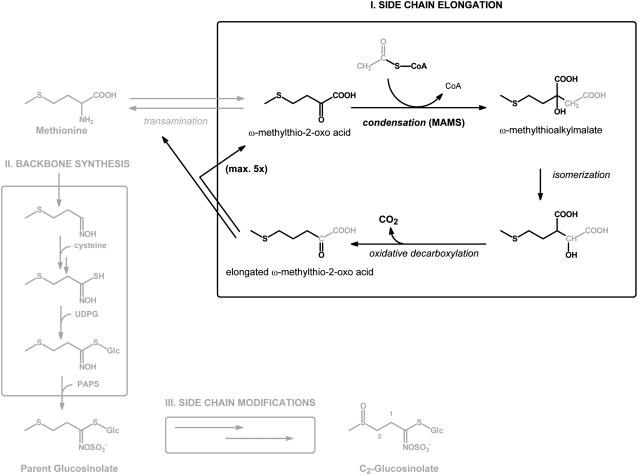
Glucosinolates are amino acid-derived plant secondary metabolites (formed mainly from Met, Trp, and Phe in Arabidopsis) that consist of a β-thio-Glc moiety, a sulfonated oxime, and a variable side chain formed from the parent amino acid. In the case of the Met-derived glucosinolates, the side chain of the amino acid is elongated by up to six methylene groups prior to biosynthesis of the glucosinolate backbone (Fig. 2), resulting in the formation of products with C3 to C8 side chains (Wittstock and Halkier, 2002; Halkier and Gershenzon, 2006). In the iterative, three-step elongation process demonstrated in in vivo feeding studies, transaminated Met (4-methylthio-2-oxobutyrate) is subject to the same type of reactions (Fig. 2) that convert 2-oxoisovalerate into Leu (Fig. 1; Chisholm and Wetter, 1964; Matsuo and Yamazaki, 1968; Serif and Schmotzer, 1968; Graser et al., 2000). Each cycle of elongation is initiated by an aldol-type condensation between acetyl-CoA and a ω-methylthio-2-oxo acid leading to the formation of a ω-methylthioalkylmalate. Both IPMS-like sequences at the Gs-Elong locus of Arabidopsis Col-0 were shown to encode proteins that possess this activity, but each had only very limited IPMS activity (Kroymann et al., 2001; Textor et al., 2004; S. Textor, unpublished data). Therefore, the genes were called methylthioalkylmalate synthase (MAM) genes, specifically, MAM1 and MAM3 (also known as MAM-L). A third MAM gene, MAM2, has been described from other Arabidopsis ecotypes (Kroymann et al., 2003). Analysis by Field et al. (2004) of a MAM3 knockout line from Arabidopsis showed the absence of long-chain glucosinolates (C6, C7, and C8), confirming the role of MAM3 in glucosinolate biosynthesis.
Figure 2.
General scheme for the biosynthesis of aliphatic glucosinolates involving Met side-chain elongation (I), backbone synthesis (II), and side-chain modifications (III). The proposed cycle for side-chain elongation of deaminated Met (I) commences with an aldol-type condensation between the respective ω-methylthio-2-oxo acid and acetyl-CoA, a reaction catalyzed by MAM. The methylthioalkylmalate product is converted through subsequent isomerization and oxidative decarboxylation into a ω-methylthio-2-oxo acid that is elongated by one methylene group. The elongated ω-methylthio-2-oxo acid is either transaminated and enters glucosinolate backbone synthesis (II) or undergoes additional cycles of side-chain elongation. Glucosinolate backbone synthesis is represented in a simplified manner without the identified enzymes and cofactors. The depicted end product, 2-methylsulfinylethyl glucosinolate, is a C2-glucosinolate that has not been side-chain elongated and does not occur naturally in Arabidopsis.
The function of the two other IPMS-like sequences, located at opposite ends of chromosome I (At1g18500 and At1g74040), has not yet been clearly determined, though cluster analysis of the deduced amino acid sequences revealed that they are more closely related to one another and to the two reported IPMS sequences from wild tomato (Lycopersicon pennellii; GenBank accession nos. AAB61598 and AAB61599) than to the MAM1 or MAM3 sequences, suggesting they probably encode true IPMSs (Kroymann et al., 2001). Just recently, a MAM sequence (GenBank accession no. DQ143886) was reported from Brassica atlantica that shares a high amino acid identity (90% and 86%) with the predicted IPMS homologs of Arabidopsis and is able to restore the growth of an IPMS-null Escherichia coli mutant in the absence of Leu (Field et al., 2006), and thus was called an IPMS (BatIMS). Such a rescue of an IPMS-null E. coli mutant was also described for At1g74040 and furthermore for both MAM-encoding genes, MAM1 and MAM3 (Junk and Mourad, 2002; S. Textor, unpublished data). An overview of these confusing, and in part contradictory, results (Table I) makes the point that it is problematic to definitely assign the in planta function of these genes based solely on complementation of an IPMS-null E. coli mutant strain. Analyses of a knockout mutant in At1g18500 also gave little information about the function of this IPMS gene since neither a significant change in Leu nor in glucosinolate content was detected, and only (pleiotropic?) alterations in Asn, Gln, His, and Val content were observed (Field et al., 2004).
Table I.
Complementation of IPMS-null E. coli mutant by IPMS-like genes of Arabidopsis
Complementation indicated by “+,” yes; “−,” no; or not determined (ND).
| AGI Code No. | Name | Junk and Mourad (2002)a | Field et al. (2004)b | S. Textor (Unpublished Data)c | This Articled |
|---|---|---|---|---|---|
| At1g18500 | IPMS1 | ND | − | ND | − |
| At1g74040 | IPMS2 | − | + | ND | + |
| At5g23010 | MAM1 | + | − | ND | ND |
| At5g23020 | MAM3 | + | − | + | ND |
Conditions not described.
Overnight at 37°C.
Three days at 28°C.
Three days at either 30°C or 37°C.
At least one of the two predicted IPMS genes at chromosome I of Arabidopsis, named IPMS1 (At1g18500) and IPMS2 (At1g74040) in this article, should encode an active IPMS because such an enzyme activity is absolutely essential to the plant for its synthesis of Leu. In this study, IPMS1 and IPMS2 were cloned and heterologously expressed in E. coli. Determination of the substrate specificity of the purified proteins in conjunction with analysis of knockout mutant lines reveals that IPMS1 and IPMS2 both encode bona fide IPMSs involved in Leu biosynthesis and do not participate in the chain elongation of glucosinolates.
RESULTS
IPMS1 and IPMS2 Have IPMS Activity
The open reading frames (ORFs) of IPMS1 (At1g18500) and IPMS2 (At1g74040) from Arabidopsis were separately cloned without the predicted N-terminal targeting sequence (ChloroP; Emanuelsson et al., 1999) into an expression vector containing a polyhistidine coding domain (His-tag) and expressed in E. coli. The recombinant protein was purified over an Ni-NTA agarose affinity column and showed on SDS-PAGE a protein band at the expected size of around 65 kD that was 90% to 95% pure (data not shown); about 4.5 mg of protein was obtained from a 100-mL bacterial culture.
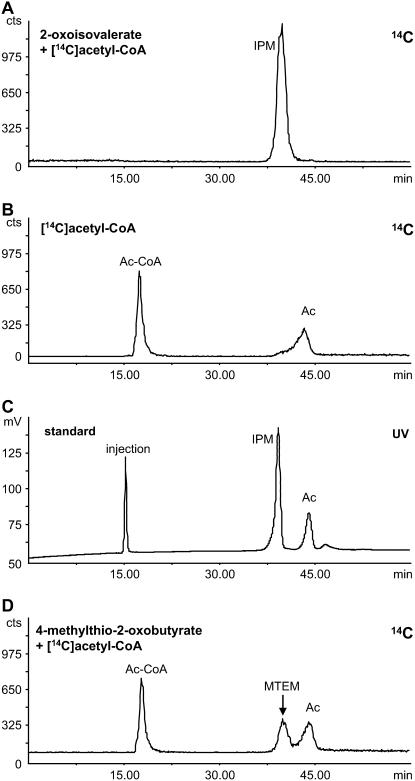
Incubation of 0.5 mm [14C]acetyl-CoA and 3 mm 2-oxoisovalerate with 50 μg of partially purified IPMS protein for 1 h gave a complete incorporation of the 14C label into a product that yielded a single peak in the radiodetector trace of the HPLC (Fig. 3A). This product peak coeluted with a synthetic standard of 2-isopropylmalate (Fig. 3C). In these measurements, no difference was detected in enzyme activity between IPMS1 and IPMS2. In the absence of 2-oxo acid substrate, [14C]acetyl-CoA was partially hydrolyzed to [14C]acetate and CoA (Fig. 3B), probably due to the pH of the enzyme assay and a minor acetyl-CoA hydrolyzing activity of the enzyme itself (see also Table III). Boiled enzyme incubated with 2-oxoisovalerate and [14C]acetyl-CoA yielded a similar chromatogram (data not shown). To check for MAM activity associated with formation of chain-elongated, Met-derived glucosinolates, the purified IPMS proteins were incubated with 4-methylthio-2-oxobutyrate and [14C]acetyl-CoA under the same conditions. However, a much smaller amount of [14C]-2-(2′-methylthio)ethylmalate was measured compared to the amount of 2-isopropylmalate formed from 2-oxoisovalerate, and most of the [14C]acetyl-CoA remained (Fig. 3D). These data suggest that the IPMS1 and IPMS2 genes are much more likely to serve as IPMSs in Leu biosynthesis than in the formation of methylthioalkylmalate compounds for glucosinolate biosynthesis.
Figure 3.
Radio-HPLC analyses of the biochemical assay for IPMS2. Results for IPMS1 had a similar pattern. A, Incubation of IPMS2 with 500 μm [14C]acetyl-CoA and 3 mm 2-oxoisovalerate shows [14C]-2-isopropylmalate (IPM) as the only radioactive labeled product. B, In the absence of 2-oxo acid substrate, a small amount of [14C]acetate (Ac) is formed, whereas most of the [14C]acetyl-CoA (Ac-CoA) remains intact and is hardly retained in the HPLC column. C, UV trace (230 nm) of the HPLC showing the elution pattern of a standard solution containing 10 mm acetic acid (Ac) and 5 mm 2-isopropylmalate (IPM), and the injection peak at 15 min. D, Incubation of the IPMS2 gene product with 4-methylthio-2-oxobutyrate and [14C]acetyl-CoA yields a small amount of [14C]2-(2′-methylthio)ethylmalate (MTEM), whereas most of the [14C]acetyl-CoA (Ac-CoA) remains.
Table III.
Substrate specificity of IPMS1 and IPMS2
Additional support for the function of the IPMS2-encoded protein comes from the observation that it is able to complement the IPMS-null E. coli strain CV512(DE3). This bacterium was not able to grow on a minimal medium without supplemented amino acids, unless it had been transformed with a construct carrying either the E. coli IPMS gene leuA or the IPMS2 gene from Arabidopsis. Transcription of the gene construct was induced with isopropyl-β-galactoside (IPTG), and both the E. coli leuA transformant (positive control) and the IPMS2 transformant grew at 30°C within 3 d, whereas no bacterial growth was observed for CV512(DE3) alone (data not shown). Growth of the IPMS2 transformant was less at 37°C. In contrast to this experiment, our attempts to complement CV512(DE3) with the IPMS1 construct were unsuccessful.
IPMS1 and IPMS2 Have Similar But Not Identical Biochemical Characteristics
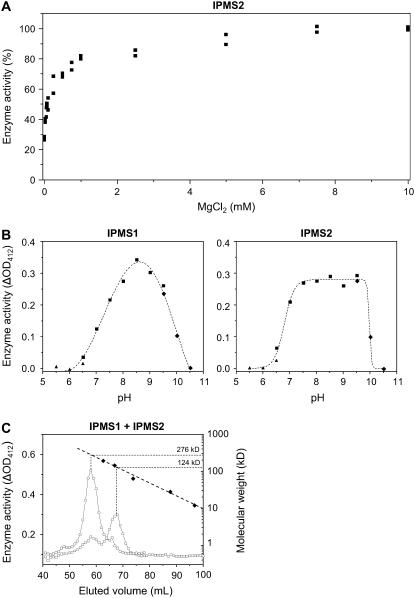
IPMS1 and IPMS2 activity not only depended upon the presence of a 2-oxo acid substrate and acetyl-CoA, but also required Mg2+ in millimolar concentrations (Fig. 4A). Hence, 4 mm Mg2+ was added routinely to the incubations. Quantitative measurements using an endpoint assay with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; see “Materials and Methods”) showed a 75% to 95% loss of enzyme activity when the His-tag purified protein was desalted into a buffer without Mg2+ and then incubated in the absence of Mg2+. Enzyme activity was completely lost in the presence of 10 mm EDTA. The residual enzyme activity in the absence of Mg2+ (monitored with the radio-HPLC assay) was also completely lost if the incubation assay was adjusted to 4 mm Ca2+, Cu2+, Ni2+, or Zn2+, whereas the residual activity was not inhibited by the presence of 4 mm Fe2+ or Co2+. While 4 mm K+ gave a slight stimulation of the residual enzyme activity and the addition of 4 mm Mn2+ restored about 50% of the initial activity, only the addition of 4 mm of Mg2+ brought the enzyme activity back to its full initial rate.
Figure 4.
Properties of Arabidopsis IPMSs. A, Effect of MgCl2 concentration on IPMS2 activity; 0 mm corresponds to a desalted enzyme preparation without addition of MgCl2. A similar graph was obtained for IPMS1. B, pH Curves for enzyme activity of IPMS1 and IPMS2 using MES (▴), BisTris-propane (▪), and 2-amino-2-methyl-1-propanol (♦). Each data point corresponds to a duplicate enzyme activity measurement with DTNB that is corrected for chemical hydrolysis of acetyl-CoA at each particular pH value. C, Determination of the molecular mass of IPMS1 (□) and IPMS2 (○) by calibrated gel-filtration chromatography. The column was calibrated by measuring the elution volume (♦) of β-amylase (200 kD), alcohol dehydrogenase (150 kD), bovine serum albumin (66 kD), carbonic anhydrase (29 kD), and cytochrome C (12.4 kD). The void volume of the column determined with Blue Dextran (2.0 × 103 kD) was 44 mL.
The pH curves of IPMS1 and IMPS2 are similar and display rather broad optima around pH 8.5 (Fig. 4B). The more flattened curve for IPMS2 with a sharp drop of enzyme activity above pH 9.5 was seen consistently in replicate experiments. A pH of 8.0 was routinely used in enzyme assays to minimize the spontaneous chemical hydrolysis of acetyl-CoA that increased noticeably above this value.
The molecular mass of the native recombinant protein was determined by gel filtration on a calibrated Superdex-200 column, assaying the activity of eluted 1-mL fractions by the DTNB method (Fig. 4C). The main activity of IPMS1 eluted at a molecular mass of 124 kD, suggesting that the functional protein is a dimer, as the monomer size calculated from the amino acid sequence is 63.1 kD (including the additional 0.8 kD from the His-tag). On the other hand, the main activity of IPMS2 eluted at approximately 280 kD, indicating this enzyme is active as a tetramer (predicted monomer size 64.1 kD). Omitting the 150 mm of NaCl from the elution buffer or adding 2 mm MgCl2 to the elution buffer did not affect these results.
Enzyme kinetics were determined in a continuous spectrophotometric assay with N-ethylmaleimide (NEM; see “Materials and Methods”) and are presented in Table II. The main difference between IPMS1 and IPMS2 was the Km obtained for acetyl-CoA: 45 μm and 16 μm, respectively. There was no significant difference observed in the Km for 2-oxoisovalerate (304 μm and 279 μm), nor any difference in specific activity between IPMS1 and IPMS2.
Table II.
Kinetic parameters for IPMS1 and IPMS2
| Enzyme | Substrate | Km (μm) ± se | Vmax (μmol min−1 g−1) ± se | kcat (s−1) | kcat/Km (m−1 s−1) |
|---|---|---|---|---|---|
| IPMS1 | 2-Oxoisovaleratea | 304 ± 68 | 2.3 × 103 ± 0.2 × 103 | 2.4 | 7.8 × 103 |
| Acetyl-CoAb | 45 ± 10 | 2.1 × 103 ± 0.1 × 103 | 2.2 | 4.7 × 104 | |
| IPMS2 | 2-Oxoisovaleratea | 279 ± 51 | 2.2 × 103 ± 0.1 × 103 | 2.3 | 8.3 × 103 |
| Acetyl-CoAb | 16 ± 4 | 1.8 × 103 ± 0.1 × 103 | 1.9 | 1.2 × 105 |
Measured in the presence of 500 μm acetyl-CoA.
Measured in the presence of 1 mm 2-oxoisovalerate.
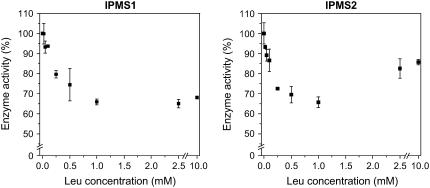
IPMS activity in plants is generally considered to be feedback inhibited by Leu (Singh, 1999; Coruzzi and Last, 2000). To test this possibility, aliquots of the incubation mixture were adjusted to Leu concentrations between 0.025 mm and 10 mm. We detected a slight inhibitory effect of Leu on the activity of IPMS1 and IPMS2 (Fig. 5). Inhibition of both enzymes reached a maximum of 30% to 35% around 1 mm Leu, but declined to 15% at higher concentrations of Leu in the case of IMPS2. Changing the pH of the incubation mixture to 7.5 did not augment the inhibitory effect of Leu as had been observed for the IPMS of yeast (Ulm et al., 1972). To account for the possible influences of posttranscriptional modifications of the IPMS proteins in planta, the effect of Leu was also tested on the IPMS activity present in crude extracts of Arabidopsis. An extract prepared from leaf material (3.5 weeks old) according to the protocol of Textor et al. (2004), but leaving out the ammonium-sulfate precipitation step, was measured in the presence of an acetyl-CoA regenerating system. The inhibitory effect of Leu on the IPMS activity present in the crude extract was similar to those values measured for the heterologously expressed IPMSs: 14% at 0.5 mm Leu, 33% at 1.0 mm Leu, and 48% at 5.0 mm Leu. No inhibitory effects on the crude IPMS enzyme were observed for Ile, Val, and Gly. In previous work, the IPMS of spinach was reported to be 100% inhibited by micromolar concentrations of Leu (Hagelstein and Schultz, 1993). However, our experiments on the IPMS activity present in a chloroplast-enriched crude extract of spinach prepared after Falk et al. (2004), without the gel-filtration step, did not show such a pronounced effect. The inhibition of IPMS activity was similar to that seen with the Arabidopsis crude extract: 32% at 0.5 mm Leu, 42% at 1 mm Leu, and 66% at 5 mm Leu.
Figure 5.
Leu inhibition of the heterologously expressed IPMS1 and IPMS2 from Arabidopsis.
Both Enzymes Accept Other Substrates and Catalyze the First Condensation Reaction of Glucosinolate Chain Elongation at Low Rates
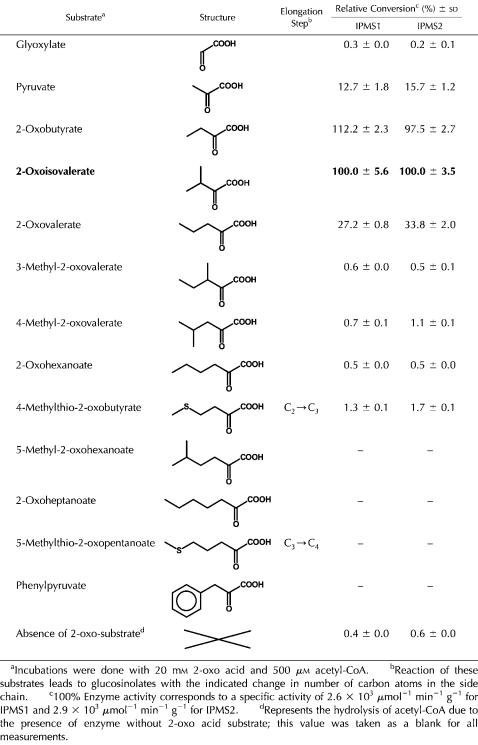
The detectable conversion of 4-methylthio-2-oxobutyrate to its alkylmalate derivative by IPMS1 and IPMS2 from Arabidopsis (Fig. 3) led us to test various 2-oxo acid substrates other than 2-oxoisovalerate. Potential substrates were incubated with [14C]acetyl-CoA at saturating concentrations and the reaction mixtures analyzed by radio-HPLC. The reaction products were identified by coelution of synthetic standards and/or liquid chromatography-mass spectrometry analyses (Kroymann et al., 2001; Textor et al., 2004; S. Textor, unpublished data). The enzymatic rates for converted substrates were determined with DTNB in a timed enzyme assay and are expressed relative to the conversion of 2-oxoisovalerate (Table III). The specific activities for IPMS determined in this way are comparable to those in Table II that were determined with NEM in the less-sensitive continuous spectrophotometric assay.
There was no major difference between IPMS1 and IPMS2 with respect to their substrate specificities. Of the tested substrates, 2-oxobutyrate seems to perform as well or even better than the true substrate 2-oxoisovalerate (common name for 3-methyl-2-oxobutyrate), while pyruvate and 2-oxovalerate were also converted in ample yield. However, 3-methyl-2-oxovalerate and 4-methyl-2-oxovalerate, i.e. transaminated products of Ile and Leu, respectively, were converted only in trace amounts, while phenylpyruvate, the 2-oxo acid of Phe, was not converted at all. The reaction between 4-methylthio-2-oxobutyrate and acetyl-CoA occurred at a relatively low yield (consistent with the data in Fig. 3), but was 3 to 4 times more efficient than the enzymatic conversion of its methylene analog, 2-oxohexanoate. As mentioned in the introduction, formation of 2-(2′-methylthio)ethylmalate represents the aldol-type condensation reaction in the first cycle of Met side-chain elongation and yields a direct precursor of C3-glucosinolate biosynthesis. However, this reaction proceeds at a far slower rate than the conversion of 2-oxoisovalerate to isopropylmalate. Estimations by the DTNB assay gave a Km value of at least 3 mm for 4-methylthio-2-oxobutyrate that results in a specificity constant (kcat/Km) of less than 13 m−1 s−1. The Km for pyruvate was in the millimolar range as well. Substrates having a carbon chain longer than 4-methylthio-2-oxobutyrate or 2-oxohexanoate were not accepted at all.
To shed further light on whether IPMS1 and IPMS2 participate in glucosinolate formation, we investigated whether or not the low activity of these enzymes with 4-methylthio-2-oxobutyrate is a general feature of IPMS enzymes. When the substrate specificity of the IPMS cloned from E. coli (the leuA gene) was measured by incubation of the purified His-tag protein with [14C]acetyl-CoA, the substrates 4-methylthio-2-oxobutyrate, pyruvate, and 2-oxovalerate were converted to their malate derivatives to the same extent relative to 2-oxoisovalerate as observed for the IPMSs of Arabidopsis (Supplemental Fig. S1).
Plant Lines Mutated in IPMS1 and IPMS2 Still Had Wild-Type Levels of Leu
To determine how the IPMS genes contribute to amino acid and glucosinolate biosynthesis, we characterized three mutant lines of Arabidopsis (Col-0), one with a T-DNA insertion in the IPMS1 gene (Salk_101771) and two with a T-DNA insertion in the IPMS2 gene (Salk_051060 and Salk_000074; Alonso et al., 2003). The genomic DNA of individuals from crosses segregating for each insert were tested for the presence of a T-DNA insert in the respective genes using oligonucleotide primer pairs derived from the T-DNA insert and the respective IPMS genes (Supplemental Table S1), and the product sizes and sequences obtained were consistent with the reported insertion sites (Supplemental Fig. S2). Individual plants were identified that were homozygous for the T-DNA insert, heterozygous, or lacking the T-DNA insert (outsegregants).
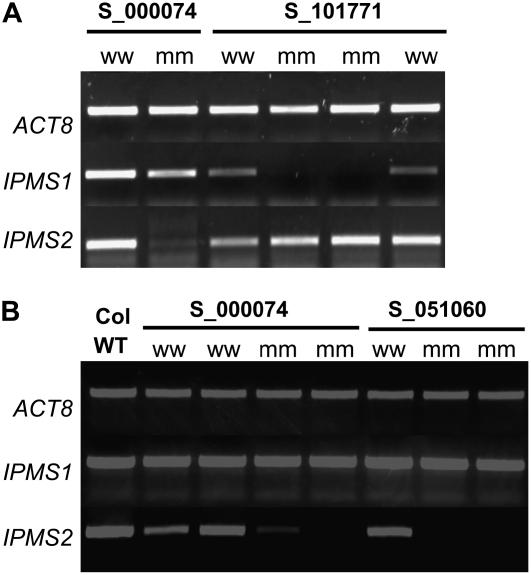
As shown in Figure 6, transcripts for IPMS1 and IPMS2 were readily detected in wild-type plants by reverse transcription (RT)-PCR, but no transcript of IPMS1 was detected in homozygotes for insertion in this gene (Salk_101771 line). In homozygotes for the IPMS2 insertions, no IPMS2 transcript was observed in the Salk_051060 line, and only a weak band for IPMS2 was detected in homozygotes for the Salk_000074 line. In the mutant lines for each IPMS gene, transcript of the other IPMS gene was present, but no compensatory effects were observed. The IPMS1 mutant line grew somewhat slower and had undulated leaves that tended to be slightly chlorotic as compared to its outsegregant. However, the IPMS2 mutants had a normal appearance.
Figure 6.
Semiquantitative RT-PCR analyses of IPMS transcript levels in leaf tissues of Salk T-DNA insertion lines, homozygous (mm) for the insertion or lacking the insertions (ww), and Col-0 wild type (WT). A, Results for the IPMS1 mutant line S_101771 compared to the IPMS2 mutant line S_000074; B, results for the IPMS2 mutant lines S_000074 and S_051060.
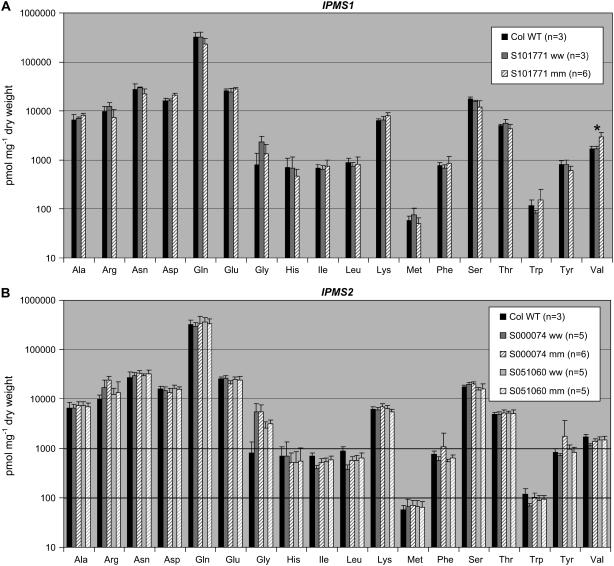
Amino acid analyses of 3- to 4-week-old rosettes of the IPMS1 mutant (Salk_101771 [mm]; Fig. 7A) showed an increase in the content of the aliphatic amino acid Val from 1,662 ± 236 pmol mg−1 dry weight to 2,919 ± 639 pmol mg−1 dry weight. No significant change in any other measured amino acid was observed compared to the controls (Col-0 wild type and the Salk_101771 outsegregant). The IPMS2 mutants (Salk_000074 and Salk_051060; Fig. 7B) showed no significant differences in amino acid content compared with their respective controls. The rosettes of the three mutant lines and their controls were also analyzed for their glucosinolate content, but no significant changes were observed either in quantity or quality (Supplemental Fig. S3).
Figure 7.
Analyses of the free amino acid content of homozygous T-DNA insertion lines for IPMS1 (A; Salk_101771 [mm]) and IPMS2 (B; Salk_051060 [mm] and Salk_000074 [mm]). The IMPS1 mutant shows a significant increase in Val content in comparison with the corresponding outsegregants (ww) and Col-0 wild type. Error bars indicate sd.
Both Genes Are Expressed Constitutively throughout the Plant
Semiquantitative RT-PCR analysis revealed that both IPMS genes are expressed throughout the plant and mRNA transcript levels do not differ extensively between various organs (Supplemental Fig. S4). These results are consistent with data from the Arabidopsis ATH1 Genome Array (Affymetrix), posted on the Internet and retrieved with the GENEVESTIGATOR program (Zimmermann et al., 2004). The collected microarray data also show that both IPMS genes are similarly expressed among different developmental stages and after specific stress treatments (Supplemental Fig. S5). An exception is the level of IPMS1 transcript in the seeds during silique ripening, which is at least twice as high as in most other plant tissues.
DISCUSSION
IPMS1 and IPMS2 Function in Leu Formation But Not in Glucosinolate Biosynthesis
Heterologous expression of the Arabidopsis IPMS1 (At1g18500) and IPMS2 (At1g74040) cDNAs in E. coli and the study of the enzymatic properties of the protein products demonstrated that both genes encode IPMSs, which catalyze the aldol-type condensation reaction between acetyl-CoA and 2-oxoisovalerate in the biosynthesis of Leu. Nonetheless, both proteins were also able to catalyze the MAM reaction between acetyl-CoA and 4-methylthio-2-oxobutyrate to some extent. However this reaction occurs with such a low specificity constant (kcat/Km < 13 m−1 s−1) in comparison with the previously measured value for MAM3 (1.4 × 103 m−1 s−1; S. Textor, unpublished data) that this MAM activity is probably of no significance in vivo. In addition, an insertion mutant for IPMS1 with undetectable transcript levels of IPMS1 (Salk_101771), an insertion mutant for IPMS2 with undetectable transcript levels of IPMS2 (Salk_051060), and an IPMS2 insertion mutant line with reduced IPMS2 transcript levels (Salk_000074) all did not display any changes in glucosinolate content (Supplemental Fig. S3). Hence, we conclude that IPMS1 and IPMS2 have a primary function in Leu biosynthesis and are not significantly involved in the Met side-chain elongation of glucosinolate biosynthesis.
In accordance with this conclusion, the IPMS2 gene in a prokaryotic expression construct rescued the IPMS-null E. coli mutant strain CV512(DE3), but curiously an IPMS1 construct did not. Rescue of the Leu-deficient E. coli strain by IPMS2, but not IPMS1, was also observed by Field et al. (2004); however, Junk and Mourad (2002) could not demonstrate any complementation with IPMS2, but did for MAM1 and MAM3 (see also Table I). The inconsistencies in demonstrating the ability of these plant genes to complement the Leu auxotrophy in E. coli provide a cautionary tale about the value of complementation assays as surrogates for a full characterization of in vitro enzyme activity.
Amino acid analysis of the IPMS1 and IPMS2 insertion mutants also gave equivocal results about in vivo function, since none of the insertion mutants showed any changes in soluble Leu content (Fig. 7). As plants require Leu for survival, a total lack of Leu accumulation was not expected in these viable mutant lines. Nevertheless, the absence of any detectable change in Leu content is noteworthy and must demonstrate that IPMS1 and IPMS2 compensate for each other's absence very well, because it is unlikely that the MAM enzymes with their very low IPMS activity can contribute to Leu biosynthesis in an efficient way (S. Textor, unpublished data).
Despite the lack of a reduction in Leu, the IPMS1 insertion mutant has a clear phenotype in its elevated soluble Val content (Fig. 7). Such an increase in Val content was also observed in an independently generated En-1 insertion mutant of IPMS1 by Field et al. (2004), but was not discussed any further. This feature may be a consequence of the fact that the IPMS1 mutant—despite the presence of IPMS2—has less IPMS activity relative to Val-aminotransferase activity than found in wild-type plants. Since both enzymes use 2-oxoisovalerate as a substrate, such an imbalance will increase the amount of Val at the expense of Leu. This will reduce the feedback inhibition of acetohydroxyacid synthase (AHAS), because Val is a less potent inhibitor of AHAS activity than Leu and, moreover, it is actually the combination of Leu and Val that has the strongest inhibitory effect on AHAS (Miflin and Cave, 1972; Lee and Duggleby, 2001). Such a scenario may account for the maintenance of normal levels of Leu in the IPMS1 insertion mutant, despite lower quantities of IPMS, and an enhancement of Val content. A higher activity of AHAS could also raise the levels of Ile, but this does not occur probably due to feedback inhibition by Ile of Thr dehydratase—the enzyme that makes 2-oxobutyrate, the substrate of AHAS for Ile synthesis (Singh, 1999; Coruzzi and Last, 2000).
IPMS1 and IPMS2 Differ Somewhat in Gene Expression Pattern and Properties of the Encoded Protein
Based on RT-PCR analyses and publicly available microarray experiments assembled with the GENEVESTIGATOR program (Supplemental Figs. S4 and S5), both IPMS genes are constitutively expressed throughout the plant and thus can complement each other, consistent with the normal Leu phenotype of the insertion mutant lines. However, the two genes are at least subfunctionalized at a regulatory level (Moore and Purugganan, 2005) since IPMS1 expression, but not that of IPMS2, is twice as high in seeds as in most other plant tissues. In the developing seed, Leu is one of the main free amino acids and may be specifically synthesized there since it is not abundant in the phloem (Baud et al., 2002). The particular role of IPMS1 during seed development might explain the 20% reduction in seed fecundity reported by Field et al. (2004) for an IPMS1 knockout line.
The major biochemical difference between IPMS1 and IPMS2 appears to be the Km value for acetyl-CoA, 45 μm and 16 μm, respectively, making IPMS2 the more efficient enzyme at comparable turnover numbers. Calibrated gel-filtration chromatography showed that IPMS1 is active as a dimer (124 kD), whereas IPMS2 is mainly active as a tetramer (approximately 280 kD; Fig. 4C); nonetheless, these data should be interpreted with care as both proteins contain a His-tag that might influence the formation of quaternary structure (Wu and Filutowicz, 1999). The IPMS enzymes previously isolated from microorganisms were characterized either as dimers (Saccharomyces cerevisiae, Corynebacterium glutamicum, Mycobacterium tuberculosis) or tetramers (Salmonella typhimerium; Leary and Kohlhaw, 1972; Roeder and Kohlhaw, 1980; Pátek et al., 1994; Koon et al., 2004). Gel filtration of the IPMS activity present in crude extracts of Arabidopsis showed a broad band of enzyme activity between 50 and 200 kD with a major peak at 95 to 120 kD (Textor et al., 2004). Assuming that IPMS1 is also present as a dimer in vivo and IPMS2 mostly as a tetramer, the main IPMS activity present in the plant crude extract is IPMS1.
Other IPMSs Have Different Kinetic Constants, Cation Preferences, and Sensitivity to Leu Feedback Inhibition
The Km of IPMS1 and IPMS2 for 2-oxoisovalerate are similar to each other (304 μm and 279 μm) and are much higher than the reported value of 75 μm for the IPMS present in spinach chloroplasts. The spinach IPMS also has a distinct Km value for acetyl-CoA, 5 μm versus 45 μm and 16 μm for IPMS1 and IPMS2, respectively (Hagelstein and Schultz, 1993).
The IPMS enzymes of Arabidopsis, just as the spinach IPMS, are dependent upon millimolar concentrations of Mg2+ (Fig. 4) for optimal enzyme activity. This cofactor can only be replaced with Mn2+, which yields about 50% of the initial enzyme activity. However, Mn2+ is clearly preferred over Mg2+ as a cofactor by the MAM enzymes (Falk et al., 2004; Textor et al., 2004). Some IPMS enzymes of fungi and bacteria also employ Mn2+ or Zn2+ as a cofactor (Stieglitz and Calvo, 1974; Wiegel, 1978; Roeder and Kohlhaw, 1980; Koon et al., 2004). The broad pH optima at 8.5 obtained for IPMS1 and IPMS2 is common for IPMSs, irrespective of their source, and the same pH dependence has also been observed for the MAMs (Falk et al., 2004; Textor et al., 2004).
Generally, enzymes at critical branchpoints in plant amino acid biosynthesis are feedback inhibited by their end-product amino acids (Coruzzi and Last, 2000). Accordingly, Leu inhibits the enzyme activity of both heterologously expressed IPMSs. A maximum of 30% to 35% inhibition was reached around a concentration of 1 mm Leu (Fig. 5), and a similar effect was detected on the IPMS activity in the crude extract of Arabidopsis. A somewhat greater effect has been reported on the IPMS present in crude extracts of maize embryos (61% inhibition at 50 μm Leu, 84% inhibition at 5 mm Leu; Oaks, 1965), but the IPMS of spinach chloroplasts was reported to be 100% inhibited by micromolar concentrations of Leu (Hagelstein and Schultz, 1993). However, in a chloroplast-enriched crude extract from spinach, we only detected a 66% inhibition of IPMS at 5 mm Leu. A 100% inhibition of IPMS activity at 1 mm concentrations of Leu is common among bacteria (Stieglitz and Calvo, 1974). The production of 2-oxoisovalerate itself is regulated through feedback inhibition of AHAS by Leu, Val, and Ile (Fig. 1; Singh, 1999). Concentrations of 1 to 5 mm of these amino acids result in a 45% to 65% inhibition of AHAS enzyme activity in Arabidopsis (Lee and Duggleby, 2001) and other plants (Miflin and Cave, 1972), i.e. an effect of similar magnitude as that of 1 to 5 mm Leu on IPMS activity.
IPMSs Produce Other Products in Vitro and in Vivo
The substrate specificity of IPMSs is not very high. For example, the reported reaction rates for 2-oxobutyrate are usually higher than those found for the true substrate 2-oxoisovalerate, despite the higher Km for 2-oxobutyrate (Webster and Gross, 1965; Strassman and Ceci, 1967; Rabin et al., 1968; Gross, 1970; Kohlhaw and Leary, 1970; Ulm et al., 1972; Wiegel and Schlegel, 1977; Wiegel, 1981; Kohlhaw, 1988). This is also true for the Arabidopsis enzymes. In addition, most reported IPMSs and, to a lesser extent, the MAMs from Arabidopsis (S. Textor, unpublished data) can catalyze the condensation between pyruvate and acetyl-CoA. The product of this condensation reaction, citramalate, has been reported as a metabolite in Arabidopsis (Fiehn et al., 2000) and tomato (27.6 μmol g−1 fresh weight in orange-colored fruits; Roessner-Tunali et al., 2003). The presence of citramalate has led to the suggestion that plants possess a tricarboxylic acid cycle bypass previously described in bacteria (Grant and Smith, 2000). Citramalate may also have a function in Ile biosynthesis by serving as a precursor of 2-oxobutyrate, a substrate for AHAS. This metabolic route has been demonstrated for a mutant of the microorganism Serratia marcescens (Kisumi et al., 1977), the halophilic archeon Haloarcula hispanica (Hochuli et al., 1999), and in Leptospira interrogans (Xu et al., 2004).
Compounds with a carbon chain length longer than 2-oxoisovalerate, like 2-oxovalerate, 4-methyl-2-oxovalerate, and 2-oxohexanoate, have usually been reported to be active-site inhibitors of IPMS (Webster and Gross, 1965; Gross, 1970; Kohlhaw and Leary, 1970; Ulm et al., 1972; Wiegel and Schlegel, 1977; Wiegel, 1981; Kohlhaw, 1988) rather than substrates, as described in this study. An exception is the paper of Rabin et al. (1968) that reports on the conversion of 2-oxovalerate, 4-methyl-2-oxovalerate, and 2-oxohexanoate by an IPMS activity from Pseudomonas aeruginosa at yields comparable to those reported here. Our results indicate that the IPMSs from Arabidopsis and E. coli can use 2-oxo acids ranging in length from glyoxylate to 2-oxohexanoate. However, the shortest and longest substrates react at rates that might have escaped detection in older work with less-sensitive methods.
IPMS and MAM Proteins Share Some But Not All Structural Features
The most striking difference between the amino acid sequences of the IPMS and MAM proteins from Arabidopsis is the absence of about 150 amino acids at the C-terminal domain of the MAMs (Fig. 8). This domain contains a conserved allosteric Leu binding site (Koon et al., 2004) whose absence is likely responsible for the lack of Leu inhibition of the E. sativa MAM synthase (Falk et al., 2004). One can expect the MAMs of Arabidopsis to show similar behavior. The only published crystal structure for an IPMS is that of the M. tuberculosis protein (Koon et al., 2004). Although Arabidopsis IPMS1 and IPMS2 share only about 25% amino acid identity with the M. tuberculosis protein (Fig. 8; 1SR9), the amino acids of the hydrophobic Leu binding pocket of the M. tuberculosis protein are either conserved in the Arabidopsis proteins (Tyr-554, Ala-558, Ala-565, Ala-567) or replaced by comparable amino acids (Leu-535 → Val, Val-551 → Ala), except for Ala-536, which is substituted by the more hydrophilic Asp. All of the amino acid residues of the M. tuberculosis IPMS involved in binding acetyl-CoA and 2-oxoisovalerate, including the GxGERxG motif, are conserved in the IPMS and MAM sequences of Arabidopsis. However, the HxH(D/N)D motif involved in binding of the metal ion is less conserved. The second His residue (His-287) is substituted by Gln in the IPMS sequences of Arabidopsis and wild tomato (but not in the MAM sequences), and there is a substitution of Ala by Ser, six amino acid residues upstream. Such changes might explain why the Arabidopsis IPMSs use Mg2+ as cofactor instead of Zn2+ as the M. tuberculosis IPMS does. The cofactor Zn2+ preferably binds to nitrogen and sulfur-containing amino acid side chains, such as those of His, while Mg2+ prefers residues with oxygen functions (such as the carbonyl group of Gln and hydroxyl group of Ser). The favored cofactor of the MAM proteins, Mn2+, behaves similarly as Mg2+ but nonetheless has higher affinity with nitrogen-containing ligands than Mg2+ (Bock et al., 1999; Glusker et al., 1999).
Figure 8.
Alignment of deduced amino acid sequences for IPMS1, IPMS2, MAM1, and MAM3 with IPMS sequences from wild tomato (GenBank accession nos. AAB61598 and AAB61599), E. coli (Swiss-Prot: P09151), and M. tuberculosis whose protein structure has been elucidated (PDB: 1SR9). Black shading indicates individual amino acids that are conserved within all sequences, dark gray shading individual amino acids that are identical in at least six out of eight sequences, and light gray shading amino acids that are identical in at least five out of eight sequences. The ChloroP-predicted cleavage sites are marked with an underscore. Amino acid residues mentioned in the text are represented below the alignment, likewise amino acid residues of the Leu binding site that are marked with an “L” and the conserved motifs GxGERXG and HxH(D/N)D. Amino acid positions are numbered relative to 1SR9.
Apart from their 60% similarity in amino acid sequence, the close similarity between the IPMSs and the MAMs of Arabidopsis can also be deduced from their overlapping substrate specificities. The IPMSs are able to catalyze the condensation reaction with the MAM substrate 4-methylthio-2-oxobutyrate (Table II). Conversely, purified MAM3 can catalyze the reaction with 2-oxoisovalerate (S. Textor, unpublished data), even though both of these reactions occur at such low rates that they probably do not play a major role in planta. In this respect, it is remarkable that ectopic overexpression of a presumptive IPMS from B. atlantica (BatIMS) in Arabidopsis caused a doubling of aliphatic glucosinolates in the leaves of the T1 generation in addition to perturbed amino acid levels (Field et al., 2006). However, these changes seem unlikely to be the result of an overlap in substrate specificity between IPMS and MAM, since this transformant displayed extensive morphological, metabolic, and transcriptional changes, including the up-regulation of at least four of the 10 known genes of the aliphatic glucosinolate biosynthetic pathway.
The ability to catalyze a reaction with 4-methylthio-2-oxobutyrate at a very low rate is not restricted to the IPMSs of glucosinolate-producing plants like Arabidopsis, but is also characteristic of the IPMS of E. coli (Supplemental Fig. S1), suggesting that it is an inherent property of this enzyme type. This overlap in substrate usage plus the approximate 60% amino acid identity between IPMSs and MAMs makes it plausible that MAMs were derived from an ancestral IPMS through gene duplication. Such duplication could have persisted if it had no negative effects on Leu homeostasis. Posttranscriptional mechanisms like Leu inhibition of IPMS activity may ensure that there are no major changes in amino acid content despite changes in IPMS gene expression, as seen in the case of the IPMS1 and IPMS2 mutants. Subsequent neofunctionalization (Moore and Purugganan, 2005) of one of the IPMS duplicate copies to a MAM would have involved selection for an increase in substrate specificity toward 4-methylthio-2-oxobutyrate and the loss of the regulatory domain for Leu feedback inhibition at some stage in this process. Either as a consequence of these changes or through additional selection, most of the original activity for 2-oxoisovalerate has been lost and some of the MAMs have developed affinities for substrates even larger than 4-methylthio-2-oxobutyrate. In future research, we will seek evidence for this scenario and try to determine what structural changes in IPMS accompanied this process.
MATERIALS AND METHODS
Plants
Seeds of Arabidopsis (Arabidopsis thaliana L. Heynh), ecotype Col-0 (CS3879 Arabidopsis Biological Resource Center), were sown densely in ordinary potting soil mixed with vermiculite (3:1). Plants were raised in a controlled growth chamber with a diurnal cycle of 10 h light and 14 h dark at 22°C. Illumination was from a mixture of Fluora (Osram) and Cool White lamps at 230 μmol m−2 s−1. Seeds of the Salk mutant lines were obtained from the the European Arabidopsis Stock Centre (Nottingham, UK).
RNA Isolation and cDNA Cloning
Total RNA was isolated from liquid nitrogen-frozen root tissue (IPSM1) or total leaf tissue (IPMS2) with Trizol reagent (Invitrogen) according to manufacturer's instructions. First-strand cDNA was synthesized with 2 μg of total RNA, 200 units of MMLV reverse transcriptase (Promega), and 0.5 μg of gene-specific oligonucleotide primer using the reagents and instructions provided.
IPMS2 was amplified from the first-strand cDNA product using the primer pair 1ipms2m/2ipms2n (for all primers used, see Supplemental Table S1), resulting in a truncated ORF lacking 138 nucleotides corresponding to a putative chloroplast transit peptide (ChloroP; Emanuelsson et al., 1999). The reaction product was gel purified using a QiaQuick gel extraction kit (Qiagen), cloned directly into the pBAD-TOPO (Invitrogen) expression vector, and transformed into TOP10 cells (Invitrogen) according to the manufacturer's instructions. The DNA of transformed colonies was purified by a miniprep (Invisorb spin plasmid mini kit; Invitek) and screened by restriction analyses and DNA sequencing on an ABI 3700 DNA sequencer with Big Dye terminators (PE Applied Biosystems).
As protein expression of the IPMS2/pBAD-TOPO construct in BL21-CodonPlus-RIL cells (Stratagene) was very poor, IPMS2 was subcloned into the pCR-T7/CT-TOPO vector (Invitrogen). The desired IPMS2 fragment was obtained from the previous IPMS2/pBAD-TOPO construct in a PCR reaction using the Expand High Fidelity PCR system (La Roche) and the primer pair 1ipms2m+atg/2ipms2n in accordance with the provided instructions. The PCR fragment produced was directly cloned into the pCR-T7/CT-TOPO vector and transformed into TOP10F′ Escherichia coli cells. The resulting bacterial colonies were screened for presence of the desired IPMS2/pCR-T7/CT-TOPO construct.
A cDNA corresponding to the entire ORF of IPMS1 was amplified from the first-strand product using the primer pair 1ipms1k/2ipms1j in a PCR with Pfu-Turbo DNA polymerase (Stratagene). The reaction product was gel purified, cloned directly into the pCR4-TOPO (Invitrogen) vector, and transformed into TOP10 E. coli cells (Invitrogen). Screening for the desired IPMS1/pCR4-TOPO construct yielded a construct that had a single C-to-T mutation at position 176. This mutation was corrected by use of primer 1ipms1i+atg that together with primer 2ipms1j was used to amplify IPMS1 without the 171 nucleotides coding for a putative ChloroP-predicted chloroplast transit peptide. The PCR product obtained from the Expand High Fidelity PCR system was directly cloned into the pCR-T7/CT-TOPO vector and transformed into TOP10F′ E. coli cells. However, none of the clones contained an IPMS1/pCR-T7/CT-TOPO construct that was free of point mutations. We attempted to restore a construct that had only one point mutation at nucleotide 169 with the QuickChange site-directed mutagenesis kit of Stratagene using primers mut1-for and mut1-for-r. An IPMS1/pCR-T7/CT-TOPO construct was isolated that had a single—though silent—point mutation at nucleotide position 169 of the truncated cDNA (codon GAC changed into GAT).
cDNA Expression in E. coli
The IPMS1/pCR-T7/CT-TOPO construct and IPMS2/pBAD-TOPO construct were both expressed in E. coli strain BL21(DE3) pLysS (Invitrogen). A fresh colony harboring the construct was picked from the plate with a sterile toothpick and grown on 25 mL of Luria-Bertani medium with chloramphenicol (34 μg/mL) and ampicillin (100 μg/mL) for 62 h at 18°C. This 25 mL of culture was used to inoculate another 500 mL of antibiotic-containing medium (2× 250 mL) that was subsequently incubated at 18°C until an OD600 of 0.6 was reached. Expression of the cDNA was then induced with 2 mm IPTG and incubation continued overnight. Cells were harvested the next morning by centrifugation in 50 mL Falcon tubes for 10 min at 6,500g and 4°C, and the bacterial pellets were stored at −80°C.
Purification of the Expressed His-Tag Protein
A bacterial pellet was homogenized in a 2-mL Eppendorf tube with 1.5 mL of lysis buffer, containing 50 mm sodium borate buffer, pH 8.0, 300 mm NaCl, 10 mm imidazole, and 1 mm MgCl2, and left on ice for 30 min after the addition of 1.0 × 105 units lysozyme (Merck). A few glass beads (∅ 2 mm) were put in the tube and the cells were disrupted in a sonicator bath (Sonorex RK100, 35 kHz; Bandelin) filled with ice water (3–4 ×, 3 min). Cell debris was precipitated by centrifugation for 7 min at 20,500g and 4°C.
A Poly-Prep chromatography column (2-mL bed volume, 10-mL reservoir; Bio-Rad) was prepared with 1.5 mL of 50% Ni-NTA agarose (Qiagen) and rinsed with 5 mL of lysis buffer. The bacterial lysate was added to the resin and the column was placed on a rotator for 1 h at 4°C, allowing the His-tag protein to bind to the resin. The column was then placed in a vertical position and washed with 12 mL of wash buffer, which had the same composition as lysis buffer except for an elevated (20 mm) concentration of imidazole. The His-tag protein was eluted with 3 mL of buffer containing 250 mm imidazole, and the eluent was immediately transferred to an Econo-Pac 10 DG column (Bio-Rad) and desalted into 4 mL of a 50 mm Tris buffer, pH 8.0, containing 1 mm MgCl2 and 10% glycerol. This desalted preparation was used for enzyme assays. The protein concentration of each preparation was determined with the BCA protein assay kit (Pierce) using bovine serum albumin as a standard; concentrations generally ranged from 300 to 600 μg/mL.
Enzyme Assays
Qualitative Assay Using Radio-HPLC Detection
The assay contained 100 μL of enzyme preparation that was incubated at 30°C with 3 mm of the 2-oxo-acid substrate, 500 μm [1-14C]acetyl-CoA (from Hartmann, diluted with unlabeled acetyl-CoA [Sigma-Aldrich] to 0.4 mCi mmol−1) and 4 mm MgCl2, in a final volume of 250 μL 100 mm Tris buffer, pH 8.0. The incubation was stopped after 1 h by adding 750 μL ethanol. The denatured protein was precipitated by centrifugation and the supernatant reduced to a volume of approximately 100 μL using a DNA Speedvac 1100 (Savant). A portion of the concentrated supernatant (20 μL) was analyzed on the radio-HPLC according to Falk et al. (2004).
Continuous Spectrophotometric Assay (NEM Assay)
The reaction mixture contained 10 to 500 μm acetyl-CoA, 50 to 2,000 μm 2-oxoisovalerate, 0.1 mm NEM (Sigma-Aldrich), 4 mm MgCl2, 100 mm Tris, pH 8.0, and 5 μL of enzyme preparation in a total volume of 500 μL. The change in optical density over time at 30°C was followed against a solution of 1 mm NEM in a UV-2501PC spectrophotometer (Shimadzu) using micro quartz cuvettes (Hellma) with black walls. The loss of the thioesterbond in acetyl-CoA and the loss of a double bond in NEM upon binding to CoA-SH result in an ΔOD232 of −1.08 × 104 (1-cm light path) per mole of CoA-SH formed (Webster and Gross, 1965).
Spectrophotometric End-Point Assay (DTNB Assay)
The enzyme preparation (1–5 μL) was incubated for 10 min at 30°C with 10 mm 2-oxoisovalerate, 500 μm acetyl-CoA, 4 mm MgCl2, and 100 mm Tris, pH 8.0, in a total volume of 150 μL. Incubations were synchronized by pipetting the enzyme solution in the lid of an Eppendorf tube containing the incubation mixture, after which the enzyme in all vessels was simultaneously spun down for a few seconds in a table-top centrifuge; reactions were stopped by freezing the vessels in liquid nitrogen. To the frozen reaction mixture, 200 μL of ethanol and 200 μL of a fresh 1 mm solution of DTNB (Sigma-Aldrich) in 100 mm Tris, pH 8.0, were added. The mixture was left at room temperature to allow the free thiol group of CoA to react with DNTB, forming a yellow-colored, 3-carboxy-4-nitrothiophenol anion with an ɛ412 of 14,140 m−1 cm−1 (Kohlhaw, 1988). When no further color developed, the mixture was centrifuged for 5 min at 16,000g and the absorbance measured against water at 412 nm. The enzyme assay was corrected for unspecific hydrolysis of acetyl-CoA by subtracting the absorbance of a blank incubation where no 2-oxo-acid substrate had been added. The assay was generally linear for the first 15 min with 2 μg of protein.
Enzyme Characterization
Substrate Specificity
The various 2-oxo acid substrates tested in the radio-HPLC assay with [14C]acetyl-CoA were either obtained from Fluka or Sigma-Aldrich, or synthesized in our laboratories (Falk et al., 2004; S. Textor, unpublished data). Reaction rates of the different substrates were determined at 20 mm (estimated to be a saturating concentration) with the DNTB assay; the amount of enzyme added was varied from 1 to 50 μL, depending on the substrate, to stay within detection limits.
Enzyme Kinetics
The Km and Vmax values for acetyl-CoA and 2-oxo-isovalerate were determined with the NEM assay. They were calculated from the recorded initial (linear) reaction rates at different substrate concentrations of three to four independent experiments using the Enzyme Kinetic Module (Version 1.1) of Sigmaplot (Version 8.0).
Other Enzyme Characteristics
The pH optima for IPMS1 and IPMS2 were determined in the pH range of 5.5 to 10.5 in 0.5-pH units using the DTNB assay in which Tris buffer was replaced by MES (pH 5.5–6.5), BisTris-propane (pH 6.5–9.5), or 2-amino-2-methyl-1-propanol (pH 9.5–10.5). The DTNB assay was also used to test the effect of Leu on enzyme activity in a range from 25 μm to 10 mm. Furthermore, the effect on enzyme activity of Mg2+ concentration, Mn2+, and K+ was investigated with the DTNB assay taking an enzyme preparation after His-tag purification that had been desalted to Tris buffer lacking MgCl2. The effect of other cations, i.e. Ca2+, Co2+, Cu2+, Fe2+, K+, Mn2+, and Zn2+, on this enzyme preparation had to be tested with the qualitative radio-HPLC assay because these ions are strong oxidizers that react with the 3-carboxy-4-nitrothiophenol anion of the DNTB assay, causing a rapid loss of the yellow color. All cations were used as their chloride salts, except Fe2+ and Zn2+ that were used as their sulfate salts.
The molecular mass of the native recombinant protein was estimated by exclusion chromatography on a Superdex 200 column (Hiload 16/60; Pharmacia Biotech) that had been calibrated with β-amylase (200 kD), alcohol dehydrogenase (150 kD), bovine serum albumin (66 kD), carbonic anhydrase (29 kD), and cytochrome C (12.4 kD). The column was loaded with 200 μL of enzyme preparation and eluted at 1 mL min−1 with a buffer consisting of 50 mm Tris, pH 8.0, 10% glycerol, and 150 mm NaCl. Sixty fractions of 1 mL were collected after discarding the first 40 mL of eluent, and 138.5 μL of each fraction was tested in a modified DNTB assay (25-min incubation at room temperature) for enzyme activity.
E. coli CV512 Complementation and Origin of E. coli IPMS
The IPMS1/pCR-T7/CT-TOPO and IPMS2/pBAD-TOPO cDNA constructs were transformed in E. coli CV512 (F+ leuY371; Somers et al., 1973). The CV512 strain lacks functional IPMS and is able to grow on M9 minimal medium with Glc as a carbon source only when it is either supplemented with Casamino acids (Difco) or transformed with a construct containing a functional IPMS. To ensure expression from a T7 polymerase-driven construct, the IPTG inducible T7-polymerase gene was introduced into strain CV512 by a λDE3 lysogenation kit (Novagen) giving CV512(DE3). As a positive control, a pET28a (Novagen) construct harboring the E. coli DH5α (Hanahan, 1983) IPMS gene leuA was used for complementation studies. The leuA gene was amplified by PCR from DH5α with the primer pair IPMFEff/IPMErv (Supplemental Table S1) and cloned into pET28a using the BamHI/XhoI restriction sites provided by the primers. Complementation efficiency was tested at two different incubation temperatures, 30°C and 37°C.
The pET28a/LeuA construct was also used as a source of E. coli IPMS that was tested in the radio-HPLC assay for its substrate specificity. BL21(DE3) E. coli cells (Invitrogen) with the LeuA construct were grown in 100 mL of Luria-Bertani medium containing 50 μg mL−1 kanamycin at 37°C until an OD600 of 0.5 was reached. Expression was induced with 1 mm IPTG and incubation continued for 2.5 h. Cells were harvested and the expressed protein isolated and purified in the same way as described for the Arabidopsis IPMSs, taking advantage of the C-terminal His-tag present in the construct.
Plant Mutant Lines
PCR of Genomic DNA
Genomic DNA was extracted from expanding leaves using an abbreviated protocol of Rogers and Bendich (1985). About 5 mg of young leaf tissue was collected in a 1.5-mL microfuge tube and homogenized with 10 μL of 2× CTAB solution (2% cetyltrimethylammonium bromide [w/v], 100 mm Tris, pH 8.0, 1.4 m NaCl, 1% polyvinylpyrrolidone [4,000 molecular weight]) using a micropestle. The sample was incubated at 65°C for 1 to 2 min, cooled briefly on ice, and extracted with 10 μL of chloroform:isoamyl alcohol (24:1, v/v). After 10 μL of water was added, the samples were subjected to centrifugation at 11,000g. The upper phase was recovered and 0.1 to 0.5 μL was used per PCR of 20-μL volume (for details, see below).
PCR of RNA Transcript
Total RNA was isolated from freshly harvested, freeze-dried whole rosettes of individual 3- to 4-week-old plants with Trizol reagent (Invitrogen) according to the manufacturer's instructions. A cDNA population was synthesized with 2 μg of RNA, either 0.5 μg of dT12-18 or 0.17 μg of gene-specific primers (Invitrogen) and 200 units of MMLV reverse transcriptase (Promega) using the reagents and instructions provided. From each of these reverse transcriptase reactions, a 20-μL PCR was prepared consisting of 1× PCR buffer (Promega), 0.2 mm dNTPs, 0.5 μm each primer (see Supplemental Table S1 for specific primer pairs for ACT8, IPMS1, and IPMS2), 0.5 units of Taq Polymerase (Promega), and an aliquot of reverse transcriptase reaction equivalent to 40 ng of template RNA. The reactions were subjected to an initial thermal denaturation of 94°C for 2 min, followed by 30 or 35 cycles at 94°C for 30 s, a primer-dependent annealing temperature for 30 s, 72°C for 2 min, and a final incubation at 72°C for 5 min. After electrophoresis and incubation in ethidium bromide, DNA fragments were analyzed and quantified (GeneGenius with GeneTools Analysis Software Version 3.02 [Synoptics]; or Gel Logic 200 with Kodak MI software). PCRs of each individual reverse transcriptase reaction with specific oligonucleotide primer pairs were done at least two times.
Amino Acid Analyses
The procedure was optimized to extract total free amino acids from Arabidopsis plant material and to quantify the individual amino acids in a single HPLC run. The amino acids present in a crude plant extract were derivatized with mercaptoethanol and O-phthaldialdehyde yielding a fluorescent isoindole (Roth, 1971; Sarwar and Botting, 1993), a method inappropriate for the detection of Cys (weakly fluorescent derivative) and Pro (contains no primary amino group for derivatization reaction). Rosettes of individual 3- to 4-week-old individual plants were harvested, and individually frozen in liquid nitrogen, lyophilized, and pulverized. Aliquots of 10 and 50 mg were transferred to 2-mL Eppendorf tubes and resuspended with 0.8 mL of 0.1 n HCl to rehydrate the tissue. After incubation for 15 min at room temperature, the samples were centrifuged at 16,000g and the supernatant transferred to a fresh tube. Samples for HPLC were prepared by adding 50 μL of the supernatant to a 350-μL glass vial insert containing 50 μL of a 1:1 (v/v) solution of 0.1 n HCl and 0.5 m potassium borate, after which the vials were capped. The autosampler of the HPLC (Agilent HP1100 series) was programmed to mix the vial content with 30 μL of reagent consisting of 0.085 m O-phthaldialdehyde (Fluka) and 1% (v/v) β-mercaptoethanol in a 0.5 m sodium borate solution immediately before injection of 50 μL of sample onto the HPLC column (Supelcosil LC-18-DB [250 × 4.6 mm, 5-μm particle size]; Supelco). The column was run with a 0.02 m citrate solution of pH 5.5 (solvent A) and methanol:acetonitrile (65:35, v/v; solvent B) at 28°C with a gradient as follows: 15% of solvent B at start, linear gradient to 38.5% of solvent B over 42 min, increase to 45% of solvent B in 1 min, linear gradient to 62.5% of solvent B over 20 min, increase to 100% of solvent B in 0.5 min, 100% of solvent B for 3 min, decrease to 15% of B in 0.5 min, and re-equilibration of the column at 15% of solvent B for 8 min (total time of run is 75 min). The derivatized amino acids were measured with a fluorescence detector (Agilent HP1100 series) at an excitation wavelength of 340 nm and an emission wavelength of 445 nm. Amino acid samples were quantified by calibration curves that were prepared with a 0.5 mm amino acid stock solution of Asn, His, and Gln and an amino acid stock solution of Fluka that contained all the other amino acids in a 0.5 mm concentration. Aliquots of 5, 10, 20, 30, 40, and 50 μL amino acid standard solution were added to a 350-μL glass vial insert and adjusted to 50 μL with 0.1 n HCl. The vials were capped after the addition of 50 μL of 0.5 m potassium borate and run in the same sequence as the plant samples.
Glucosinolate Analyses
Glucosinolates were extracted from 10 mg of lyophilized plant material and converted into their desulfoglucosinolate counterparts according to Brown et al. (2003). The desulfoglucosinolates were identified and quantified by HPLC analyses on a C18 reversed-phase column (LiChrospher RP-18, 250 × 4.6 mm i.d., 5-μm particle size; Chrompack) by comparison of retention times and UV spectra to those of purified standards and by measuring the A229 relative to an internal standard (Reichelt et al., 2002; Brown et al., 2003).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Radio-HPLC analyses of the biochemical assay for the IPMS of E. coli.
Supplemental Figure S2. Predicted positions of the T-DNA inserts in the Salk knockout lines for IPMS1 (Salk_101771) and IPMS2 (Salk_051060 and Salk_000074).
Supplemental Figure S3. Analyses of the glucosinolate content of homozygotic T-DNA insertion lines for IPMS1 (Salk_101771 [mm]) and IPMS2 (Salk_051060 [mm] and Salk_000074 [mm]).
Supplemental Figure S4. Semiquantitative RT-PCR analysis of IPMS1 and IPMS2 gene expression in various plant organs.
Supplemental Figure S5. Expression levels of IPMS1 and IPMS2 in different plant tissues and various growth stages according GENEVESTIGATOR.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank Axel Schmidt for his advice on cloning and expression procedures, Kimberly Falk for her help and advice in the isolation of IPMS activities from crude plant extracts, and Michael Reichelt for his assistance with the HPLC analyses. We thank the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre for providing Arabidopsis plant lines.
This work was supported by the Max Planck Society and a Marie Curie Individual Fellowship (MCFI–2002–01677) to J.-W.d.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jonathan Gershenzon (gershenzon@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40 151–160 [Google Scholar]
- Bock CW, Katz AK, Markham GD, Glusker JP (1999) Manganese, a replacement for magnesium and zinc: functional comparison of the divalent ions. J Am Chem Soc 121 7360–7372 [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481 [DOI] [PubMed] [Google Scholar]
- Campos de Quiros H, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Mithen R (2000) α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet 101 429–437 [Google Scholar]
- Chisholm MD, Wetter LR (1964) Biosynthesis of mustard oil glucosides. IV: the administration of methionine-C14 and related compounds to horseradish. Can J Biochem 12 1033–1040 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Last R (2000) Amino acids. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 358–411
- Ellerstrom M, Josefsson LG, Rask L, Ronne H (1992) Cloning of a cDNA for rape chloroplast 3-isopropylmalate dehydrogenase by genetic complementation in yeast. Plant Mol Biol 18 557–566 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neutral network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk KF, Vogel C, Textor S, Bartram S, Hick A, Pickett JA, Gershenzon J (2004) Glucosinolate biosynthesis: demonstration and characterization of the condensing enzyme of the chain elongation cycle in Eruca sativa. Phytochemistry 65 1073–1084 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18 1157–1161 [DOI] [PubMed] [Google Scholar]
- Field B, Furniss C, Wilkinson A, Mithen R (2006) Expression of a Brassica isopropylmalate synthase gene in Arabidopsis perturbs both glucosinolate and amino acid metabolism. Plant Mol Biol 60 717–720 [DOI] [PubMed] [Google Scholar]
- Field B, Guillermo C, Traka M, Botterman J, Vancanneyt G, Mithen R (2004) Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol 135 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusker JP, Katz AK, Bock CW (1999) Metal ions in biological systems. The Rigaku Journal 16 8–17 [Google Scholar]
- Grant M, Smith S (2000) Meeting report; communal weeding. Genome Biol I reports 4024.1–4024.3 [Google Scholar]
- Graser G, Schneider B, Oldham NJ, Gershenzon J (2000) The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch Biochem Biophys 378 411–419 [DOI] [PubMed] [Google Scholar]
- Gross SR (1970) α-Isopropylmalate synthase (Neurospora). Methods Enzymol 17A 777–790 [Google Scholar]
- Hagelstein P, Schultz G (1993) Leucine synthesis in spinach chloroplasts: partial characterization of 2-isopropylmalte synthase. Biol Chem Hoppe Seyler 374 1105–1108 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57 303–333 [DOI] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166 557–580 [DOI] [PubMed] [Google Scholar]
- Hochuli M, Patzelt H, Oesterhelt D, Wuthrich K, Szyperski T (1999) Amino acid biosynthesis in the halophilic archaeon Haloarcula hispanica. J Bacteriol 181 3226–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, Sonnewald U, Willmitzer L (1993) Cloning and expression analysis of β isopropylmalate dehydrogenase from potato. Mol Gen Genet 236 309–314 [DOI] [PubMed] [Google Scholar]
- Junk DJ, Mourad GS (2002) Isolation and expression analyses of the isopropylmalate synthase gene family of Arabidopsis thaliana. J Exp Bot 53 2453–2454 [DOI] [PubMed] [Google Scholar]
- Kisumi M, Komatsubara S, Chibata I (1977) Pathway for isoleucine formation from pyruvate by leucine biosynthetic enzymes in leucine-accumulating isoleucine revertants of Serratia marcescens. J Biochem (Tokyo) 8 95–103 [DOI] [PubMed] [Google Scholar]
- Kohlhaw GB (1988) α-Isopropylmalate synthase from yeast. Methods Enzymol 166 414–423 [DOI] [PubMed] [Google Scholar]
- Kohlhaw GB, Leary TR (1970) α-Isopropylmalate synthase (Salmonella typhimurium). Methods Enzymol 17A 771–777 [Google Scholar]
- Koon N, Squire CJ, Baker EN (2004) Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc Natl Acad Sci USA 101 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis resistance quantitative trait locus. Proc Natl Acad Sci USA 100 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Leary TR, Kohlhaw GB (1972) α-Isopropylmalate synthase from Salmonella typhimurium: analysis of the quaternary structure and its relation to function. J Biol Chem 247 1089–1095 [PubMed] [Google Scholar]
- Lee Y-T, Duggleby RG (2001) Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxy acid synthase and reconstitution with its catalytic subunit. Biochemistry 40 6836–6844 [DOI] [PubMed] [Google Scholar]
- Matsuo M, Yamazaki M (1968) Biosynthesis of sinigrin. VI: Incorporation from homomethionine (2-14C, 15N) and some labelled compounds into sinigrin. Chem Pharm Bull (Tokyo) 16 1034–1039 [DOI] [PubMed] [Google Scholar]
- Miflin BJ, Cave PR (1972) The control of leucine, isoleucine, and valine biosynthesis in a range of higher plants. J Exp Bot 23 511–516 [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8 122–128 [DOI] [PubMed] [Google Scholar]
- Oaks A (1965) The synthesis of leucine in maize embryos. Biochim Biophys Acta 111 79–89 [DOI] [PubMed] [Google Scholar]
- Pátek M, Krumbach K, Eggeling L, Sahm H (1994) Leucine synthesis in Corynebacterium glutamicum: enzyme activities structure of leuA, and effect of leuA inactivation on lysine synthesis. Appl Environ Microbiol 60 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin R, Salomon II, Bleiweis AS, Carlin J, Ajl SJ (1968) Metabolism of ethylmalic acids by Pseudomonas aeroginosa. Biochemistry 7 377–388 [DOI] [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59 663–671 [DOI] [PubMed] [Google Scholar]
- Roeder PR, Kohlhaw GB (1980) The α-isopropylmalate synthase from yeast, a zinc metalloenzyme. Biochim Biophys Acta 613 482–487 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5 69–76 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M (1971) Fluorescence reaction for amino acids. Anal Chem 43 880–882 [DOI] [PubMed] [Google Scholar]
- Sarwar G, Botting HG (1993) Evaluation of liquid chromatographic analysis of nutritionally important amino acids in food and physiological samples. J Chromatogr 615 1–22 [DOI] [PubMed] [Google Scholar]
- Serif GS, Schmotzer LA (1968) Biosynthesis of the aglycones of plant thioglucosides—I: precursor studies of the aglycone of progoitrin. Phytochemistry 7 1151–1157 [Google Scholar]
- Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In BK Singh, ed, Plant Amino Acids. Dekker, New York, pp 227–248
- Singh BK, Shaner DL (1995) Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers JM, Amzallag A, Middleton RB (1973) Genetic fine structure of the leucine operon of Escherichia coli K-12. J Bacteriol 113 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz BI, Calvo JM (1974) Distribution of the isopropylmalate pathway to leucine among diverse bacteria. J Bacteriol 118 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassman M, Ceci N (1967) A study of acetyl-CoA condensation with α-keto acids. Arch Biochem Biophys 119 420–428 [DOI] [PubMed] [Google Scholar]
- Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA, Gershenzon J (2004) Biosynthesis of methionine derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensation enzyme of the chain elongation-cycle. Planta 218 1026–1035 [DOI] [PubMed] [Google Scholar]
- Ulm EH, Bohme R, Kohlhaw G (1972) α-Isopropylmalate synthase from yeast: purification, kinetic studies, and effect of ligands on stability. J Bacteriol 110 60–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RE, Gross SR (1965) The α-isopropylmalate synthase of Neurospora. I: The kinetic and end product control of α-isopropylmalate synthetase function. Biochemistry 4 2309–2318 [Google Scholar]
- Wiegel J (1978) Mn++-specific reactivation of EDTA inactivated α-isopropylmalate synthetase from Alcaligenes eutrophus H16. Biochem Biophys Res Commun 82 907–912 [DOI] [PubMed] [Google Scholar]
- Wiegel J (1981) α-Isopropylmalate synthetase as a marker for the leucine biosynthetic pathway in several Clostridia and in Bacterioles fragiles. Arch Microbiol 130 385–390 [DOI] [PubMed] [Google Scholar]
- Wiegel J, Schlegel HG (1977) α-Isopropylmalate synthase from Alcaligenes eutrophus H16: I. purification and general properties. Arch Microbiol 112 239–246 [DOI] [PubMed] [Google Scholar]
- Wittenbach VA, Teaney PW, Hanna WS, Rayner DR, Schloss JV (1994) Herbicidal activity of an isopropylmalate dehydrogenase inhibitor. Plant Physiol 106 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7 263–270 [DOI] [PubMed] [Google Scholar]
- Wu J, Filutowicz M (1999) Hexahistidine (His6)-tag dependent protein dimerization: a cautionary tale. Acta Biochim Pol 46 591–599 [PubMed] [Google Scholar]
- Xu H, Zhang Y, Guo X, Ren S, Staempfli AA, Chiao J, Jiang W, Zhao G (2004) Isoleucine biosynthesis in Leptospira interrogans Serotype lai Strain 56601 proceeds via a threonine independent pathway. J Bacteriol 186 5400–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsh-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.