Abstract
The basal defenses important in curtailing the development of the phloem-feeding silverleaf whitefly (Bemisia tabaci type B; SLWF) on Arabidopsis (Arabidopsis thaliana) were investigated. Sentinel defense gene RNAs were monitored in SLWF-infested and control plants. Salicylic acid (SA)-responsive gene transcripts accumulated locally (PR1, BGL2, PR5, SID2, EDS5, PAD4) and systemically (PR1, BGL2, PR5) during SLWF nymph feeding. In contrast, jasmonic acid (JA)- and ethylene-dependent RNAs (PDF1.2, VSP1, HEL, THI2.1, FAD3, ERS1, ERF1) were repressed or not modulated in SLWF-infested leaves. To test for a role of SA and JA pathways in basal defense, SLWF development on mutant and transgenic lines that constitutively activate or impair defense pathways was determined. By monitoring the percentage of SLWF nymphs in each instar, we show that mutants that activate SA defenses (cim10) or impair JA defenses (coi1) accelerated SLWF nymphal development. Reciprocally, mutants that activate JA defenses (cev1) or impair SA defenses (npr1, NahG) slowed SLWF nymphal development. Furthermore, when npr1 plants, which do not activate downstream SA defenses, were treated with methyl jasmonate, a dramatic delay in nymph development was observed. Collectively, these results showed that SLWF-repressed, JA-regulated defenses were associated with basal defense to the SLWF.
Plants defend themselves from pathogens and herbivores using constitutive and induced resistance mechanisms (Karban and Baldwin, 1997; Thomma et al., 1998; Vorwerk et al., 2004). Emerging research has shown that plants utilize induced defense mechanisms that are dependent on the attacker, and, in certain interactions, a subset of responses are species specific (McDowell and Dangl, 2000; Walling, 2000; Kaloshian and Walling, 2005). The study of salicylic acid (SA)- and jasmonic acid (JA)/ethylene (ET)-dependent signaling pathways in defense against pathogens, pests, and wounding have identified the key genes involved in defense gene regulation and have uncovered complex signaling networks and cross-talk between the pathways (Walling, 2000; Glazebrook, 2001; Rojo et al., 2003). In Arabidopsis (Arabidopsis thaliana), activation of the SA pathway has been shown to be important in both basal and resistance gene (R)-mediated biotrophic pathogen defense, while the JA/ET pathway is activated in response to necrotrophic pathogens, feeding by tissue-damaging herbivores, and wounding (Kessler and Baldwin, 2002; Glazebrook, 2005), although exceptions do exist (Thaler et al., 2004; Glazebrook, 2005; Musser et al., 2005).
The SA-dependent signaling pathway regulates the expression of a wide array of defense-response genes, including the PATHOGENESIS-RELATED PROTEIN (PR) genes. In addition, the SA-dependent pathway confers a broad-spectrum resistance known as systemic acquired resistance, which is a long-acting, induced resistance mechanism against a wide variety of invading pathogens (Ryals et al., 1996; Durrant and Dong, 2004). In contrast, the JA/ET pathway induces genes whose protein products have antimicrobial and antifungal activity, such as PLANT DEFENSIN PROTEIN1.2 (PDF1.2) and THIONIN2.1 (THI2.1), and accumulate in response to necrotrophic pathogens (Rojo et al., 2003). In addition, in caterpillar-infested tomato (Solanum lycopersicum), JA signaling is important in the regulation of wound-response genes, such as proteinase inhibitors, polyphenol oxidases, Thr deaminase, and arginase, which have roles in antagonizing caterpillar growth and development (Felton et al., 1989; Ryan, 2000; Chen et al., 2005). In addition, the JA-regulated VEGETATIVE STORAGE PROTEIN1 (VSP1) has been shown to have anti-insect phosphatase activity against Drosophila melanogaster (fruit fly), Diabrotica undecimpunctata howardi (corn root worm), and Callosobruchus maculatus (cow pea weevil) (Liu et al., 2005). Along with the aforementioned direct defenses, the JA/ET pathway induces indirect defenses through the production and release of plant volatiles that attract both predators and parasitoids of the herbivore (Kessler and Baldwin, 2002).
Unlike chewing insects, less is known about molecular responses to insects from other feeding guilds. Phloem-feeding insects are intriguing due to their “stealthy” feeding mechanisms that cause little damage to the plant tissue as they establish direct access to amino acids and carbohydrates through the vascular tissue. To date, most studies of phloem-feeding insects have examined aphid interactions, including Myzus persicae (green peach aphid) with tomato or Arabidopsis, Myzus nicotianae (tobacco aphid) with tobacco (Nicotiana spp.), Macrosiphum euphorbiae (potato aphid) with tomato, and Schizaphis graminum (greenbug aphid) with sorghum (Sorghum bicolor) (Fidantsef et al., 1999; Moran and Thompson, 2001; Ellis et al., 2002; Martinez de Ilarduya et al., 2003; Voelckel et al., 2004; Zhu-Salzman et al., 2004; De Vos et al., 2005; Li et al., 2006; Thompson and Goggin, 2006). By monitoring the RNA levels of sentinel defense genes after aphid feeding, studies in Arabidopsis show that SA-regulated transcripts increase (Moran and Thompson, 2001; Moran et al., 2002; De Vos et al., 2005). Wound- and JA/ET-regulated genes are induced transiently or at lower levels during M. persicae-Arabidopsis and M. euphorbiae-tomato aphid feeding (Moran and Thompson, 2001; Martinez de Ilarduya et al., 2003).
Transcriptome analysis after aphid feeding on Arabidopsis further confirmed the trends observed by Moran and Thompson (Moran et al., 2002; De Vos et al., 2005). These changes in RNA levels suggest that responses to aphid feeding are more similar to “pathogen” defense responses than “chewing insect” defenses. While SA-regulated transcripts are induced, the role of SA in Arabidopsis basal defense to aphids remains controversial (Moran and Thompson, 2001; Mewis et al., 2005; Pegadaraju et al., 2005). In addition, recent experiments have shown mutations in PHYTOALEXIN DEFICIENT4 (PAD4), which is regulated by SA, increase susceptibility to M. persicae; pad4 susceptibility is correlated with a delayed aphid-induced senescence (Pegadaraju et al., 2005). In contrast to M. persicae-Arabidopsis interactions, basal SA defenses decrease M. euphorbiae longevity in tomato, and SA is important in Mi1.2-mediated resistance to potato aphids (Li et al., 2006).
Like aphids, the silverleaf whitefly (SLWF; Bemisia tabaci type B; Bemisia argentifolii) is an obligate phloem-feeding pest. Although these animals share membership in the same feeding guild, aphid and whitefly feeding are not synonymous. Unlike aphids, which probe extensively and are more mobile in their feeding habits, whitefly nymphs feed continuously from the same location throughout their 28+-day nymphal development (Gill, 1990; Byrne and Bellows, 1991; Johnson and Walker, 1999; Freeman et al., 2001). The continuous and long-term interaction between whiteflies and their host results in an intimate relationship and possibly pronounced and distinct defense responses when compared to aphids. In addition, whiteflies and aphids are likely to have different salivary components that may elicit different responses from their host (Walling, 2000).
The response of crop plants to SLWF feeding suggests that the JA/ET and novel defense pathways are induced (van de Ven et al., 2000; Walling, 2000). In tomato, JA/ET-regulated basic PR genes accumulate to higher levels than SA-regulated acidic PR gene transcripts (D.P. Puthoff, C.S. LeVesque, T.M. Perring, and L.L. Walling, unpublished data). Genes identified through differential RNA display in response to SLWF feeding in tomato and squash (Cucurbita pepo), Whitefly Induced1 and SILVERLEAF WHITEFLY INDUCED1 (SLW1), respectively, have also been shown to be JA inducible (van de Ven et al., 2000; Walling, 2000). Novel pathways appear to contribute to SLWF defense in crop plants (van de Ven et al., 2000; Walling, 2000). For example, SLW3 transcripts do not accumulate after application of known defense-response chemicals or in response to feeding by a closely related whitefly biotype (B. tabaci type A; van de Ven et al., 2000). Although these studies demonstrate the complexity and dynamic interactions between crops and the SLWF, these plants lack the powerful genetic and genomic resources afforded by the plant model system Arabidopsis. Further studies that examine the role of both the JA- and SA-defense pathways in Arabidopsis in response to SLWF are necessary to allow comparisons with aphid-induced responses.
The SLWF is a generalist and infests a wide variety of crop plants, including members of the Brassicaceae. Infestations of Brassica oleracea in the field have been reported as high as 10 nymphs/cm2, indicating that members of this family are natural hosts for this phloem-feeding pest (Liu, 2000). Therefore, it is timely to harness the genetic resources in the model plant Arabidopsis, a member of the Brassicaceae, to provide insights into the mechanisms that contribute to basal resistance to phloem-feeding whiteflies. Here, a foundation for Arabidopsis responses to SLWF feeding is provided by comparing RNA levels of well-characterized SA-, JA-, and ET-defense genes. We show that SLWFs induced SA-signaling pathways and suppressed or did not alter expression of JA/ET-regulated genes. To test for the role of these defense pathways in basal resistance, five Arabidopsis SA and JA mutant/transgenic lines (nonexpressor of PR1 [npr1], constitutive immunity10 [cim10], coronatine insensitive1 [coi1], constitutive expression of vsp1 [cev1], and NahG) and wild-type Columbia were utilized to monitor SLWF nymphal development and sentinel SA- and JA-defense gene RNAs. These experiments and infestation studies with methyl jasmonate (MeJA)-treated npr1 plants demonstrated that basal defenses suppressed by SLWF feeding were critical for constraining nymphal development.
RESULTS
Regulation of Known SA-, JA-, and ET-Defense Transcripts in Response to SLWF Instar Feeding
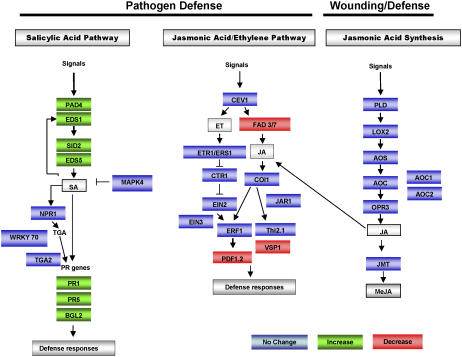
To date, more than 30 defense genes have been aligned into complex SA-, JA-, and ET-signaling cascades (Glazebrook, 2001; Devoto and Turner, 2003; Shah, 2003). Other defense genes have been identified, but their role or placement in defense signaling has yet to be determined. Transcriptome analysis after SLWF feeding in Arabidopsis ecotype Columbia has implicated that the SA-dependent pathway is induced, while the JA-dependent pathway shows no change or is repressed (Kempema et al., 2007). These transcript profile studies suggest that Arabidopsis perceives and responds to SLWF more like a pathogen than a tissue-damaging herbivore. Figure 1 summarizes the trends in known defense gene expression after SLWF nymph feeding gleaned from Kempema et al. (2007).
Figure 1.
Summary of Arabidopsis defense gene expression patterns after SLWF second and third instar feeding. Genes involved in SA- and JA/ET-defense signaling and SA and JA biosynthesis pathways are shown as colored boxes. Green and red denote an increase or decrease in RNA levels, respectively (<1.5-fold). Blue indicates no change in expression. Several ethylene receptor genes have been identified, including ETR1, ERS1, EIN4, ERS2, and ETR2; only ETR1 and ERS1 are illustrated. These gene expression trends are based on microarray studies reported by Kempema et al. (2007).
The microarray data indicated that increases in SA-regulated defense gene RNAs are detected by 21 d after SLWF feeding (Kempema et al., 2007). To assess the timing of defense gene activation in response to SLWF nymph feeding, the levels of two sentinel defense gene RNAs were assessed at 0, 7, 14, 21, and 28 d after SLWF infestation (Fig. 2A). Transcripts for the SA-regulated PR1 gene were first detected at 14 d after infestation and increased to highest levels by 28 d. In contrast, the levels of the JA-regulated PDF1.2 RNAs were not detected in control, noninfested, or SLWF-infested leaves within the 28-d period.
Figure 2.
Defense gene transcript accumulation after infestation with SLWF nymphs. Total RNA was extracted from SLWF-infested or control noninfested rosette leaves. Infestations were performed at 22°C. cDNAs were synthesized and used in PCR reactions with gene-specific primers for defense genes involved in SA-dependent pathway (SID2, EDS5, PAD4, PR1, BGL2, PR5) and JA/ET-dependent pathway (ERS1, ERF1, THI2.1, VSP1, PDF1.2, FAD3). ACTIN7 was used as a control (20 cycles). A, RT-PCRs were performed on RNAs isolated from SLWF-infested and noninfested plant leaves collected at 0, 7, 14, 21, and 28 d. SA- and JA-regulated gene RNAs were detected after 25 cycles of PCR. B, Leaves from 21-d infested (I) and control noninfested (C) plants were collected. SA- and JA-regulated gene RNAs were detected after 25 and 27 cycles of PCR, respectively. RT-PCRs were performed on the RNAs used in the microarray experiments (Kempema et al., 2007) and RNAs from two additional infestation experiments (see “Materials and Methods”). A representative experiment is displayed.
To confirm the microarray data reported by Kempema et al. (2007), RNAs control, noninfested, and 21-d SLWF-infested plants from three independent biological experiments were used (Fig. 2B). Transcripts for the SA-regulated genes PR1, PR5, and β-1,3-GLUCANASE2 (BGL2; PR2) increased after 21 d of nymph feeding compared to noninfested control plants. The RNAs for genes important in events upstream of SA or for the synthesis of SA, such as SALICYLIC ACID INDUCTION DEFICIENT2 (SID2), ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5), and PAD4 were also elevated after nymphal feeding (Fig. 2B). These results indicated that, like biotrophic pathogens, the SA-defense pathway was activated. If similar to pathogen-plant interactions, this pathway could have a role in basal defense to SLWFs.
In contrast, RNAs encoded by genes known to be involved in JA biosynthesis, such as OMEGA-3 FATTY ACID DESATURASE3 (FAD3), or that respond to JA, such as PDF1.2, decreased in infested leaves relative to control noninfested leaves (Figs. 1 and 2B). Unlike FAD3 and PDF1.2, THI2.1 RNAs were not detected in noninfested controls or after SLWF nymph feeding (Figs. 1 and 2B). Two ethylene-responsive genes were also examined. ETHYLENE RESPONSE SENSOR1 (ERS1) RNA levels declined in SLWF-infested leaves relative to control leaves, while ETHYLENE RESPONSE FACTOR1 (ERF1) RNA levels were at similar levels in both infested and control leaves (Fig. 2B).
Local and Systemic Induction of Defense Genes in Response to SLWF Infestation
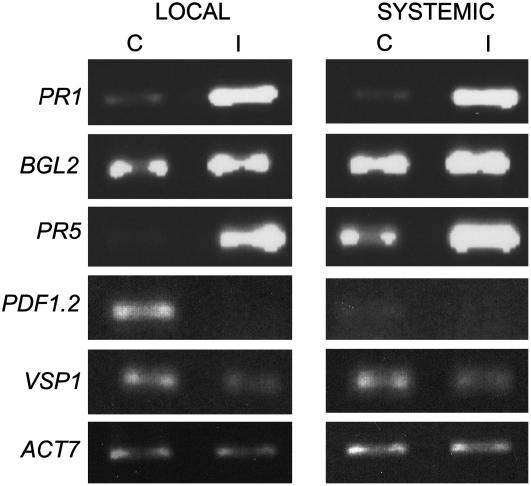
To evaluate if Arabidopsis mounts a systemic response to SLWF feeding, the change in SA and JA sentinel gene RNAs was examined both in local infested leaves and apical noninfested leaves (systemic) after a 21-d infestation. Reverse transcription (RT)-PCR with gene-specific primers showed that, unlike responses to aphids, the trends identified in SLWF-infested Arabidopsis leaves were also observed in apical, noninfested leaves. SA-regulated gene transcripts (PR1, PR5, and BGL2) accumulated both locally and systemically after nymph feeding (Fig. 3). JA-responsive RNAs (VSP1 and PDF1.2) were not present or were at lower levels in both local and systemic leaves. Collectively, the whole-plant response to SLWF infestation was distinctive from what has been observed with other phloem feeders in Arabidopsis.
Figure 3.
Local and systemic accumulation of SA- and JA-defense gene RNAs. Infested leaves (local) and noninfested, apical leaves (systemic) from 21-d SLWF-infested plants (I) were collected. Control tissue (C) was collected from developmentally matched leaves on noninfested plants. PCR was performed on cDNA using gene-specific primers (25 cycles for SA-regulated genes and 27 cycles for JA-regulated genes). ACTIN7 was used as a control (20 cycles). Three biological replicate infestations at 22°C were performed. One representative experiment is shown.
Repression of JA Responses Enhances SLWF Development
To assess the role of SA- and JA-signaling pathways in defense against SLWFs, lines with impaired SA (npr1 and NahG) and JA (coi1) signaling were examined (Cao et al., 1994; Feys et al., 1994; Lawton et al., 1995). This was complemented with lines that constitutively activated SA (cim10) and JA (cev1) defenses (Ellis and Turner, 2001; Maleck et al., 2002). Although cim10 is less well characterized than some mutants that constitutively express SA defenses, it was chosen for these studies because it does not display a dwarf phenotype, nor does it produce the spontaneous lesions that are commonly observed in SA overexpression lines (Maleck et al., 2002).
Mutant and wild-type plants were infested with SLWFs (>100 nymphs/plant) to assess impacts on nymphal development using a no-choice bioassay (Fig. 4). SLWF development was assayed by scoring the total number of insects at each developmental stage (first, second, third, or fourth instars) on each of the eight replicate plants. The percentage of insects that had reached advanced stages of development (fourth instars) by day 24 was calculated and compared between all six lines using Tukey's multiple comparison test (Fig. 4; Supplemental Fig. S1). In addition, to assess defense pathway activation during SLWF feeding, the changes in levels of marker genes PR1, BGL2, PDF1.2, and VSP1 RNAs were monitored in all lines. Most SA- and JA-defense mutants have not been utilized in long-term infestation or infection studies. The examination of defense gene transcripts in these defense mutants provided further characterization of both SA- and JA-dependent gene expression at later times in plant development. Some defense genes are expressed at higher basal levels in older plants (Kus et al., 2002). Therefore, it was important that the defense gene transcripts were monitored in the mutant noninfested plants and after challenge with SLWFs to interpret bioassay results.
Figure 4.
SLWF nymph development on mutant, transgenic, and wild-type plants. At 24 d postinfestation, the numbers of total nymphs and nymphs in their first (gray), second (dotted), third (white), and fourth (black) instars were counted and the percentage of insects in each instar on wild-type Columbia (WT) and SA- and JA-signaling mutant/transgenic lines determined. Defense signal mutants and lines are described within “Results” and include the following: SA-deficient (npr1-1, NahG), JA-deficient (coi1-1), SA overexpression (cim10), and JA overexpression (cev1) plant lines. Three biological replicate infestations were performed at 22°C and analyzed, and a representative experiment is shown. The infestation level and biological variation within the replicate plants of each genotype can be found in Supplemental Figure S1. Significance of variation in percentage of fourth instars across genotypes was determined using Tukey's multiple comparison analysis at the 99.6% individual confidence level. The significance is indicated by the following: a, b, or c. On average, infested plants had approximately 107 nymphs.
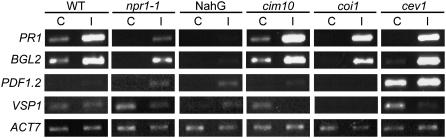
At the time of infestation, PR1 RNAs were abundant in noninfested cim10 plants and at lower levels in wild-type plants, confirming the cim10 constitutive immunity phenotype (Supplemental Fig. S2). After 24 d of infestation, 76% of total insects that developed on cim10 plants were in their fourth instar (Fig. 4; Supplemental Fig. S1). This contrasted to the slower development of SLWF nymphs on wild-type plants, where approximately 45% of insects were fourth instars. These data indicated that SLWF development was significantly accelerated on the SA overexpression mutant cim10. Similarly, insects on coi1 mutant plants, which do not perceive JA, showed accelerated development trends (65% fourth instars). After 24 d of infestation, the JA/ET-regulated transcripts PDF1.2 and VSP1 accumulated to lower levels in the cim10 and coi1 plants relative to infested wild-type plants (Fig. 5). In addition, SA-dependent transcripts (PR1 and BGL2) in cim10 and coi1 mutants were at similar or elevated levels relative to the wild-type plants. Collectively, these data suggested that either elevated SA and/or reduced JA responses compromised Arabidopsis basal resistance to the SLWF, as reflected by enhanced nymphal development.
Figure 5.
Local accumulation of defense gene RNAs. RNAs for sentinel SA (PR1, BGL2) and JA (VSP1, PDF1.2) defense-response genes were monitored by RT-PCR using gene-specific primers and 25 and 27 cycles, respectively. cDNAs were synthesized from RNAs from leaves from uninfested (C) mutant and control plants or 24-d infested leaves (I) from mutant and control plants. ACTIN7 was used as a control (20 cycles). Each infested plant had approximately 107 feeding nymphs. Three biological replicate infestations at 22°C were performed. One representative experiment is shown.
This hypothesis was further supported by the development rates of SLWF nymphs on the SA mutant lines npr1 and NahG, which impair SA signaling and catabolize SA, respectively. The percentage of fourth instars on npr1 and NahG plants was significantly different from wild-type plants. Only 18% and 16% of SLWF nymphs were in their fourth instar on npr1 and NahG plants, respectively, when compared to wild-type plants (Fig. 4). In accordance with this finding, the percentage of SLWFs in their second and third instars rose. In these mutants, the SA-regulated RNAs PR1 and BGL2 accumulated to lower levels than in wild-type in both noninfested and infested leaves, and, in a reciprocal fashion, JA-dependent PDF1.2 and VSP1 transcripts increased compared to wild type (Fig. 5). These data indicated that by abolishing SA defenses and/or enhancing JA defenses in npr1 and NahG plants, enhanced defenses active against SLWF nymphs, as reflected in significant delays in nymphal development, were displayed.
Similarly, on the JA-pathway overexpression mutant cev1, significantly fewer nymphs reached the fourth instar (13%) than on wild-type plants (Fig. 4). Consistent with the constitutive activation of JA defenses in cev1 plants (Ellis and Turner, 2001), the JA-dependent transcripts PDF1.2 and VSP1 accumulated to high levels in uninfested cev1 than wild-type leaves (Fig. 5). Despite elevated JA defenses, SLWF nymph feeding caused SA transcripts (PR1, BGL2) to accumulate in infested cev1 leaves; in fact, PR1 and BGL2 RNAs accumulated to similar levels in RNA-blot analysis in the cev1 mutant and wild-type plants (data not shown). PDF1.2 transcripts increased, while VSP1 transcripts decreased after SLWF infestation. Collectively, these data indicated that the SLWF nymphs provided signals that allowed for strong expression of SA-regulated genes and repression of VSP1 in the cev1 mutant. The facts that nymphs feeding on cev1 plants exhibited delayed development relative to wild-type plants and the SA-pathway gene RNAs accumulated in both cev1 and wild-type plants suggested that enhanced JA responses, and not SA defenses, were responsible for delaying the development of SLWF nymphs.
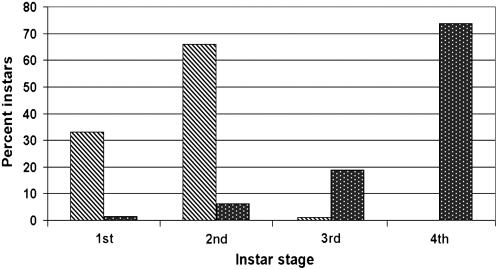
As cross-talk between JA- and SA-defense pathways is commonly associated with responses to biotic threats and displayed in defense mutant studies, it was important to further dissect the relative importance of the suppressed JA and induced SA defenses in SLWF basal resistance. npr1 plants uncouple the cross-talk between SA and JA signaling. For example, during Pseudomonas syringae pv tomato infection, npr1 plants have reduced levels of SA and PR1 RNAs, but JA signaling is preserved (Spoel et al., 2003). Therefore, comparisons of untreated and MeJA-treated npr1 plants should allow the role of JA-regulated defenses to be assessed. npr1 plants were treated with MeJA or served as controls. Unlike previous experiments (Fig. 4), these infestations were performed at 24°C, which significantly accelerated nymphal development (Fig. 6); the temperature dependence of whitefly nymph development is well established (Nava-Camberos et al., 2001). After 17 d of infestation at 24°C, over 74% of the SLWF nymphs feeding on control npr1 plants were in their fourth instar. Smaller numbers of first, second, and third instars were also noted in control plants (Fig. 6). In contrast, the MeJA-treated npr1 plants had no fourth-instar nymphs. Nymphs were primarily in their first (33%) and second (65%) instars (Fig. 6). MeJA had a dramatic effect on SLWF nymphal development on npr1 plants, clearly demonstrating the importance of JA-regulated defenses in basal resistance and curtailing SLWF nymphal development.
Figure 6.
SLWF development on MeJA-treated or control npr1-1 plants. At 17 d postinfestation, the numbers of nymphs in each instar were counted on npr1-1 plants treated with 100 μm MeJA/0.001% ethanol (diagonal bars) or 0.001% ethanol (black bars; control). The percentage of nymphs at each developmental stage (first through fourth instar) was determined. Infestations were performed at 24°C, which accelerated SLWF nymph development. Each infested plant had approximately 225 feeding nymphs. Two biological replicate infestations were performed. One representative experiment is shown.
DISCUSSION
The SA- and/or JA/ET-regulated defense pathways are important in basal and gene-for-gene resistance to pathogens and herbivores. After perception of a biotic threat, plants fine-tune the balance of defense pathways to orchestrate the “best” defense response to its intruder (Reymond and Farmer, 1998; Walling, 2000; Kunkel and Brooks, 2002). The cross-talk between the SA and JA pathways is thought to minimize expression of costly and ineffective defenses that divert carbon and nitrogen resources from plant vegetative growth, thereby avoiding compromises to plant vitality and reproduction. This view is supported by the facts that SA-induced defenses are important in the induced basal and gene-for-gene defenses against biotrophic pathogens (Glazebrook, 2005). Similarly, JA-induced defenses confer resistance to necrotrophic pathogens and insects.
Pests and pathogens have leveraged this molecular communication mechanism to enhance their success on host plants (Mudgett, 2005; Chisholm et al., 2006). While some pathogens evade host defenses by actively catabolizing antimicrobial compounds (Bouarab et al., 2002), there is a growing evidence that plant pathogens produce effectors that antagonize defense signaling networks (Hammond-Kosack and Parker, 2003; Kamoun, 2006). The complexity of the arms race between host and attacker is exemplified by P. syringae, which uses an array of effectors to suppress expression of defense genes and secondary metabolites, suppress programmed cell death, avoid R gene-mediated resistance, suppress cell wall remodeling, and potentially alter gene expression programs and turnover of defense regulatory proteins (He et al., 2004; Cui et al., 2005; Mudgett, 2005; Chisholm et al., 2006; Janjusevic et al., 2006). By simultaneously evaluating SLWF nymph development on mutants from both SA- and JA-defense pathways and after exogenous MeJA treatments, it appears that SLWFs should be added to the set of pathogens and pests that manipulate host-plant defense responses to their own advantage.
SLWF nymphs have an intimate and long-term interaction with their host plants. With the exception of the crawler, which emerges from the egg, SLWF nymphs are immobile and feed almost continuously for approximately 28 d under optimal Arabidopsis conditions. SLWF nymphs provided strong and reproducible signals that were perceived by Arabidopsis, resulting in increases in SA-regulated defenses and suppression of JA-regulated defenses (Fig. 2). The accumulation of PR gene RNAs after SLWF feeding in Arabidopsis was SA and NPR1 dependent, as transcripts did not accumulate to wild-type levels in NahG and npr1 plants (Fig. 5). SA-dependent defense gene RNAs accumulated both in local, infested leaves and systemically in noninfested apical leaves (Fig. 3). Previous studies in squash and tomato also show local and systemic induction of defense genes after SLWF feeding (van de Ven et al., 2000; Walling, 2000). In contrast, systemic activation of defenses was not observed in compatible M. persicae-Arabidopsis and M. euphorbiae-tomato interactions (Moran and Thompson, 2001; Martinez de Ilarduya et al., 2003). This suggests that SLWFs may provide more potent signals, more mobile signals, or larger quantities of signals (due to their prolonged feeding habits) to their host plant.
By using mutant and transgenic lines that alter SA and JA defenses, the branch of Arabidopsis defense signaling that antagonizes SLWF nymph development was identified. There was a strong correlation of SLWF success (as measured by the rate of nymphal development) with the absence of JA defenses and presence of SA defenses (Fig. 4). For example, SLWF nymph development was more rapid on cim10 and coi1 than wild-type plants (Fig. 4); coi1 and cim10 plants accumulated the SA-regulated PR1 and BGL2 RNAs and displayed reduced JA defenses (PDF1.2 and VSP1 RNAs; Fig. 5). Reciprocally, cev1, NahG, and npr1 mutants had an enhanced basal resistance to SLWFs; the delayed SLWF nymph development was correlated with enhanced JA-regulated defenses in these lines (Fig. 5). The fact that SA-dependent RNAs were abundant in cev1, cim10, and wild-type plants, but only cev1 displayed an increased basal resistance, suggested that JA-dependent defenses, and not SA defenses, were responsible for the delays in nymph development observed on cev1 and cim10 plants (Figs. 4 and 5).
The importance of JA-regulated defenses in basal resistance to SLWFs was also supported by comparing SLWF development on untreated and MeJA-treated npr1 plants. npr1 mutants lack the ability to activate SA defenses (Spoel et al., 2003), and MeJA treatments accentuated the npr1 delay in SLWF nymph development relative to the untreated npr1 plants (Fig. 6). Collectively, these data and those reported above indicated that the suppressed JA-regulated defenses were important in slowing SLWF nymphal development. Furthermore, the highly induced SA defenses did not appear to significantly contribute to the basal resistance to SLWFs in Arabidopsis, although SA-dependent defenses may have a role in other aspects of the SLWF-Arabidopsis interaction, such as host choice, fecundity, or longevity. These data contrasted to the preferential induction of SA defenses observed in biotrophic pathogen-plant interaction and the importance of SA defenses in both basal and R gene-mediated resistance (Glazebrook, 2005).
The data presented here support the idea that SLWFs enhance their success on Arabidopsis plants by failing to activate or suppressing the effectual JA-regulated defenses. It is possible that SLWFs evade activation of the JA pathway since SLWFs cause little tissue damage (intracellular punctures) until they establish feeding sites at minor veins of the phloem (Cohen et al., 1996; Walling, 2000). SLWFs could also prevent the activation of JA defenses by introducing inhibitors that directly or indirectly antagonize JA-signaling pathway activation or action. Finally, SLWFs strongly activated SA defenses, even in cev1 plants. Therefore, it is possible that SLWFs down-regulated the effectual JA defenses via SA cross-talk in wild-type plants. The SLWF effector(s) that induces SA defenses and/or suppresses JA defenses is unknown, but is presumed to be a salivary component synthesized by the whitefly or one of its endosymbionts (Walling, 2000). While whitefly saliva is not well characterized biochemically (Funk, 2001), the watery and sheath salivas of other hemipterans, such as aphids, are rich in potential defense signaling molecules, including pectinases, complex carbohydrates, proteins, peroxidases, phospholipids, amylases, lipases, and/or phosphatases (Miles, 1999; Walling, 2000).
Additional evidence for herbivore manipulation of plant defenses (the “decoy” hypothesis) to enhance insect performance is accumulating from studies with both tissue-damaging herbivores and phloem-feeding aphids (Zhu-Salzman et al., 2005; Thompson and Goggin, 2006). For example, Helicoverpa zea larvae egest saliva containing Glc oxidase into their feeding sites to suppress the JA-regulated defenses that deter larval growth (Musser et al., 2002). Glc oxidase uses Glc to produce hydrogen peroxide to activate SA defenses, such as PR1 protein accumulation; however, a SA-independent mechanism is responsible for suppressing the effectual JA-mediated defenses of tobacco, such as nicotine production (Musser et al., 2002, 2005).
Several studies from the molecular plant-aphid interaction literature also support the “decoy” hypothesis. It should be noted that changes in JA- or SA-defense gene RNA levels and aphid population dynamics on defense mutants have varied, presumably due to the differences in aphid-infestation experimental design (Moran and Thompson, 2001; Ellis et al., 2002; Moran et al., 2002; De Vos et al., 2005; Mewis et al., 2005; Pegadaraju et al., 2005). In general, rises in PR RNAs have been noted, and, like SLWFs, JA-defense gene RNAs are often suppressed or not highly induced after aphid feeding on Arabidopsis (Moran and Thompson, 2001; Ellis et al., 2002; Moran et al., 2002; De Vos et al., 2005). More variation is observed in defense mutant studies. The clear reciprocal phenotypes of SA- and JA-defense mutants, as were seen for SLWFs, have not been documented previously in the Arabidopsis-aphid literature. While several studies have shown that M. persicae population growth is slowed in cev1, npr1, and NahG lines or after MeJA treatments (Moran and Thompson, 2001; Zhu-Salzman et al., 2004; Mewis et al., 2005), other studies showed neither NahG, npr1, nor coi1 changed M. persicae population dynamics relative to wild-type plants (Moran and Thompson, 2001; Ellis et al., 2002; Mewis et al., 2005; Pegadaraju et al., 2005).
Given the variability in the aphid-plant interactions studies to date, the simultaneous analyses of five defense mutants were crucial in providing a comprehensive and reproducible picture establishing the importance of JA-regulated defenses in deterring SLWF nymphal development. While the specific JA-dependent genes important in SLWF defense have yet to be identified, basal defense toward SLWF in Arabidopsis appeared to be antibiotic. Preliminary no-choice egg-deposition and choice bioassays show that SLWF exhibits no preference for any of the mutants altered in constitutive defenses, including cell wall composition, secondary metabolites, and trichome density (data not shown). Both generalist (M. persicae) and specialist (Brevicoryne brassicae) aphid interactions with Arabidopsis suggest that JA-dependent defenses have antibiotic effects on aphids (Mewis et al., 2005). However, in tomato, Mi1.2-mediated resistance toward SLWF feeding is antixenotic in that it acts to deter the whitefly from establishing a feeding site (Nombela et al., 2003). If SLWFs establish a feeding site on Mi1.2 plants, nymphs have feeding behaviors similar to plants lacking Mi1.2 (Jiang et al., 2001).
If viewed in the broadest terms, the SLWF-Arabidopsis interactions bear a semblance to Arabidopsis interactions with fungal biotrophs like Erysiphe spp. (Reuber et al., 1998; Kempema et al., 2007); both sets of organisms induce SA-dependent defenses. However, when basal resistance mechanisms are investigated, the SLWF and fungal biotrophs are distinct. While SA defenses are essential for the basal and induced resistance mechanisms for control of fungal biotrophs, SA-induced defenses did not appear to contribute to the mechanisms that dictate basal resistance to SLWFs. Interestingly, Arabidopsis appeared to mount a completely ineffectual response to SLWF feeding as effective JA-dependent defenses were not induced in wild-type plants. Further experiments that examine the role of cross-talk, SLWF salivary components, and downstream responses will allow identification of elicitor(s) and mechanism(s) for retarding nymphal development, which contribute to the basal resistance in Arabidopsis. The JA-mediated delays in SLWF nymph development could be used in the future to engineer resistance to SLWFs. Delays in insect development are considered important resistance mechanisms impacting insect population dynamics and providing a longer period of time for natural enemies, such as parasitoid wasps, to attack the insects (Pechan et al., 2000; Dicke and Hilker, 2003).
MATERIALS AND METHODS
Plant Growth and Insect Maintenance
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (wild-type) plants used in the local and systemic defense gene transcript studies (Figs. 1–3) were grown and infested as described by Kempema et al. (2007). Plants used in the bioassays wild type, coi1-1, npr1-1, cev1, cim10, and transgenic NahG plants, were grown in 4-inch pots in Sunshine Mix Number 1 soil (SunGro) supplemented with fertilizer (Osmocote 14–14–14; Scott Horticulture Solutions). All plants were grown under fluorescent and incandescent lights (180 μE m−2 s−1) at 22°C under long-day (16-h light:8-h dark) conditions for 2 weeks (with the exception of cev1) before infestation for 21 or 24 d under short-day (8-h light:16-h dark) conditions.
Plant size (rosette diameter) influenced SLWF oviposition preference (L.A. Kempema, S.I. Zarate, and L.L. Walling, unpublished data). At 2 weeks, all plants used in these studies had the same rosette diameter and number of leaves at the time of infestation and at the completion of the experiment. Due to its slower growth, cev1 plants were planted 7 d prior to other genotypes to allow an additional week of growth before infestation. The rosette diameter and number of leaves on 3-week-old cev1 plants was approximately the same as 2-week-old plants from the other lines.
coi1 plants were identified from a F2 seed pool on one-half-strength Murashige and Skoog medium (10 g L−1 Suc and 0.8% [w/v] agar content) containing 30 μm MeJA/0.01% ethanol (Bedoukian Research). At 7 d, homozygous coi1 seedlings were identified by elongated roots and normal aboveground organ morphology (Feys et al., 1994). The coi1 plants were transferred to pots containing soil.
cim10 mutants have wild-type stature, do not display necrotic lesions, and constitutively overexpress SA and SA-regulated defense genes (Maleck et al., 2002). These features distinguish cim10 relative to other constitutive immunity mutants and made cim10 an excellent choice for SLWF infestations. The levels of PR1 RNAs in cim10 and wild-type plants were determined using RT-PCR and gene-specific primers in noninfested 2-week-old and 3-week-old plants to confirm the cim10 constitutive immunity phenotype prior to the time of SLWF infestation (Supplemental Fig. S2).
A virus-free SLWF colony (Bemisia tabaci type B; Bemisia argentifolii Bellows and Perring) was maintained on Brassica napus var ‘Florida Broad Leaf’ (W. Atlee Burpee & Co.) grown at 27°C, 55% relative humidity under long-day (16-h light:8-h dark) conditions in the Insectory and Quarantine Facility at the University of California, Riverside. Adults were collected by aspiration. B. napus plants were germinated under the same conditions in a growth chamber for 4 weeks before being transferred to the Insectory and Quarantine Facility.
Whitefly Infestations
Adult male and female whiteflies (totaling 30–100 depending on the experiment) were collected from SLWF-infested B. napus leaves by aspiration into 15-mL falcon tubes. A tube containing male and female SLWFs was placed upright in each pot. This number of whiteflies per plant resulted in infestation levels similar to the infestation levels experienced by Brassica plants in the field (Liu, 2000). Arabidopsis plants typically had >100 feeding nymphs/plant. Nylon bags (5 × 10 inch) were placed around each pot and secured with a rubber band. Whiteflies were released by unscrewing the falcon tube. Control pots were bagged but not infested. After 7 d, all adult flies were removed from the plants by aspiration and the pots rebagged. For the time-course experiment, rosette leaves were collected from infested and control plants at 0, 7, 14, 21, and 28 d. Three biological replicates of these experiments were performed. For most defense gene expression studies, rosette leaves were collected after 21 d, when second and third instars were observed on wild-type Columbia plants. To identify the noninfested apical leaves (systemic), leaves were examined under the microscope after 21 d. Leaves were considered “local” if nymphs or eggs were observed on a rosette leaf. Leaves were considered “systemic” if no nymphs or eggs were observed on the rosette leaf. Developmentally matched systemic and local leaves were collected from noninfested plants as controls.
In each no-choice bioassay experiment, eight replicate plants/line were grown and infested as described above. However, adults were removed 2 d after infestation. The number of whiteflies and their developmental stages (first through fourth instars) were recorded after 24 d. Developmental progression was estimated by calculating the percentage of fourth instars (red-eye stage) on each plant (number of fourth instars/total nymphs). The no-choice bioassays were repeated for a total of 24 replicate plants/line. To assure an unbiased reporting of insect numbers and developmental stages, infested plants were randomly assigned letters to conceal the genotype identity. Immediately after nymphs were counted, infested leaves were placed into liquid N2 and stored at −80°C until used for RNA isolation.
MeJA Treatment of npr1 Plants
npr1 plants were grown under long-day conditions for 2 weeks at 24°C. At this time, the growth chambers were changed to short-day conditions and plants were infested with SLWF and/or treated with MeJA. Ten replicate plants were treated with 25 μL/leaf of 100 μm MeJA/0.001% ethanol or 0.001% ethanol 3 h prior to infestation and every 3 d after infestation. Solutions were added to the adaxial side of the leaves, where whiteflies tend not to feed or deposit eggs. Plants were caged with nylon bags as previously described and were infested with 30 adult whiteflies. Adults were removed after 2 d of infestation. MeJA-treated and 0.001% ethanol-treated plants were placed in separate but comparable growth chambers (24°C) to control for volatiles. Because these no-choice experiments were performed at 24°C, the SLWF nymphs developed more rapidly; the temperature dependence of insect development and, specifically, SLWF development is well established (Nava-Camberos et al., 2001). Therefore, after 17 d, plants were scored for number of nymphs at each developmental stage as described for the developmental bioassays above. The experiment was replicated twice.
Data Analysis
Defense genes induced or repressed 1.5-fold by microarray analysis were identified by Kempema et al. (2007). Briefly, microarray data was background adjusted using robust multiarray analysis and differential analysis performed using significant analysis of microarray. Data from the no-choice bioassay was analyzed using a one-way unstacked ANOVA and Tukey's multiple comparison test with Minitab software. Data for the npr1 MeJA treatment experiment were analyzed using Student's t test.
RT-PCR
Total RNA was extracted from rosette leaves using TRIzol reagent (Invitrogen). The quality of the RNA was checked on a 1% agarose denaturing gel (0.5% MOPS, 0.8% formaldehyde). Before the reverse transcriptase reaction, 1 μg of RNA was treated with TURBO DNase as indicated in the manufacturer's instructions (Ambion). Oligo(dT)20 primer (0.5 μg) was added and RNA denatured for 5 min at 70°C. RT was performed using ImpromII reverse transcriptase and RNasin ribonuclease inhibitor as indicated in the manufacturer's instructions (Promega).
PCRs (95°C 5 min, 95°C 35 s, 55°C–64°C 35 s, 72°C 2 min, 72°C 10-min final extension time) using ACTIN7 primers were used to check the cDNA synthesis and equalize cDNA amounts between reactions (25 mm MgCl2, 8 μm forward primer, 8 μm reverse primer, 1 unit Taq polymerase, 8 mm dNTPs). ACTIN7 primers were designed to span an intron to verify that no genomic DNA contamination was amplified during RT (ACT7/2, At5g09810: forward 5′-CTCATGAAGATTCTCACTGAG-3′, reverse 5′-ACAACAGATAGTTCAATTCCCA-3′; genomic 753 cDNA 652 bp). For ACTIN7, 20 PCR cycles were used. For JA/ET- and SA-regulated defense genes, transcripts were amplified using 27 and 25 cycles, respectively, and numbers of cycles are indicated in the figure legends. For the analysis of PR1 RNAs in the noninfested leaves of cim10 and wild-type plants, 22 PCR cycles were used (Supplemental Fig. S2). The following primer sequences were designed: PDF1.2, At5g44420: forward 5′-TTCTCTTTGCTGCTTCGAC-3′, reverse 5′-GTCATAAAGTTACTCATAGAGTGACAG-3′ (258-bp product); THI2.1, At1g72260: forward 5′-TCTGGTCATGGCACAAGTTC-3′, reverse 5′-GAGTGTTCATGGCACCACAC-3′ (260-bp product); VSP1, At5g24780: forward 5′-TTTTACGCCAAAGGACTTGC-3′, reverse 5′-TCAATCCCGAGTTCCAAGAG-3′ (223-bp product); FAD3, At2g29980: forward 5′-GGACACACCACCAGAACCAT-3′, reverse 5′-AGGCAACTTCTCATCGTGACC-3′ (399-bp product); ERF1, At3g23240: forward 5′-CTATCGGATCTTCTCCAGATTCTTTC-3′ (453-bp product), reverse 5′-GAGTGTTTCCTCTTCAACGCCA-3′; ERS1, At2g40940: forward 5′-GAGGAATGTGCGTTGTGATG-3′ (820-bp product), reverse 5′-CATTGGCTTTATCAAAGAGATGA-3′; SID2, At1g74710: forward 5′-GCCTATGGTGGTATGCGTTT-3′, reverse 5′-AAGCCTTGCTTCTTCTGCTG-3′ (852-bp product); BGL2, At3g57260: forward 5′-TCAAGGAAGGTTCAGGGATG-3′, reverse 5′-CAAAACTTCTCATACGTTGGTT-3′ (460-bp product); PR1, At2g14610: forward 5′-GTAGGTGCTCTTGTTCTTCCC-3′, reverse 5′-CAGATAATTCCCACGAGGATC-3′ (420-bp product); PR5, At1g75040: forward 5′-CGTACAGGCTGCAACTTTGA-3′, reverse 5′-GCGTTGAGGTCAGAGACACA-3′ (245-bp product); EDS5, At4g39030: forward 5′-TACGAGGAACTGCGTCAGAA-3′, reverse 5′-TTTGAGCAACCAATCCAACA-3′ (520-bp product); and PAD4, At3g52430: forward 5′-TTGTCGATTCGAGACGAGTG-3′, reverse 5′-TGGCTCGGCTAAGAGTTGAT-3′ (1,174-bp product). PCR products were fractionated on a 1% agarose, 0.5× Tris-borate-EDTA gels. Gels were imaged using LabWorks (UVP) and scanned using Adobe Photoshop 6.0. There were two to three biological replications for all experiments (see above). RT and PCR reactions for all experimental replications were repeated twice.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Box plot of the percent fourth instar data.
Supplemental Figure S2. PR1 RNA levels in 2- and 3-week-old cim10 plants.
Supplementary Material
Acknowledgments
We thank R. Dietrich and Syngenta Biotechnology (cim10), J. Turner (cev1 and coi1), and the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH) for providing materials used in this study. We gratefully acknowledge Frances Holzer and Yun-Shu (Angel) Chen for their help in tissue collection and SLWF colony rearing. We also thank T. Eulgem, I. Kaloshian, P. Springer, and our colleagues in the Walling, Springer, and Kaloshian laboratories for insightful discussions.
This work was supported in part by the California Agricultural Experiment Station, the U.S. Department of Agriculture National Research Initiative (Cooperative State Research, Education, and Extension Service award no. 99–35301–8077 to L.L.W.), and the Southwest Consortium (grant to L.L.W. and G. Thompson [University of Arkansas]). A Department of Education Graduate Assistance in Areas of National Need fellowship (DE P200A030254 to R. Cardullo, Department of Biology, University of California, Riverside) provided partial support for L.A.K. A National Science Foundation predoctoral fellowship provided partial support to S.I.Z.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Linda L. Walling (linda.walling@ucr.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bouarab K, Melton R, Peart J, Baulcombe D, Osbourn A (2002) A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418 889–892 [DOI] [PubMed] [Google Scholar]
- Byrne DN, Bellows TS (1991) Whitefly biology. Annu Rev Entomol 36 431–457 [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Cohen AC, Henneberry TJ, Chu CC (1996) Geometric relationships between whitefly feeding behavior and vascular bundle arrangements. Entomol Exp Appl 78 135–142 [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM (2005) Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA 102 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux J-P, Van Loon LC, Dicke M, et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18 923–937 [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG (2003) Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot (Lond) 92 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicke M, Hilker M (2003) Induced plant defences: from molecular biology to evolutionary ecology. Basic Appl Ecol 4 3–14 [Google Scholar]
- Durrant WE, Dong X (2004) Systemic required resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Turner JG (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15 1025–1030 [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Donato K, Del Vecchio RJ, Duffey SS (1989) Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. J Chem Ecol 15 2667–2694 [DOI] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidantsef AL, Stout MJ, Thaler JS, Duffey SS, Bostock RM (1999) Signal interactions in pathogen and insect attack: expression of lipoxygenase, proteinase inhibitor II, and pathogenesis-related protein P4 in the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol 54 97–114 [Google Scholar]
- Freeman TP, Buckner JS, Nelson DR, Chu CC, Henneberry TJ (2001) Stylet penetration by Bemisia argentifolii (Homoptera: Aleyrodidae) into host leaf tissue. Ann Entomol Soc Am 94 761–768 [Google Scholar]
- Funk CJ (2001) Alkaline phosphatase activity in whitefly salivary glands and saliva. Arch Insect Biochem Physiol 46 165–174 [DOI] [PubMed] [Google Scholar]
- Gill R (1990) The morphology of whiteflies. In D Gerling, ed, Whiteflies: Their Bionomics, Pest Status and Management. Intercept, Andover, UK, pp 13–46
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14 177–193 [DOI] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen ZY, Zhu LH, Kunkel BN, Alfano JR, Tang XY, Zhou JM (2004) Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J 37 589–602 [DOI] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 222–226 [DOI] [PubMed] [Google Scholar]
- Jiang YX, Nombela G, Muniz M (2001) Analysis by DC-EPG of the resistance to Bemisia tabaci on an Mi-tomato line. Entomol Exp Appl 99 295–302 [Google Scholar]
- Johnson D, Walker GP (1999) Intracellular punctures by the adult whitefly Bemisia argentifolii on DC and AC electronic feeding monitors. Entomol Exp Appl 92 257–270 [Google Scholar]
- Kaloshian I, Walling LL (2005) Hemipterans as plant pathogens. Annu Rev Phytopathol 43 491–521 [DOI] [PubMed] [Google Scholar]
- Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44 41–66 [DOI] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (1997) Induced Responses to Herbivory. Chicago University Press, Chicago
- Kempema LA, Cui X, Holzer FM, Walling LL (2007) Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol 143 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53 299–328 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Kus JV, Zaton K, Sarkar R, Cameron RK (2002) Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8 863–870 [DOI] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 19 655–664 [DOI] [PubMed] [Google Scholar]
- Liu T-X (2000) Population dynamics of Bemisia argentifolii (Homoptera: Aleyrodidae) on spring collard and relationship to yield in the lower Rio Grande valley of Texas. J Econ Entomol 93 750–756 [DOI] [PubMed] [Google Scholar]
- Liu YL, Ahn JE, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, Pittendrigh B, Murdock LL, Koiwa H, Zhu-Salzman K (2005) Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol 139 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Neuenschwander U, Cade RM, Dietrich RA, Dangl JL, Ryals JA (2002) Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics 160 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Ilarduya O, Xie Q, Kaloshian I (2003) Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol Plant Microbe Interact 16 699–708 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL (2000) Signal transduction in the plant immune response. Trends Biochem Sci 25 79–82 [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol 138 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles PW (1999) Aphid saliva. Biol Rev 74 41–85 [Google Scholar]
- Moran PJ, Cheng Y, Cassell JL, Thompson GA (2002) Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch Insect Biochem Physiol 51 182–203 [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56 509–531 [DOI] [PubMed] [Google Scholar]
- Musser RO, Cipollini DF, Hum-Musser SM, Williams SA, Brown JK, Felton GW (2005) Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in Solanaceous plants. Arch Insect Biochem Physiol 58 128–137 [DOI] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW (2002) Herbivory: Caterpillar saliva beats plant defences—a new weapon emerges in the evolutionary arms race between plants and herbivores. Nature 416 599–600 [DOI] [PubMed] [Google Scholar]
- Nava-Camberos U, Riley DG, Harris MK (2001) Temperature and host plant effects on development, survival, and fecundity of Bemisia argentifolii (Homoptera: Aleyrodidae). Environ Entomol 30 55–63 [Google Scholar]
- Nombela G, Williamson VM, Muniz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16 645–649 [DOI] [PubMed] [Google Scholar]
- Pechan T, Ye LJ, Chang YM, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS (2000) A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other lepidoptera. Plant Cell 12 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegadaraju V, Knepper C, Reese J, Shah J (2005) Premature leaf senescence modulated by the Arabidopsis PHYTOALEXIN DEFICIENT4 gene is associated with defense against the phloem-feeding green peach aphid. Plant Physiol 139 1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J 16 473–485 [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1 404–411 [DOI] [PubMed] [Google Scholar]
- Rojo E, Solano R, Sanchez-Serrano JJ (2003) Interactions between signaling compounds involved in plant defense. J Plant Growth Regul 22 82–98 [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477 112–121 [DOI] [PubMed] [Google Scholar]
- Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6 365–371 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Owen B, Higgins VJ (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol 135 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57 755–766 [DOI] [PubMed] [Google Scholar]
- van de Ven WTG, LeVesque CS, Perring TM, Walling LL (2000) Local and systemic changes in squash gene expression in response to silverleaf whitefly feeding. Plant Cell 12 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Weisser WW, Baldwin IT (2004) An analysis of plant-aphid interactions by different microarray hybridization strategies. Mol Ecol 13 3187–3195 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9 203–209 [DOI] [PubMed] [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19 195–216 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Bi J-L, Liu T-X (2005) Molecular strategies of plant defense and insect counter-defense. Insect Sci 12 3–15 [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn J-E, Koiwa H (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.