Abstract
We have used laser-capture microdissection to isolate RNA from discrete tissues of globular, heart, and torpedo stage embryos of Arabidopsis (Arabidopsis thaliana). This was amplified and analyzed by DNA microarray using the Affymetrix ATH1 GeneChip, representing approximately 22,800 Arabidopsis genes. Cluster analysis showed that spatial differences in gene expression were less significant than temporal differences. Time course analysis reveals the dynamics and complexity of gene expression in both apical and basal domains of the developing embryo, with several classes of synexpressed genes identifiable. The transition from globular to heart stage is associated in particular with an up-regulation of genes involved in cell cycle control, transcriptional regulation, and energetics and metabolism. The transition from heart to torpedo stage is associated with a repression of cell cycle genes and an up-regulation of genes encoding storage proteins, and pathways of cell growth, energy, and metabolism. The torpedo stage embryo shows strong functional differentiation in the root and cotyledon, as inferred from the classes of genes expressed in these tissues. The time course of expression of the essential EMBRYO-DEFECTIVE genes shows that most are expressed at unchanging levels across all stages of embryogenesis. We show how identified genes can be used to generate cell type-specific markers and promoter activities for future application in cell biology.
Embryogenesis represents a critical stage of the sporophytic life cycle, transforming the fertilized egg cell via a precise sequence of events into a multicellular organism (Mayer et al., 1991; Scheres et al., 1994; Franzmann et al., 1995; Laux et al., 2004). The establishment of shoot and root stem-cell systems (meristems) provides the capacity for the increasingly complex architecture of postembryonic development, which gives rise to the species-specific characteristics of the adult plant. The shoot meristem is the source of all above-ground organs generated postembryonically, maintaining a fine balance between proliferation of the stem-cell population and differentiation. The primary root meristem enables the primary root to grow through extension.
Embryogenesis in Arabidopsis (Arabidopsis thaliana) is a continuous process, although for convenience it can be separated into three major phases, described as early, mid, and late. The early phase is one of pattern formation and morphogenesis, during which the axes of the plant body plan are defined and organ systems formed. The mid phase is that of maturation, with a characteristic accumulation of storage reserves. In its late phase, the embryo prepares for developmental arrest. Arabidopsis embryogenesis is rapid, with the early and mid phases completed 11 to 12 d post fertilization and only 14 d to the completion of the late phase and the production of desiccated mature seed (Lindsey and Topping, 1993).
Understanding the molecular mechanisms underlying embryogenesis can provide insight into developmental and metabolic regulation and the signaling systems integrating these processes. A great deal of research has been invested into analyzing the genetic control mechanisms, exploiting a range of techniques to isolate genes of importance. The construction and screening of cDNA libraries from isolated RNA (Goldberg et al., 1989) and promoter/enhancer trapping (Topping et al., 1994) are just two of the techniques employed. A great deal of success has been achieved through mutational screens, highlighting genes that produce a knockout phenotype in the seed (Meinke and Sussex, 1979; Mayer et al., 1991).
Following the completion of the sequencing of the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000), research has focused on functionally characterizing the 27,000 genes predicted (Ausubel and Benfey, 2002; Wortman et al., 2003). Mutational studies have continued to play a major role in this analysis (Parinov et al., 1999; Sessions et al., 2002; Alonso et al., 2003). Mayer et al. (1991) estimated that approximately 4,000 genes are required for embryogenesis, with about 40 of these essential for pattern formation. Insertional mutagenesis screens have since demonstrated the requirement for a larger number of essential genes based on the resulting frequency of embryonic lethality (Franzmann et al., 1995; McElver et al., 2001; Tzafrir et al., 2004).
The use of DNA microarray technology potentially allows a global analysis of the expression of a large proportion of the Arabidopsis genome and has been used to study transcriptional changes in seed development in Arabidopsis. For example, Girke et al. (2000) have used DNA chips with approximately 30% genome coverage, with RNA isolated from whole seeds. More recently, we used laser-capture microdissection (LCM) in combination with DNA microarray analysis to identify genes that are differentially expressed in apical and basal domains of globular and heart stage Arabidopsis embryos (Casson et al., 2005). LCM is a powerful tool allowing the rapid and precise isolation of specific populations of cells or even individual cells from a heterogeneous tissue based on established histological identification (Emmert-Buck et al., 1996; Bonner et al., 1997; Simone et al., 1998). Quick fixation or freezing of tissue samples for LCM minimizes any undesirable changes in gene expression that could occur during sample preparation (Gillespie et al., 2002).
In this article, we extend the studies carried out previously on LCM-isolated tissues of globular and heart stage embryos (Casson et al., 2005) to include three tissues of the torpedo stage and present a time course global analysis of expression of embryonic genes.
RESULTS
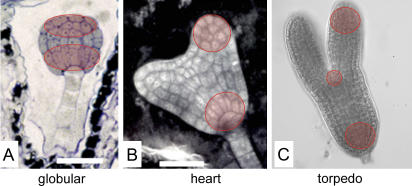
The embryonic tissues sampled by LCM for RNA profiling are indicated in Figure 1. These comprise the apical and basal domains of the globular stage embryo, cotyledonary, and root pole tissues of the heart stage embryo, and cotyledonary, root pole, and shoot apical meristem (SAM) tissues from the torpedo stage embryo following cryosectioning. For each sample, approximately 10 to 15 cells were captured, and samples from approximately 15 embryo sections were pooled, providing approximately 100 to 200 cells for RNA extraction, amplification, and analysis. For illustration, the LCM of a torpedo stage embryo is presented in Figure 2.
Figure 1.
LCM of embryonic tissues. Tissues captured by LCM at globular (A), heart (B), and torpedo (C) stages of embryogenesis, as indicated by highlighted areas.
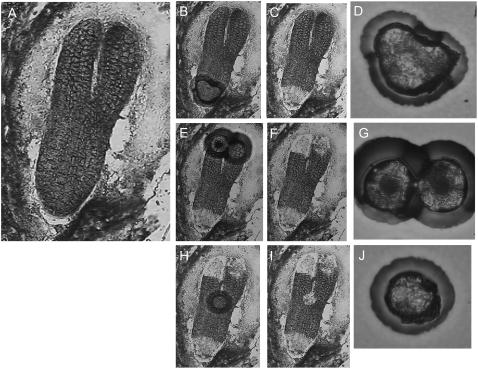
Figure 2.
LCM of cryosections of torpedo stage Arabidopsis embryos. A, Torpedo stage embryo. B, Basal region after targeting with laser, after removal of the cap (C), and basal cells captured on cap (D). E, Targeting of the cotyledonary region, after removal of the cap (F), and cotyledonary cells on the cap (G). H, Targeting of the SAM region, after removal of the cap (I), and SAM cells on the cap (J).
Amplified RNA (aRNA) samples were labeled and analyzed for expression profiles using the Affymetrix ATH1 GeneChip, which contains probes for approximately 22,800 genes of Arabidopsis. The aim was to investigate the changing gene expression patterns through the embryonic time course in both the apical and basal domains. For an additional comparative time point, GeneChip data were utilized from the nonembryonic cotyledon and root tissue of a seedling, 7 d post germination (dpg; Schmid et al., 2005; AtGenExpress Consortium, http://www.affymetrix.arabidopsis.info). aRNA samples were shortened and showed a 3′ bias (Supplemental Fig. S1). This potential problem was taken account of during microarray analysis by reducing the number of probe pairs used (from 11 to 8) and restricting them to those designed toward the 3′ end of transcripts, as discussed previously (Casson et al., 2005). The reduced probe pairs were used for the analysis of all aRNA samples. For each tissue sampled, we calculated a mean and sd signal value across the replicates based on the signal values (Supplemental Tables S1–S9). We have also described elsewhere a detailed validation of the microarray data by both reverse transcription-PCR and promoter-β-glucuronidase (GUS) fusion experiments of candidate genes (Casson et al., 2005).
Estimation of the Number of Genes Expressed in the Torpedo Stage Embryo
The use of the ATH1 GeneChip allows an estimation to be made of the number of genes that are being expressed in the tissue under analysis and also a comparison to other analyses performed on the same microarray platform. The use of the term estimate refers to the fact that a cutoff point of a minimal Affymetrix signal must be imposed; genes deemed to be expressed are those with a value equal to or greater than this value. The application of a minimal Affymetrix signal value of 75 has been applied to a study of the root transcriptome (Birnbaum et al., 2003). Recently, we applied a minimal signal value of 40 to a study of globular and heart stage embryonic tissue (Casson et al., 2005), and we have extended that study to calculate an estimated number of expressed genes for torpedo stage embryos, using both of these cutoff values.
The replicates for each tissue type (torpedo SAM, cotyledon, and root) were collated, and the mean signal values were ranked highest to lowest, thus revealing the number of genes with an equal or greater value than the designated cutoffs (Supplemental Table S10). Using the cutoff value of 75 for the mean value of the replicates analyzed, it was determined that between 8,353 and 11,690 genes (approximately 37%–51%) are expressed in the three tissue types of the torpedo stage embryo. If the lower mean signal threshold value of 40 is applied, up to approximately 77% of the genes are deemed to be expressed.
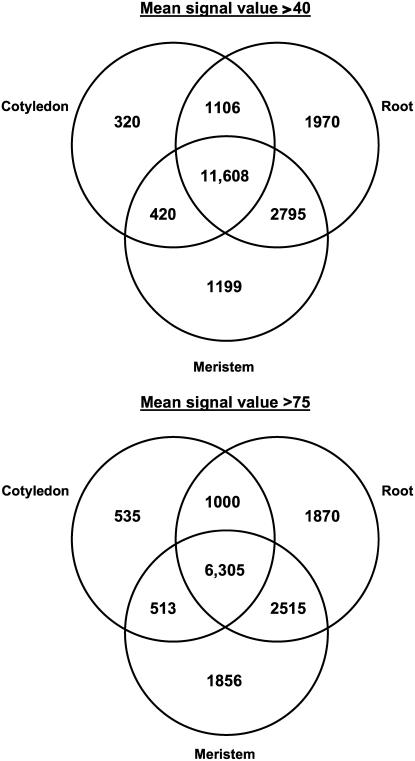
A spatial analysis was then performed on genes predicted to be expressed to ascertain what degree of overlap exists between the different tissue types. This analysis is summarized in Figure 3 as a Venn diagram for each cutoff value. The majority of expressed genes are present in all tissue types (60% when using the signal threshold value of 40 and 43% when using the signal threshold value of 75). There are also significant numbers of genes present in single tissue types only (18% when using the signal threshold value of 40 and 29% when using the signal threshold value of 75), suggesting distinct spatial transcriptional profiles are present.
Figure 3.
Venn diagrams showing overlapping expression of genes between the tissue regions of the torpedo stage embryo. A, Venn diagram is presented for signal cutoff values of 40 and 75, with the overlapping regions corresponding to the number of expressed genes present in more than one tissue type. The central region corresponds to the expressed genes present in all tissue types.
Cluster Analysis of Developmental Stages
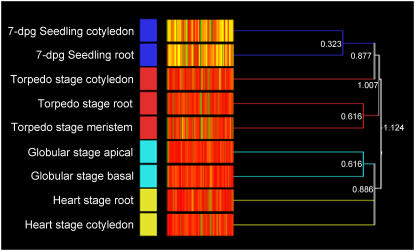
Cluster analysis of the entire transcriptional profile of roots (basal) and cotyledons (apical) of the globular, heart, torpedo stage embryos and seedlings, and torpedo stage SAM revealed that the transcriptional profiles of the apical regions are more closely related to their respective developmental stage basal region than they are to the other apical regions (Fig. 4). These clusters demonstrate that there is a strong correlation between biological replicates from each tissue sampled (see also Supplemental Tables S1–S9), and the data support the hypothesis that each developmental stage has its own distinct transcriptional profile. This further suggests that genes with specifically apically and basally localized expression patterns contribute only a minority of the overall profile. The condition clustering analysis also identifies a clear separation between the early embryonic globular and heart stages, and the later torpedo stage, which is calculated to be closer to the seedling in terms of its transcriptional profile. Interestingly, the torpedo stage SAM clusters closer to the torpedo stage root than it does to the torpedo stage cotyledon, indicating a clear transcriptional difference between these adjacent regions.
Figure 4.
Cluster analysis of the entire transcriptional profile of roots (basal) and cotyledons (apical) of the developmental stages of globular, heart, torpedo, and seedling and torpedo stage SAM. All samples were initially normalized together to a per-gene median value. Clustering analysis was then performed using condition tree clustering on all samples. The branch values are calculated as distance, i.e. dissimilarity of expression between data clusters; low values represent relatively low dissimilarity, or relatively high similarity (GeneSpring version 7.2).
Transcriptional Changes along an Embryonic Developmental Time Course
To characterize in more detail the transcriptional changes taking place between stages, a separate analysis was undertaken for both the apical (cotyledon) regions and the basal (root) regions. While not directly targeting genes of potential importance in apical-basal polarity, it was hoped that such an analysis would provide an insight into potential differences in the functional gene classes of importance in the different regions. A clustal analysis was also undertaken with the aim of elucidating potentially important groups of genes with similar expression profiles across the three stages.
Apical Developmental Time Course
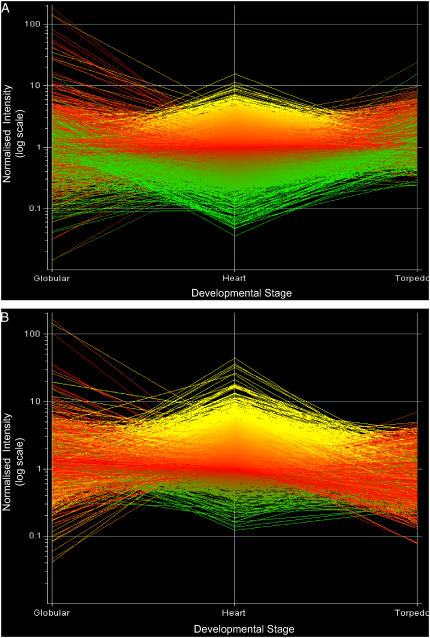
All the apical (cotyledon) samples were normalized together to a per-gene median value using robust multi array average. A graphical view of the normalized data is shown in Figure 5A, with the expression value of each gene plotted on a log scale against the developmental time course. The three developmental stages each has a distinct profile around a core of genes centered on the default expression value of 1. The globular stage displays the highest degree of variability with a number of genes of very high and very low expression values. At the heart stage, there is a similar general spread of high and low expression values but without the extreme outliers present at the globular stage. In comparison, the torpedo stage data show a much smaller range of expression levels. Figure 5A includes data for all the genes present on the GeneChip, not all of which are likely to be expressed at a given stage. Additionally, no statistical significance has been attributed to the expression values displayed, but it nevertheless provides a useful measure of the potential differences that could be present between the stages.
Figure 5.
Transcriptional changes across a developmental time course (globular, heart, and torpedo stage embryos) in apical (A) and basal (B) domains. All samples were normalized together to a per-gene median value, indicated in red. Genes expressed to levels higher than the median values are indicated in yellow, while genes expressed to lower levels are indicated in green. All genes on the GeneChip are represented.
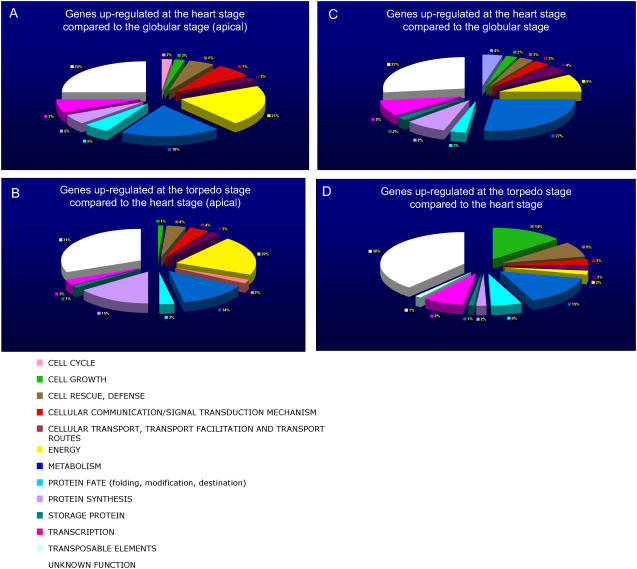
To assess changes in gene expression patterns on a functional level, the data were filtered by significance using a Student's t test with a maximum confidence level of 95% (P ≤ 0.05) for genes whose expression was significantly different from a value of 1. A total of 1,872 genes satisfied this criterion in at least one of the three developmental stages. Further filtering was accomplished by calculating a fold-change between the expression values at different developmental stages. Comparisons were made between globular and heart, and heart and torpedo stages. In each case, the 100 most up-regulated genes passing the significance filter were selected. We chose the 100 most up-regulated genes for illustrative purposes, but all our data are available at the NASCArray Web site (http://affymetrix.arabidopsis.info/) for further interrogation by the community. These genes were assigned functional annotation using information from http://mips.gsf.de/proj/thal/db/. Figure 6, A and B display the functional classifications of the 200 genes (100 in apical tissues, 100 in basal tissues) most up-regulated between developmental stages.
Figure 6.
Functional annotations of the 100 most up-regulated genes (passing the significance filter: P < 0.05 in at least one stage) between developmental stages on the apical developmental (A and B) and basal (C and D) time course, respectively. The color keys show the designated gene class annotations.
The data reveal during the transition from globular and heart stages the up-regulation of genes involved in energy production; for example, the photosystem, which comprise 21% of the up-regulated genes during that developmental phase. Other significant functional groups up-regulated are metabolism (19%), cellular communication/signal transduction (7%), and transcription (7% at heart stage). The transition from heart stage to torpedo stage is associated with the up-regulation of genes related to the production of energy (20% of the up-regulated genes), and, once again, these are heavily biased toward the photosystem. Also of note is that 15% of the up-regulated genes are involved in protein synthesis, perhaps reflecting a change in emphasis as the embryo progresses toward late embryogenesis. Metabolic genes also comprise 14% of the total. Through all the functional comparisons, those genes of unknown function represented between 22% and 31% of the total.
K-means clustal analysis was performed on the 1,872 genes satisfying the significance criteria, using Pearson correlation (GeneSpring version 7.2). Here, the user defines the maximum number of clusters formed; in this case, 10 cluster experiments are illustrated (Supplemental Fig. S11A). It was found that approximately seven distinct expression patterns are present within the sampled genes (Supplemental Fig. S11B).
An analysis was carried out to determine whether any of the clusters obtained were specifically enriched for particular families of predicted transcription factors or receptor kinases. A database of approximately 1,400 predicted transcription factors and receptor kinases (Davuluri et al., 2003; Shiu and Bleecker, 2003; http://Arabidopsis.med.ohio-state.edu/AtTFDB/) was used to probe the clusters. Due to the low numbers of gene family members of interest present in the filtered gene list, no valid statistical significance could be attributed to the numbers appearing in individual clusters. Despite this limitation, no individual cluster showed any notable enrichment for particular families of transcription factors or receptor kinases.
Basal Developmental Time Course
As with the apical time course, all the basal (root) samples were normalized together to a per-gene median value, and all genes on the GeneChip are graphically represented (Fig. 5B). The respective stages have distinct profiles, which are broadly similar to those observed for the apical region. However, in contrast, there appears to be a considerably more compact profile centered around the expression value range 1 to 3, with a reduced number of genes that have relatively extreme expression values. A similarity of expression profile is shown between the developmental stage datasets and between the overall apical and basal time course profiles.
As for the apical time course analysis, the data were filtered by significance using a Student's t test with a maximum confidence level of 95% (P ≤ 0.05) for genes whose expression was significantly different from a value of 1. For the basal developmental time course, 1,226 genes satisfied this criterion in at least one of the three developmental stages. Further filtering was again accomplished by calculating a fold-change between the expression values at different developmental stages.
Four main functional groups are up-regulated in the basal tissue between the globular and heart stages (Fig. 6, C and D): metabolism (27%), energy (9%), protein synthesis (8%), and transcription (8%). Over the heart stage to the torpedo stage transition, five main functional groups are up-regulated: metabolism (15%), cell growth (14%), cell rescue/disease (8%), transcription (8%), and protein fate (6%). As with the apical time course functional analysis, a change in pattern occurs between the heart and torpedo stages, possibly reflecting the approach of late embryogenesis. The fraction accounted for by genes with an unknown function is higher than that of the apical region, comprising between 27% and 38% of the total.
K-means clustal analysis was performed on the 1,226 genes satisfying the significance criteria. The 10-cluster analysis (Supplemental Fig. S12) found that approximately seven distinct expression patterns are present within the sampled genes, similar to the results for the apical tissue analysis. As was also found for apically expressed genes, no individual cluster showed any notable enrichment for particular families of transcription factors or receptor kinases.
Transcriptional Profiles of Apical versus Basal Domains
To study further the transcriptional profiles of apical and basal regions, we compared data for the apical and basal region at each stage along the developmental time course. In addition, a comparison was also made between GeneChip data for the cotyledon and root tissue of a 7-dpg seedling, produced by the AtGenExpress Consortium and provided by NASCArrays at http://www.affymetrix.arabidopsis.info. As well as analyzing the most differentially expressed genes between the regions of a particular developmental stage, we also identified the up-regulated genes of each region/developmental stage to assess the degree of overlap and therefore the degree to which these genes were specifically apical or basal throughout development.
Globular Stage Apical versus Basal Domain
The globular stage apical and basal domain tissue samples were normalized together to a control sample, which in this case was the basal sample. Therefore, genes in the apical sample had an expression value of either >1 (up-regulated), <1 (down-regulated), or 1 (identical expression in both tissue samples). The resulting data were then filtered by significance using a Student's t test with a maximum confidence level of 95% (P ≤ 0.05) for genes whose expression was significantly different from a value of 1. To correct for the occurrence of false positives, a Benjamini and Hochberg false discovery rate multiple testing corrections was used to adjust the P values (Benjamini and Hochberg, 1995; GeneSpring version 7.2).
A total of 585 genes satisfied these criteria and were sorted into those up-regulated and down-regulated in the apical region compared to the basal region. Further filtering was accomplished by calculating a fold-change between the expression values of the two regions for a particular gene. All genes showing significant up- or down-regulation (280 and 305 genes, respectively) in the apical versus basal regions passing the significance filter at P ≤ 0.05 are shown in Supplemental Tables S11 and S12. It is interesting to note that while the highest fold-change observed in the apical sample is approximately 37.5 times in the basal sample, the highest is for At1g04410, a cytosolic, malate dehydrogenase that is only 6.7 times more highly expressed in the basal sample compared to the apical sample. A range of functional groups is represented as differentially regulated, including transcription factors such as At2g21320, a CONSTANS B-box zinc finger family protein that is up-regulated by approximately 37-fold; and At4g16430, a βHLH protein that is up-regulated approximately 6-fold. Also of note in the apical sample are At5g42220, a ubiquitin family protein (with a predicted role in protein fate) up-regulated approximately 26-fold, and At5g10480 (PEPINO), an EMBRYO-DEFECTIVE (EMB) gene encoding a putative antiphosphatase, which is up-regulated approximately 6-fold.
Heart Stage Cotyledon versus Root
As previously done, heart stage cotyledon and root samples were normalized together using the root sample as the control and filtered. A total of 532 genes satisfied these filtering criteria and were sorted into those up-regulated and down-regulated in the cotyledon compared to the root (or vice versa for the root compared to the cotyledon).
All genes showing significant up- or down-regulation (345 and 187, genes respectively) in the cotyledon and root passing the significance filter (P ≤ 0.05) are shown in Supplemental Tables S12 and S14. The cotyledon gene sample is enriched with putative transcriptional regulators, with three out of the 10 most up-regulated falling in this class. However, of these, only At4g37750 (AINTEGUMENTA), which is approximately 35-fold up-regulated in the cotyledon sample, displays a high fold-change. Of the others, At4g02840, a small nuclear riboprotein, and At4g07950, a DNA-directed RNA polymerase, are up-regulated only approximately 3-fold. In contrast, the root gene samples display notably high fold-changes. In this tissue, metabolic genes show the three highest fold-changes, from approximately 30-fold up to the approximately 44-fold increase displayed by At5g01870, a putative lipid transfer protein. The root sample also includes one putative transcriptional regulator in the 10 most up-regulated genes, At1g32790, a predicted RNA-binding protein that is up-regulated approximately 8-fold in the root compared to the cotyledon.
Torpedo Stage Cotyledon versus Root
The torpedo stage cotyledon and root samples were normalized and filtered, as previously. A total of 1,834 genes satisfied these criteria and were sorted into those up-regulated and down-regulated in the cotyledon compared to the root. All 1,834 genes showing significant up- and down-regulation in the cotyledon versus root passing the significance filter (P ≤ 0.05) are shown in Supplemental Tables S15 and S16. Among the genes most up-regulated in the cotyledonary tissues are a number of transcriptional regulators and signaling components, as well as the expected storage components typical of this stage of embryogenesis.
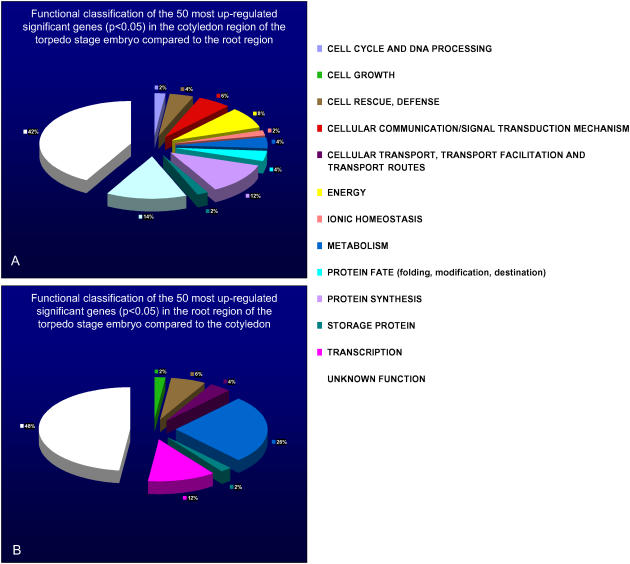
For illustrative purposes, the 100 most up-regulated genes in the cotyledon and root (50 genes for each tissue) passing the significance filter were assigned functional annotation (http://mips.gsf.de/proj/thal/db/; Fig. 7). In general, the cotyledon sample has a larger range of functional groups represented compared to the root sample. The most enriched groups in the cotyledon sample are transcription (14%) and protein synthesis (12%); additional groups enriched at a lower level include energy (8%) and signal transduction (6%). In contrast, only two main groups are enriched in the root sample, namely metabolism (26%) and transcription (12%). Cell rescue and defense response genes also comprise 6% of the root sample and 4% of the cotyledon sample. In both samples, the largest group is that of unknown function, which comprises 42% of the cotyledon sample and 48% of the root sample.
Figure 7.
Functional annotation of the 50 most differentially expressed (by fold-change) significant genes (P < 0.05) between the cotyledon and root regions of the torpedo stage embryo, respectively. A, Genes relatively highly expressed in the cotyledon. B, Genes relatively highly expressed in the root. The color key shows the designated gene class annotations.
Globular Stage Apical Domain versus Torpedo Stage SAM
In addition to comparisons between the apical and basal regions of the different stages of embryogenesis, we compared the apical region of the globular stage embryo to the SAM region of the torpedo stage. The reasoning behind this comparison is that in addition to being the region from which the cotyledons are eventually derived, it has also been shown that, despite no morphologically recognizable structure, expression of essential SAM genes is centered in this region (Barton and Poethig, 1993; Long et al., 1996; Mayer et al., 1998). The clonal destiny of this region to form the SAM is also predicted to be determined in the early globular stage, therefore making this a valid comparison between a presumptive SAM at the globular stage and its more developed form at the torpedo stage (Christianson, 1986; Poethig et al., 1986).
The globular stage apical region and the torpedo stage SAM samples were normalized and filtered. A total of 921 genes satisfied these criteria and were sorted into those up-regulated and down-regulated in the torpedo stage SAM compared to the globular stage apical region (or vice versa). All genes showing significant up- and down-regulation genes in the torpedo stage SAM versus the globular stage apical region passing the significance filter (P ≤ 0.05) are shown in Supplemental Tables S17 and S18. In the torpedo stage meristem sample, the meristem transcription factor At1g622360 (STM) is up-regulated approximately 21-fold. Two out of the 10 most expressed genes in the torpedo stage meristem sample are cytoskeleton protein genes: At1g04820 (TUA4), a tubulin α-2/α-4 chain, and At2g35630 (MICROTUBULE ORGANISATION PROTEIN 1 [MOR1]), both of which are up-regulated more than 5-fold in the torpedo stage SAM sample.
Seedling Cotyledon versus Root
RNA used for the analysis of seedling tissues was produced by the AtGenExpress Consortium (see “Materials and Methods”) and had not been amplified. The seedling cotyledon and root sample data (7 dpg) were normalized and filtered. A total of 13,357 genes satisfied these criteria and were sorted into those up-regulated and down-regulated in the cotyledon compared to the root. All genes showing up- and down-regulation in the cotyledon versus root passing the significance filter (P ≤ 0.05) are shown in Supplemental Tables S19 and S20. In the cotyledon, three major functional classes are represented in the 10 most up-regulated genes, namely chloroplastic metabolism/biosynthesis, cell rescue and defense response, and energy. In the root sample, only two functional classes are represented in the 10 most up-regulated genes, i.e. cell rescue and defense response, and metabolism. All the genes in these samples are backed by very high fold-changes. A number of transcription factors are found to be differentially regulated.
Expression Patterns of Known EMB Genes
Genes that under normal conditions are required for viability, and when disrupted cannot be passed on to subsequent generations, can be considered essential. The precise number of such genes expressed during embryogenesis has not yet been established, but it is estimated that 500 to 1,000 genes in Arabidopsis produce an emb phenotype when mutated (Franzmann et al., 1995; McElver et al., 2001). Tzafrir et al. (2004) have described a collection of 220 EMB genes required for normal embryo development. The embryonic expression patterns for many of the genes in this collection have not been characterized, and so we analyzed the GeneChip data to determine temporal and spatial patterning of these transcripts.
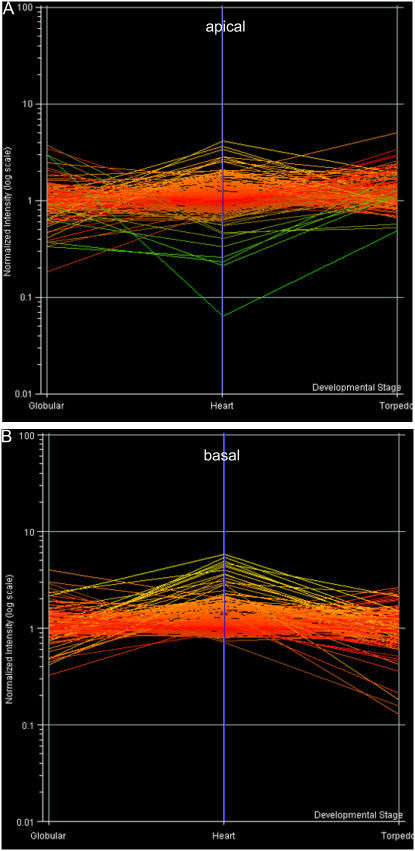
Applying the two arbitrary signal cutoff values as before, it was found that approximately 84% of the EMB genes were expressed in at least one of the three tissue types at a signal threshold of 75, rising to almost 96% at a signal threshold of 40. An analysis into the spatial expression of these genes (Fig. 8) revealed that approximately 76% were present in all tissue types at the lower signal threshold. The 4% of EMB genes deemed not to be expressed at the lower signal threshold could potentially be expressed in the hypocotyl region, which was not sampled or at another stage of embryogenesis. In support of this possibility, comparison with GeneChip data presented by Casson et al. (2005) showed that two of the nonexpressed genes were present at earlier stages of embryogenesis, At4g21130 at the globular stage and At2g45690 at both the globular and heart stages, albeit at threshold levels.
Figure 8.
Transcriptional changes of 222 EMB genes across a developmental time course (globular, heart, and torpedo stage embryos) in apical (A) and basal (B) domains. All samples were normalized together to a per-gene median value, indicated in red. Genes expressed to levels higher than the median values are indicated in yellow, while genes expressed to lower levels are indicated in green.
It was possible to identify EMB genes with differential expression patterns between tissue types (Supplemental Table S21). Significant examples include At2g34650 (PID), which is approximately 15-fold more abundant in the cotyledons than the root, and At1g62360 (STM), which is approximately 62-fold more abundant in the SAM than in cotyledonary tissue.
Embryonically Active Gene Promoters
The modification of seed development using genetic engineering, such as for the manipulation of embryonic storage product accumulation, requires gene promoters that are active in seed tissues to drive the transcription of transgenes. Such promoters are also useful as cell type markers for developmental studies. To characterize the activity promoters associated with genes identified as being differentially regulated on the basis of the GeneChip data, four promoter-GUS constructs were created and introduced into Arabidopsis plants by Agrobacterium tumefaciens-mediated transformation.
Genes selected for promoter∷GUS analysis are shown in Table I. The expression patterns based on the GeneChip data during embryogenesis are also shown. Following growth in soil, siliques were removed from plants and analyzed for GUS expression by histology.
Table I.
Genes selected for promoter-GUS analysis
The mean expression determined by LCM and GeneChip analysis is shown.
| Gene | Description | Globular Apical | Globular Basal | Heart Cotyledon | Heart Root | Torpedo Cotyledon | Torpedo Root |
|---|---|---|---|---|---|---|---|
| At5g45600 | Putative YEATS domain/transcriptional activator | 481.8 | 413.3 | 244.8 | 2,952.4 | 518.8a | 455 |
| At2g31510 | Putative RING zinc finger protein | 91.4 | 60.6 | 73.4 | 834.6a | 63.7 | 65.7 |
| At5g14610 | DRH1 DEAD box protein-like protein | 331.9a | 403.7a | 2,710.6 | 395.7a | 799.8 | 259.4 |
| At5g50810 | Small zinc finger-like protein | 682 | 266.5 | 165.1 | 504.1 | 798.3a | 175.4 |
Indicates an abnormally high mean signal value due to a large signal variation in one replicate.
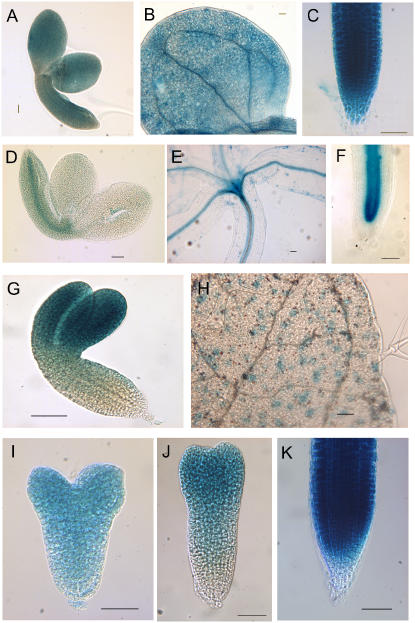
GeneChip data for At5g45600, encoding a predicted YEATS domain transcriptional activator protein, indicate that it is expressed relatively strongly in both apical and basal domains of globular, heart, and torpedo stage embryos (Table I). Figure 9, A to C shows promoter-GUS activity for this gene, which is also expressed throughout the cotyledonary stage embryo (Fig. 9A) and the cotyledon (Fig. 9B) and root tip (Fig. 9C) of the seedling. At2g31510, encoding a predicted RING zinc finger protein, shows relatively low levels of transcript abundance in globular embryos, with higher expression in the heart stage root (Table I). GUS activity from the promoter is observed most strongly in the root provascular strand during embryogenesis (Fig. 9D) and in the vascular tissues of aerial parts and roots in seedlings (Fig. 9, E and F). For At5g14610, encoding a DRH1 DEAD box protein-like protein, the GeneChip data suggest expression is found in both apical and basal tissues throughout the embryonic stages, with strongest expression in the cotyledons of heart stage embryos. Promoter-GUS activity was most strongly found in the cotyledons and hypocotyl of the cotyledonary stage embryo (Fig. 9G), with occasional diffuse staining observed in the root. In the seedling, expression appears to be restricted to the stomatal guard cells of the cotyledons and leaf (Fig. 9H). Finally, At5g50810, encoding a predicted small zinc finger-like protein, is also expressed in all embryonic tissues studied (Table I), and promoter-GUS analysis showed a changing pattern of GUS staining during embryogenesis; constitutive staining was observed at the heart stage of embryogenesis (Fig. 9I), but expression was then lost in the root as the embryo entered the torpedo stage (Fig. 9J). Postembryonically, expression is seen most strongly in the seedling root tip (Fig. 9K). These results therefore show a good correlation between the GeneChip analyses and the promoter-GUS fusion studies.
Figure 9.
Promoter-GUS fusion activities for genes identified as either constitutively expressed in embryos (A–C) or preferentially expressed in either basal (D–F), apical (G and H) domains, or changing pattern during development (I–K). A to C, At5g45600. GUS activity in cotyledonary stage embryo (A), cotyledon at 3 dpg (B), and primary root tip at 3 dpg (C). D to F, At2g31510. GUS activity in cotyledonary stage embryo (D), hypocotyl and young shoot at 3 dpg (E), and primary root tip at 3 dpg (F). G and H, At5g14610. GUS activity in cotyledonary stage embryo (G) and leaf stomatal guard cells (H). I to K, At5g50810. GUS activity in heart stage (I) and torpedo stage (J) embryo, and primary root tip at 3 dpg (K).
DISCUSSION
Microarray analysis is a very powerful technique allowing the expression profiles of thousands of genes to be monitored simultaneously. In combination with LCM, we show it is possible to gain a high resolution picture of global gene transcription during the development of the Arabidopsis embryo. In this article, we use a bioinformatics approach to characterize gene expression changes in different tissue domains across a developmental time course from the globular to torpedo stage embryo.
Gene Expression in the Torpedo Stage Embryo
The imposition of an Affymetrix MAS 5.0 signal cutoff value has been employed in two recent studies as a means to providing an estimation of the number of genes expressed in a tissue of interest: roots (Birnbaum et al., 2003) and early embryos (Casson et al., 2005). Using a number of genes with previously documented expression patterns, Birnbaum et al. (2003) calculated a signal cutoff value that represented a minimum value to confer presence over the level of background noise, and this was set at 75. Using this cutoff, they estimated that 10,492 genes were expressed in the root, and this corresponds to approximately 46% of the genes represented on the ATH1 GeneChip. Casson et al. (2005) also applied this signal cutoff value to GeneChip data obtained from the apical and basal regions of globular and heart stage embryos, finding between 8,027 and 10,591 genes (36%–47%) to be expressed. Using the cutoff value of 75 for the mean value of the replicates analyzed, we find that between 8,353 and 11,690 genes (37%–51%) are expressed in the three tissue types of the torpedo stage embryo analyzed. These data are in a comparable range to these previous studies and partially bridge the temporal gap between them, demonstrating that at this arbitrary cutoff value a similar number of expressed genes are estimated throughout embryogenesis and in the mature Arabidopsis root. Using a lower signal cutoff value of 40, Casson et al. (2005) predicted that up to 65% of genes are expressed. This compares with a predicted range of 59% to 77% expressed genes in the three tissue regions analyzed at this cutoff value in the torpedo stage embryo, indicating that there is no major decline in total transcript abundance at this late stage of embryogenesis.
Although RNA amplification causes a shortening of the products, we avoided potential bias between samples by using 3′ probe pairs for microarray analysis, as previously discussed (Casson et al., 2005), and treating all tissue samples identically, so that data (e.g. in terms of fold-changes in gene expression) between tissues should be relatively comparable. The P values may be affected by the use of reduced probe pairs, but we treated all samples in the same way following RNA amplification, so the data should be comparable. Nakazono et al. (2003) compared T7-aRNA from laser microdissected material from maize (Zea mays) coleoptiles with a comparable amount (40 ng) of non-aRNA using cDNA microarray and found a highly linear relationship, demonstrating reproducibility among samples. These studies only assessed changes to the expression profile after two rounds of amplification rather than the three used here. However, the results of Scheidl et al. (2002) suggested that further rounds of amplification would produce no significant increase in variability. We would, however, emphasize that the data for any given gene should not be taken at face value but should be checked by further experimentation.
Embryonic Developmental Stages Show Distinct Transcriptional Profiles
To test the hypothesis that each developmental stage under investigation has a distinct transcriptional profile, a condition tree clustering analysis was performed. All tissue types sampled from the same developmental stage clustered together, as opposed to all the apical regions clustering separately from the basal regions. This demonstrates that, in terms of overall transcriptional profile, there appears to be a greater input from the temporal expression patterns than the spatial expression patterns. These data fit with an established model based on RNA hybridization studies in tobacco (Nicotiana tabacum), which suggest that while there are significant populations of organ-specific transcripts, 60% to 77% of plant genes are expressed in heterologous organs (Goldberg, 1988). Goldberg et al. (1989) demonstrated that distinct mRNA sets are temporally regulated during embryogenesis, with expression restricted to specific developmental stages.
Scheidl et al. (2002) have suggested that any difference in expression profile observed between amplified and nonamplified samples was the consequence of a global reduction in transcript length resulting from priming with random hexamers. It might therefore be expected that the embryonic samples, which underwent amplification, would cluster separately from the seedling samples. However, the condition tree does not show this distinction; instead, the torpedo stage samples cluster with the seedling samples. Given that all the embryonic samples underwent an identical amplification procedure, this result suggests a distinction in transcriptional profile between early and mid/late embryogenesis rather than any technical bias.
The torpedo stage is characterized by maturation and the accumulation of storage reserves in preparation for developmental arrest (Lindsey and Topping, 1993). Given that the plant body pattern is already established and that the seedling continues to undergo a maturation process, it would be expected to have a transcriptional profile more similar to the torpedo stage than the early embryonic stages, as confirmed by the condition tree analysis.
Transcriptional Changes along an Embryonic Developmental Time Course
The time course data (Fig. 5) show the majority of genes to be expressed at the same level throughout the developmental stages, centered on the default expression value of 1. However, each developmental stage is represented by a distinct transcriptional profile in both the apical and basal region, as highlighted by the condition tree analysis. Although the absolute level of transcription does not in itself indicate functional importance, because of the possibility of posttranscriptional regulation for a given gene product, our data do demonstrate clear transcriptional changes during embryogenesis.
Functional annotation of genes that show up-regulation along the developmental time course provides new information but must be considered with caution, taking into account statistical significance and confidence of annotation. In order not to be prohibitively restrictive, the significance filter was relaxed with a 95% confidence required at only one of the three developmental stages. This allows a general overview of the time course but has the potential to allow nonstatistically significant results. Second, only approximately 10% of genes in the Arabidopsis genome (approximately 2,500) have had their function deduced or confirmed by direct experimental analysis; therefore, the vast majority of functions are putative and assigned on the basis of sequence similarity (The Arabidopsis Genome Initiative, 2000; Martinoia et al., 2002; Hilson et al., 2003).
Along the apical time course, genes with a functional role in energy, predominantly those involved in photosynthesis and carbon fixation, were up-regulated at every stage. This is in accordance with in situ hybridization studies that show an increasing abundance of chloroplastic gene transcripts progressively from the proembryo stage of embryogenesis, with the highest concentration observed in the cotyledons of the mature embryo (Degenhardt et al., 1991). The peak in transcript abundance in the mature embryo corresponds with the fate of the cotyledons as the initial photosynthetic organs of the seedling and allows photosynthesis to commence promptly postgermination (Degenhardt et al., 1991; Raghavan, 1997). Chloroplastic gene transcripts are also present as up-regulated in the basal time course, which corresponds with the constitutive pattern of expression observed in developing embryos of Gossypium hirsutum (Borroto and Dure, 1986), Glycine max (Chang and Walling, 1991, 1992), and Arabidopsis (Degenhardt et al., 1991).
Another significant change in the apical time course is the up-regulation of genes involved in protein synthesis between the heart stage and the torpedo stage. This could be representative of the transition from early embryogenesis to the maturation and protein accumulation characteristic of mid/late embryogenesis (Lindsey and Topping, 1993).
Along the basal time course, the transition from heart to torpedo stage is accompanied by the significant up-regulation of genes encoding proteins involved in cell growth, specifically the growth of cell walls. This group included a substantial number of Hyp-rich glycoproteins, including expansins, which are regarded as key regulators of wall extension and cell expansion (Cosgrove, 2000; Li et al., 2003). The up-regulation of this group of genes may reflect the elongation that the embryo undergoes between the heart stage and the torpedo stage. Extensins have also been implicated in desiccation tolerance, and the up-regulation at the torpedo stage of embryogenesis may also represent a stage in the preparation for developmental arrest and desiccation (Jones and McQueen-Mason, 2004).
K-means clustering was performed on the filtered apical and basal time course gene sets. In both cases, seven distinct dynamic expression patterns were observed along the time course. These represent all but one of the eight possible patterns of change, with the missing pattern for apical samples being a reduction in expression from globular to heart stage followed by unchanged expression from heart to torpedo stage (Supplemental Fig. S2B), and for the basal samples, the continued reduction in expression from globular to heart to torpedo stage (Supplemental Fig. S3B). The clustering program used assigned genes to clusters based on a user-defined cluster number and did not create these clusters if the number was defined as seven. Hennig et al. (2004) predicted nine models of dynamic expression pattern for genes involved in reproductive development of Arabidopsis and used an alternative clustering package to assign their selected genes to the model class of best fit. This would appear to be an improvement in terms of selecting genes, as it would not create multiple clusters of genes showing essentially similar expression profiles. The number of time points available is also of critical importance in cluster analysis; the three included here are the minimum and, as can be seen, produce clusters with considerable associated noise. Beemster et al. (2005) conducted cluster analysis on significantly modulated genes during leaf development from 9 to 31 d after sowing. Ten time points were utilized, and 16 very well defined clusters of 20 or more genes were produced. The acquisition and inclusion of a further time point for the cotyledonary stage of development would be predicted to greatly enhance the fidelity of clustering.
Apical-Basal Tissue Comparisons
GeneSpring analysis was used to uncover statistically significant genes, which show differential expression between the apical (cotyledon) and basal (root) samples along the developmental time course as a resource for future analysis. An additional aim was to deduce whether some of these apical and basal genes represented an organ-specific gene set throughout embryogenesis.
Casson et al. (2005) presented an analysis of spatially expressed putative transcription factors at the globular and heart stages of embryogenesis. They identified a gene (At2g21320) encoding a CONSTANS-like B-box zinc finger protein, expressed predominantly in the apical region. This gene emerged from our GeneSpring analysis of the Casson et al. (2005) data as the most differentially expressed significant gene in the globular apical region, providing a level of validation to the analysis. A number of other putative transcription factors emerged from this analysis, which could be investigated further.
As a nonembryonic comparison, data from 7-dpg seedling cotyledons and roots were analyzed. Compared to the embryonic samples, the seedling analysis produced extremely high fold-changes between the tissue samples. This suggests that differential spatial gene expression is more pronounced in the seedling than the embryo.
A comparison was also made between the apical region of the globular stage embryo and the SAM region of the torpedo stage embryo. The presence of STM in the up-regulated genes of the SAM indicates that the SAM was successfully captured. Caution is required when analyzing this data, as the initial LCM step was not precise enough to achieve specific capture of the SAM, and therefore some degree of contamination from the surrounding tissue is expected. Interestingly, two known cytoskeletal genes were up-regulated in the SAM compared to the globular stage apical region, namely TUA4 (Kopczak et al., 1992) and MOR1 (Whittington et al., 2001). The Affymetrix MAS 5.0 mean signal values indicate that TUA4 is constitutively expressed throughout the torpedo stage embryo (mean signal values between 193 and 295) but is probably not expressed in the globular stage apical region (mean signal value 24), thus accounting for the high fold-change observed. MOR1 appears to be expressed in the globular stage apical region (mean signal value 119) and at a similar level in the torpedo stage cotyledons and root (mean signal values 142 and 151) but appears much more abundant in the SAM (mean signal value 802). The expression across tissues and developmental stages is consistent with the conclusion by Whittington et al. (2001) that MOR1 is required at all stages of development and in all organs.
Functional analysis of the 100 most differentially expressed genes in the cotyledon versus root provides a very different perspective to the temporal function analysis conducted for the apical and basal time courses. Basing predicted function for unknown genes on sequence similarity, approximately 30% of the genome remains without putative functional classification (The Arabidopsis Genome Initiative, 2000). Strikingly, between 42% and 48% of the most up-regulated genes uncovered have no predicted function, indicating a possible preferential role for such genes during embryogenesis, though given the relatively small number of genes included in this analysis, this may not be statistically significant.
The range of functions represented in the up-regulated root genes is limited compared to those of the cotyledon. The most significant functional group represented in the root is that of metabolism, and this correlates well with the analysis performed by Yamada et al. (2003) on mature root tissue, which showed metabolism to be the largest functional group, comprising approximately 13% (plus 5.5% designated as protein metabolism) of the 549 root-specific transcripts identified. The cotyledon shows up-regulation of a wide range of functional groups, including energy and protein synthesis, correlating with the increase in chloroplastic gene transcripts to reach a peak in the cotyledons of the mature embryo (Degenhardt et al., 1991). A number of the genes functionally classified as protein synthesis are chloroplast and plastid ribosomal protein precursors. Knockout mutants of plastid ribosomal proteins (S21 and L11) have been shown to impair photosynthesis and thus show an overlap with the up-regulation of energy function (Pesaresi et al., 2001; Morita-Yamamuro et al., 2004).
A significant number of putative transcription factors are present in both the cotyledon and root gene lists, providing potential targets for further analysis into spatial control mechanisms.
Analysis of the overlap between significant apical and basal genes at each developmental stage did not reveal the existence of a population of genes with a distinct spatial expression pattern throughout development. Expression pattern analysis of genes such as PIN4 show that defined spatial expression along the embryonic time course does exist (Friml et al., 2002). The analysis did show a significant overlap between the torpedo stage of embryogenesis and the seedling, not just in root- and cotyledon-specific transcripts, but also in transcripts changing their spatial localization. This may reflect a more consistent population of genes expressed in the late embryo, whose maturation processes continue into the seedling, in contrast to early embryogenesis, where more dynamic sets of genes may be required for pattern formation and morphogenesis. The existence of genes with changing spatial localization through embryogenesis, and into the seedling, is highlighted by the small zinc finger-like protein (At5g50810) analyzed in this work, which changes from being predominantly localized in the cotyledons of the torpedo stage embryo to being predominantly localized in the root of the seedling.
EMB Gene Expression
Of the collection of 220 EMB genes described by Tzafrir et al. (2004), our results indicate that approximately 96% are expressed in at least one of the embryonic tissues sampled when a signal value cutoff of 40 is imposed, and the remaining 4% are either expressed at a different stage of embryogenesis (Casson et al., 2005) or are predicted to be expressed in a tissue region not analyzed in this study (e.g. the hypocotyl). This percentage of EMB genes predicted to be expressed at each signal cutoff value is higher for the torpedo stage than that observed for the globular and heart stage data (Casson et al., 2005). It is possible that several EMB genes are important at later stages of embryogenesis, a view supported by the observed greater number of terminal phenotypes observed at later (cotyledonary) stages of embryogenesis than very early (preglobular) stages (Tzafrir et al., 2004).
Many of the EMB genes appear to be of low abundance, as indicated by low signal values in the GeneChip data, and so care must be taken when assessing some of the fold-changes observed between different embryonic regions. Nevertheless, a number of EMB genes show distinct spatial patterns of differential expression in the torpedo stage embryo. For example, the Ser-Thr protein kinase PINOID is highly up-regulated (15-fold more abundant) in the cotyledon compared to the root correlating with the defective cotyledon morphology observed in the pinoid mutant (Christensen et al., 2000). In the root and SAM, two members of the TITAN (TTN) family, TTN6 and TTN9, show up-regulation compared to the cotyledons (Liu and Meinke, 1998). The ttn9 mutant shows developmental arrest at a very early stage, reaching a maximum of four cells (Liu et al., 2002; Tzafrir et al., 2002). TTN9 appears to be expressed in both the root and SAM region but not in the cotyledons. TTN6 encodes a deubiquitinating enzyme and was shown to be more abundant in the root compared to the cotyledon, where there is likely no expression. Embryonic cells in the ttn6 mutant are rounded and disorganized, culminating in developmental arrest at the globular stage, in addition to a disruption of endosperm cellularization (Doelling et al., 2001; Tzafrir et al., 2002). TTN6 did not emerge as significantly spatially distributed in the analysis of EMB gene differential expression conducted by Casson et al. (2005) on globular and heart stage embryos. In light of the developmental arrest observed at the globular stage of embryogenesis in ttn6 mutants, this may suggest that constitutive expression is required at these critical early stages of embryogenesis with a contrasting spatial expression pattern required for functions during late embryogenesis.
CONCLUSION
Bioinformatics tools provide a powerful approach to identify changing patterns of gene expression during development. In combination with relatively new tools such as LCM, a new level of resolution can be achieved. In turn, this allows the identification of specific classes of genes for further study. It must be remembered that this analysis represents a starting point for detailed functional studies, and further experimental research is required to expand on the findings obtained to define the molecular mechanisms underpinning the cellular patterning and biochemical differentiation of the plant embryo and the complex networks of interactions involved.
MATERIALS AND METHODS
Sample Preparation and LCM
Embryonic tissues at globular, heart, and torpedo stage were embedded in OCT embedding medium (RA Lamb), frozen, and cryosectioned, as described (Casson et al., 2005). LCM was performed using a PixCell II system using CapSureHS LCM caps (Arcturus), as described (Casson et al., 2005). For globular, heart stage, and the SAM of torpedo stage embryos, three independent biological replicates were sampled for RNA amplification and analysis. For cotyledon and root tissues of torpedo stage embryos, six independent biological replicates were used.
RNA Extraction and RNA Amplification
RNA from LCM cells was extracted using the Absolutely RNA Nanoprep kit (Stratagene) and amplified using the MessageAmp aRNA kit (Ambion Europe), as described (Casson et al., 2005).
DNA Microarray Analysis
The Affymetrix GeneChip Arabidopsis ATH1 Genome Array (approximately 22,800 genes) was used for DNA microarray analysis, as described (Casson et al., 2005). The Arabidopsis Microarray and Bioinformatics service at GARNET performed probe labeling, hybridization, and analysis (Craigon et al., 2004). The raw GeneChip data were normalized using robust multiarray average, a log scale measurement of expression developed by Irizarry et al. (2003). All the samples were initially normalized together to a per-gene median value. Clustering analysis was then performed using condition tree clustering on all samples. Similarity was measured using Spearman correlation (GeneSpring version 7.2). K-means clustal analysis was performed on the 1,226 genes satisfying the significance criteria using Pearson correlation (GeneSpring version 7.2). GeneChip data for the cotyledon and root tissue of a 7-dpg seedling was produced by the AtGenExpress Consortium (Schmid et al., 2005) and provided by NASCArrays at http://www.affymetrix.arabidopsis.info. All our embryonic GeneChip data can also be accessed through this NASCArray Web site.
Gene Constructs and Plant Transformation
Approximately 2.5-kb genomic sequences were cloned upstream of the ATG codon of the genes of interest by PCR. The primer pairs used for promoter amplification are as follows: At5g45600, forward GTAGTGATGATACTCAAGCACACC, reverse, CTCGGCTTAACTTCAACAGATCTGCTTC; At2g31510, forward TTGGATCCCATGGAGTGCACGTTTCCTCTCG, reverse TTGGATCCGATCAGAGAAAACGAAATGGC; At5g14610, forward CCAACTGTCATAGGCATATAAGTCC, reverse CCTCAGGAGCGTAACGAATTGCAG; At5g50810, forward GCGGATTCTGCTTTTCCTTTAG, reverse GCAATTCCGGGTTGTTTGCC. Each promoter fragment was initially cloned into the TOPO vector and then transferred to the binary vector pΔGUS-CIRCE, which contains the GUS reporter gene, for transformation into plants. Plant transformation was carried out using the floral dip method of Clough and Bent (1998), using Agrobacterium tumefaciens strain C58C1 (Koncz and Schell, 1986). Tissue localization of GUS enzyme activity was performed as described (Topping and Lindsey, 1997). To stain embryos, siliques were dissected open using a fine-pointed needle and each seed coat pierced individually. Penetration of the seed by the histochemical buffer and substrate was achieved by subjecting dissected siliques to a 20-min vacuum infiltration prior to incubation overnight at 37°C. All constructs were validated by sequencing.
Microscopy
Tissues were cleared and mounted for light microscopy in chloral hydrate (Topping and Lindsey, 1997) or 20% glycerol. Photographs were taken using a CoolSNAP compare with digital camera (Photometrics, Roper Scientific) with Openlab 3.1.1 software (Improvision) on Leica MZ125 (Leica Microsystems), Olympus SZH10 (Olympus), or Zeiss Axioskop (Carl Zeiss) microscopes. Images were processed in Adobe Photoshop 5.0.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Agilent Bioanalyser scans of aRNA from embryonic tissues.
Supplemental Figure S2. Cluster analysis of synexpressed genes during development of the apical domain of the embryo.
Supplemental Figure S3. Cluster analysis of synexpressed genes during development of the basal domain of the embryo.
Supplemental Tables S1 to S9. Signal values for all replicate ATH1 GeneChip data, from globular stage, heart stage, and torpedo stage embryo tissues and seedling tissues, showing means and sds of the replicate data based on the use of eight probe pairs.
Supplemental Table S10. Estimate of the number of genes expressed based on a signal value cutoff based on the use of eight probe pairs.
Supplemental Table S11. Differential expression of genes in the apical and basal region of the globular stage embryo (1).
Supplemental Table S12. Differential expression of genes in the apical and basal region of the globular stage embryo (2).
Supplemental Table S13. Differential expression of genes in the apical (cotyledon) region compared to the basal (root) region in the heart stage embryo (1).
Supplemental Table S14. Differential expression of genes in the apical and basal region of the heart stage embryo (2).
Supplemental Tables S15. Differential gene expression in the cotyledon region compared to the root region in the torpedo stage embryo (1).
Supplemental Table S16. Differential gene expression in the cotyledon region compared to the root region in the torpedo stage embryo (2).
Supplemental Table S17. Differential gene expression in the SAM of the torpedo stage embryo compared to the apical region of the globular stage embryo (1).
Supplemental Table S18. Differential gene expression in the SAM of the torpedo stage embryo compared to the apical region of the globular stage embryo (2).
Supplemental Table S19. Differential gene expression in the 7-dpg seedling (1).
Supplemental Table S20. Differential gene expression in the 7-dpg seedling (2).
Supplemental Table S21. EMB genes determined to show spatial differential expression during the torpedo stage of embryogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Sean May of the Nottingham Arabidopsis Seed Centre for assistance with microarray analysis.
This work was supported by the Biotechnology and Biological Sciences Research Council (funding to K.L.; a BBSRC Cooperative Awards in Science and Engineering studentship in collaboration with Syngenta to M.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Keith Lindsey (keith.lindsey@durham.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ausubel F, Benfey P (2002) Arabidopsis functional genomics. Plant Physiol 129 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton KM, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831 [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 132 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57 289–300 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bonner RF, Emmert-Buck MR, Cole K, Pohida T, Chuaqui RF, Goldstein SR, Liotta LA (1997) Cell sampling-laser capture microdissection: molecular analysis of tissue. Science 278 1481–1483 [DOI] [PubMed] [Google Scholar]
- Borroto KE, Dure L III (1986) The expression of chloroplast genes during cotton embryogenesis. Plant Mol Biol 7 105–113 [DOI] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- Chang YC, Walling LL (1991) Abscisic acid negatively regulates expression of chlorophyll a/b binding protein genes during soybean development. Plant Physiol 97 1260–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Walling LL (1992) Chlorophyll a/b binding protein genes are differentially expressed during soybean development. Plant Mol Biol 19 217–230 [DOI] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478 [DOI] [PubMed] [Google Scholar]
- Christianson ML (1986) Fate map of the organizing shoot apex in Gossypium. Am J Bot 73 947–958 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407 321–326 [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's Transcriptomics Service. Nucleic Acids Res 32 D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Fiebig C, Link G (1991) Chloroplast and nuclear transcripts for plastid proteins in Arabidopsis thaliana: tissue distribution in mature plants and during seedling development and embryogenesis. Bot Acta 104 455–463 [Google Scholar]
- Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD (2001) The ubiquitin-specific protease UBP14 is essential for early embryo developmental in Arabidopsis thaliana. Plant J 27 393–405 [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang ZP, Goldstein SR, Weiss RA, Liotta LA (1996) Laser capture microdissection. Science 274 998–1001 [DOI] [PubMed] [Google Scholar]
- Franzmann LH, Yoon ES, Meinke DW (1995) Saturating the genetic map of Arabidopsis thaliana with embryonic mutations. Plant J 7 341–350 [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al (2002) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673 [DOI] [PubMed] [Google Scholar]
- Gillespie JW, Best CJM, Bischel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, et al (2002) Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol 160 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB (1988) Plants: novel developmental processes. Science 240 1460–1467 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L (1989) Regulation of gene expression during plant embryogenesis. Cell 56 149–160 [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Small I, Kuiper MTR (2003) European consortia building integrated resources for Arabidopsis functional genomics. Curr Opin Plant Biol 6 426–429 [DOI] [PubMed] [Google Scholar]
- Jones L, McQueen-Mason S (2004) A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett 559 61–65 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP (1992) The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell 4 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Wurschum T, Breuninger H (2004) Genetic regulation of embryonic pattern formation. Plant Cell 16 S190–S202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6 603–610 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Topping JF (1993) Embryogenesis: a question of pattern. J Exp Bot 44 359–374 [Google Scholar]
- Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AAM, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29 405–415 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16 21–31 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kokukisaoglu Ü, Müller-Röber B, Schulz B (2002) Multifunctionality of plant ABC transporters more than just detoxifiers. Planta 214 345–355 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815 [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G (1991) Mutations affecting body organization in the Arabidopsis embryo. Nature 353 402–407 [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Sussex IM (1979) Embryo-lethal mutants of Arabidopsis thaliana: a model system for genetic analysis of plant embryo development. Dev Biol 72 50–61 [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Tanaka A, Yamaguchi J (2004) Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol 45 781–788 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qui F, Borsuk LA, Schnable PA (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D (2001) Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J 27 179–189 [DOI] [PubMed] [Google Scholar]
- Poethig RS, Coe EH, Johri MM (1986) Cell lineage patterns in maize embryogenesis: a clonal analysis. Dev Biol 117 392–404 [Google Scholar]
- Raghavan V (1997) Molecular Embryology of Flowering Plants. Cambridge University Press, Cambridge, UK
- Scheidl SJ, Nilsson S, Kalén M, Hellström M, Takemoto M, Håkansson J, Lindahl P (2002) mRNA expression profiling of laser microbeam microdissected cells from slender embryonic structures. Am J Pathol 160 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P (1994) Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120 2475–2487 [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA (1998) Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet 14 272–276 [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Topping JF, Agyeman F, Henricot B, Lindsey K (1994) Identification of molecular markers of embryogenesis in Arabidopsis thaliana by promoter trapping. Plant J 5 895–903 [DOI] [PubMed] [Google Scholar]
- Topping JF, Lindsey K (1997) Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, McElver JA, Liu CM, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke D (2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128 38–51 [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickermann A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington AT, Vugrek O, Wei KJ, Hasenbein NG, Sugimoto K, Rashbrooke MC, Wasteneys GO (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411 610–613 [DOI] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.