Abstract
In the establishment of symbiosis between Medicago truncatula and the nitrogen-fixing bacterium Sinorhizobium meliloti, the lipopolysaccharide (LPS) of the microsymbiont plays an important role as a signal molecule. It has been shown in cell cultures that the LPS is able to suppress an elicitor-induced oxidative burst. To investigate the effect of S. meliloti LPS on defense-associated gene expression, a microarray experiment was performed. For evaluation of the M. truncatula microarray datasets, the software tool MapMan, which was initially developed for the visualization of Arabidopsis (Arabidopsis thaliana) datasets, was adapted by assigning Medicago genes to the ontology originally created for Arabidopsis. This allowed functional visualization of gene expression of M. truncatula suspension-cultured cells treated with invertase as an elicitor. A gene expression pattern characteristic of a defense response was observed. Concomitant treatment of M. truncatula suspension-cultured cells with invertase and S. meliloti LPS leads to a lower level of induction of defense-associated genes compared to induction rates in cells treated with invertase alone. This suppression of defense-associated transcriptional rearrangement affects genes induced as well as repressed by elicitation and acts on transcripts connected to virtually all kinds of cellular processes. This indicates that LPS of the symbiont not only suppresses fast defense responses as the oxidative burst, but also exerts long-term influences, including transcriptional adjustment to pathogen attack. These data indicate a role for LPS during infection of the plant by its symbiotic partner.
Plants and animals have the capacity to recognize and attack pathogenic microorganisms such as fungi, viruses, or bacteria. But, in contrast to animals, plants lack a system of circulating fluids, like blood and lymph, as well as specialized immune cells. Therefore, each plant cell has to possess the ability to recognize different pathogenic molecules and to respond with a broad range of defense reactions. In recent years, it has become more and more evident that there are two general systems of plant immunity (Gómez-Gómez and Boller, 2002; Nürnberger et al., 2004). Cultivar-specific resistance is realized on a gene-for-gene basis. In this context, certain plant cultivars carrying a specific resistance gene are able to mount defense reactions against pathogenic microorganisms that carry a corresponding avirulence gene. These defense reactions usually include induction of a hypersensitive response (HR) to inhibit the spread of a pathogen as fast as possible (Dixon and Harrison, 1994). Cultivar-specific resistance is discussed to resemble the animal's adaptive immune system, although it does not involve specialized immune cells or recombinatory events on the genetic level. The second type of plant immunity is the ability to distinguish between self and nonself. This strategy is known from animals as innate immunity and this term is also applied to the plant system (Gómez-Gómez and Boller, 2002). Recognition of foreign organisms in the plant's surrounding often does not lead to such vigorous responses like the activation of a HR, but rather induces general preparations to a possible invasion by microorganisms. These defense mechanisms include induction of oxidative burst, expression of defense-related genes, production of antimicrobial compounds, like phytoalexins, and reinforcement of the plant cell wall (Benhamou, 1996; Ebel and Mithöfer, 1998; Heath, 2000). Although these responses are also activated during cultivar-specific resistance, hypersensitive cell death is absent during basal defense. The plant's innate immunity is based on recognition of microbial signal molecules, termed general elicitors or pathogen-associated molecular patterns (PAMPs). Unlike the products of avirulence genes in cultivar-specific resistance, PAMPs are expressed by a wide range of microorganisms and are usually indispensable for their biological fitness. Among the PAMPs are long-known and well-characterized elicitors like (1,3-1,6)-hepta-β-glucoside isolated from cell walls of Phytophthora sojae (Sharp et al., 1984a, 1984b), chitin from cell walls of higher fungi (Bartnicki-Garcia, 1968; Ren and West, 1992), yeast (Saccharomyces cerevisiae) invertase (Basse et al., 1992), flagellin as the first general elicitor isolated from Gram-negative bacteria (Felix et al., 1999), or lipopolysaccharides (LPS; Meyer et al., 2001; Erbs and Newman, 2003). The potential of LPS to induce defense reactions is well characterized for the animal system, but also described for plants. Pretreatment with LPS from different bacteria has been shown to prevent HR and to accelerate the production of salicylic acid in host as well as nonhost plants (Graham et al., 1977; Newman et al., 1997, 2002). Furthermore, Xanthomonas campestris pv campestris LPS is able to induce defense-related gene expression, oxidative burst, as well as calcium signaling in tobacco (Nicotiana tabacum) suspension cultures (Meyer et al., 2001; Braun et al., 2005; Silipo et al., 2005). Using LPS of Burkholderia cepacia, the induction of several early signaling and defense pathways in tobacco has been confirmed (Gerber et al., 2004). Recent studies have identified several tobacco proteins altered in their phosphorylation status in response to elicitation with B. cepacia LPS, giving valuable insights into possible signaling processes that follow the perception of the PAMP (Gerber et al., 2006).
LPS are also important signal molecules in symbiotic signaling. In the Rhizobium-legume symbiosis, evidence for the involvement of LPS in the establishment of symbiosis has been gained from mutants defective in LPS synthesis. In different kinds of Rhizobia-legume associations, alterations in the LPS structure lead to delayed nodulation, abortion of infection threads, formation of nonfixing nodules, and induction of plant defense reactions (Carlson et al., 1987; Lagares et al., 1992; Perotto et al., 1994; Niehaus et al., 1998; Campbell et al., 2002). Albus et al. (2001) showed that LPS of the microsymbiont Sinorhizobium meliloti is able to suppress elicitor-induced oxidative burst in suspension cultures of the alfalfa (Medicago sativa) host plant. Because this LPS does induce oxidative burst in cell cultures of the tobacco nonhost plant, the suppression effect is not a general feature of the rhizobial LPS, but is speculated to be a very specific feature in the establishment of a symbiotic relationship. Recent results identified the LipidA substructure of the S. meliloti LPS to be the suppressor-active portion (Scheidle et al., 2005). However, the extent to which rhizobial LPS is able to suppress the host's defense mechanisms has remained poorly characterized. To gain more information on the capacity of S. meliloti LPS to interfere with the plant's defense machinery, we performed microarray experiments to monitor the global expression of defense-related genes. Defense reactions were induced by yeast invertase (Basse et al., 1992). By treatment of suspension-cultured Medicago truncatula cells with either S. meliloti LPS and yeast invertase or yeast invertase alone, the effect of S. meliloti LPS on the level of gene expression could be addressed. Comparison of defense-related gene expression in the two kinds of treatment showed that induction of many genes strongly correlated to plant defense (e.g. Phe ammonia lyase [PAL] and isoflavone reductase [IFR]) was significantly suppressed after LPS pretreatment. Using MapMan software (Thimm et al., 2004; Usadel et al., 2005), it was furthermore possible to make a functional correlation between the suppression effect calculated for all genes displayed on the microarray and their specific function inside the cell.
RESULTS AND DISCUSSION
Invertase Induces Defense-Associated Gene Expression in M. truncatula Cell Suspension Cultures
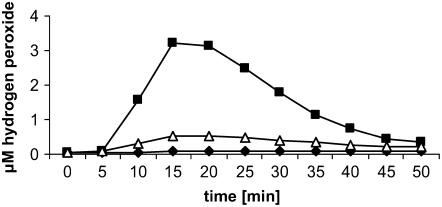
In this study, M. truncatula suspension-cultured cells derived from roots were used as a test system for the perception of microbial signal molecules. To verify the responsiveness of the cell culture, invertase was used as a well-established elicitor of plant defense reactions. In all four biological replicates used, the addition of invertase to a final concentration of 50 μg mL−1 led to the production of hydrogen peroxide (H2O2), with an average maximal concentration (±se) of 3.54 μm H2O2 (±0.15) 15 min after elicitation (Fig. 1). The oxidative burst assay was used as a test system for the uniform perception of the elicitor in different experiments. Only responsive cell cultures were used for subsequent expression profiling.
Figure 1.
Induction and suppression of oxidative burst in M. truncatula suspension-cultured cells upon treatment with invertase and invertase + S. meliloti LPS, respectively. M. truncatula cell suspensions were treated with 50 μg mL−1 invertase (black squares), 50 μg mL−1 invertase + 50 μg mL−1 S. meliloti LPS (white triangles), or water as a negative control (black diamonds). The production of H2O2 was determined utilizing the luminol-based chemiluminescence assay. Oxidative burst measured for one of the four biological replicates is shown.
For transcriptome-profiling studies, M. truncatula cell suspensions were treated for 4 h with either invertase, S. meliloti LPS, invertase + S. meliloti LPS, or water as a control. Cells were harvested, frozen, and processed for transcriptome profiling. Based on analysis of all four biological replicates in comparison to water-treated suspension cells, LPS treatment alone hardly showed any effect on gene expression in M. truncatula cell cultures. A complete list of genes meeting the prerequisites P ≤ 0.05, A ≥ 5.5, and n ≥ 6 (P, A, and n describe the statistical relevance, signal intensity, and the number of agreeing replicates, respectively) is included in Supplemental Table S1. No gene in this list was identified as induced 2-fold or more and only two genes (tentative consensus [TC]88927 and TC78069) were repressed more than 2-fold. These results indicate that M. truncatula cell cultures do not respond strongly to the LPS of the symbiotic partner. In contrast, in suspension cells treated with invertase, 336 genes were identified to be at least 2-fold induced with a statistical significance of P ≤ 0.05, whereas 43 genes were identified to be repressed at least 2-fold. Table I lists the 20 genes that are most strongly induced or repressed, respectively, and the complete list of genes meeting the prerequisites P ≤ 0.05, A ≥ 5.5, and n ≥ 6 is included in Supplemental Table S1.
Table I.
Overview of the effect of invertase treatment on M. truncatula suspension cells
Listed are the 20 most strongly induced and repressed genes obtained from transcriptomic profiling of invertase-treated cells in comparison to water-treated cultures. Suspension-cultured M. truncatula cells were treated for 4 h with either 50 μg mL−1 invertase or water as a reference. Oligo ID, Identifier of M. truncatula 70-mer oligunucleotides; TIGR ID, identifier in the TIGR M. truncatula gene index 7.0; annotation, annotation in the TIGR M. truncatula gene index 7.0; INV and INV + LPSSm, log2 expression ratios in invertase and invertase + S. meliloti LPS-treated suspension cells in comparison to a water-treated control.
| Oligo ID | TIGR ID | Annotation | INV | INV + LPSSm |
|---|---|---|---|---|
| MT014241 | TC85478 | IFR | 3.970 | 3.029 |
| MT014270 | TC85521 | CHR | 3.541 | 1.912 |
| MT015080 | TC85174 | CHS4 | 3.457 | 2.094 |
| MT000150 | TC85477 | IFR | 3.373 | 2.313 |
| MT015632 | TC87485 | Germin-like protein | 3.361 | 2.410 |
| MT015147 | TC76769 | CHS4 | 3.352 | 2.234 |
| MT008395 | TC78488 | Germin-like protein | 3.290 | 1.951 |
| MT014469 | TC86777 | Cyanogenic β-glucosidase | 3.289 | 1.967 |
| MT015072 | TC85146 | CHS2 | 3.271 | 2.018 |
| MT015146 | TC85138 | CHS8 | 3.260 | 2.059 |
| MT015144 | TC76770 | Chain A CHS | 3.243 | 2.064 |
| MT015149 | TC76765 | Putative CHS | 3.213 | 1.932 |
| MT015480 | TC77750 | Chitinase | 3.084 | 2.059 |
| MT015145 | TC76767 | CHS2 | 3.045 | 1.853 |
| MT015083 | TC85174 | CHS4 | 3.042 | 1.885 |
| MT015076 | TC85150 | CHS9 | 2.795 | 1.802 |
| MT000333 | TC85502 | PAL | 2.762 | 1.986 |
| MT000259 | TC76871 | Hypothetical protein | 2.723 | 1.529 |
| MT007784 | TC86455 | IFR | 2.671 | 1.415 |
| MT000494 | TC86191 | Hypothetical protein | 2.634 | 1.477 |
| MT000936 | TC78069 | Oligo-TC | −2.539 | −2.715 |
| MT007017 | TC85263 | Putative plasma membrane intrinsic protein | −1.786 | −1.283 |
| MT007036 | TC76601 | Aquaporin-like protein PIP2 | −1.751 | −0.993 |
| MT008134 | TC78229 | α-Xylosidase precursor | −1.678 | −1.427 |
| MT007395 | TC85871 | Pectate lyase | −1.669 | −0.959 |
| MT014301 | TC76880 | Endoxyloglucan transferase | −1.662 | −1.131 |
| MT016122 | Autointerpro | MYB-related protein… | −1.563 | −0.681 |
| MT009717 | TC79773 | AtHVA22a 65476-64429 | −1.472 | −0.863 |
| MT013556 | TC80306 | Hypothetical protein | −1.462 | −0.784 |
| MT015781 | TC81234 | Unknown | −1.460 | −0.329 |
| MT010917 | TC82525 | Unknown | −1.442 | −0.517 |
| MT014300 | BQ136812 | Xyloglucan endotransglycosylase | −1.372 | −0.902 |
| MT011472 | TC82617 | AT5g64260/MSJ110 | −1.315 | −1.191 |
| MT002409 | TC88503 | Endo-β-1,4-glucanase | −1.281 | −0.804 |
| MT001311 | TC87188 | At2g17230/T23A1.9 | −1.277 | −0.929 |
| MT016449 | Autointerpro | Leu-rich repeat-containing protein | −1.266 | −1.093 |
| MT015278 | TC76902 | MtN5 | −1.234 | −1.031 |
| MT001226 | TC87263 | Unknown | −1.233 | −0.880 |
| MT007159 | TC85575 | Protein F3M18.16 | −1.230 | −1.083 |
| MT013339 | TC84195 | Putative β-galactosidase | −1.218 | −0.838 |
Fourteen of the 20 most strongly induced genes are involved in plant secondary metabolism (e.g. IFR, chalcone synthase [CHS], chalcone reductase [CHR], and PAL). The induction of the secondary metabolism is a plant defense response well described for alfalfa, a very close relative of M. truncatula as well as for other plant systems (Kuhn et al., 1984; Dalkin et al., 1990; Ni et al., 1996; Suzuki et al., 2005; for a recent review, see Dixon et al., 2002). PAL is the branch point enzyme transforming l-Phe to trans-cinnamic acid, a precursor molecule for the formation of lignin as well as the isoflavonoid-derived phytoalexin medicarpin (Dalkin et al., 1990). Biosynthesis of the latter compound also involves the activity of CHS, CHR, and IFR.
Another gene strongly induced after elicitation with invertase was a chitinase involved in the defense against fungal pathogens. Furthermore, transcripts encoding two germin-like proteins were highly abundant in the elicited cell cultures. Germin-like proteins are ubiquitous plant proteins encoded by diverse multigene families. Although the function of most germin-like proteins could not be elucidated so far, nectarin I, a germin-like protein present in the nectar of tobacco plants, was found to exhibit superoxide dismutase activity (Carter and Thornburg, 2000). Additionally, a gene encoding a cyanogenic β-glucosidase was strongly induced after elicitation. Cyanogenesis has been described in the context of protection against herbivores, but, due to the high toxicity of HCN, other roles in plant defense were postulated (Poulton, 1990).
Six of the 20 most repressed genes encode proteins involved in cell wall degradation. Because reinforcement of the plant cell wall is one of the major responses upon pathogen attack, it was not unexpected to find the expression of degradative enzymes repressed.
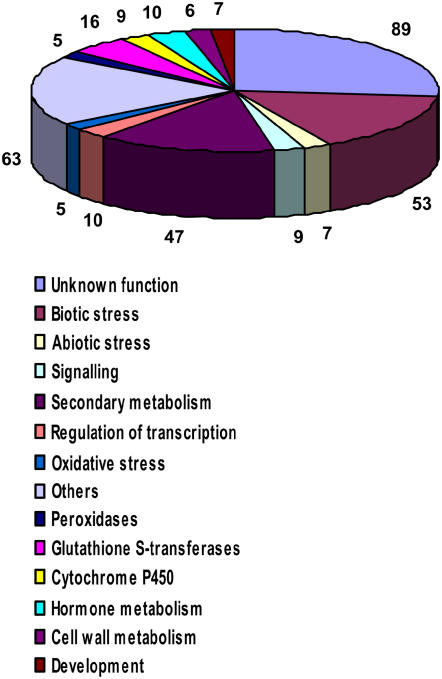
Manual classification of transcription-profiling data according to published literature revealed that, among the 336 genes induced 2-fold or more after elicitation with invertase, more than 170 have been described to be involved in plant defense or induced under biotic stress conditions. Grouping of all genes induced at least 2-fold into functional categories enables the illustration of different plant responses upon elicitation (Fig. 2). These include expression of several signaling components, activation of different hormone pathways, expression of proteins involved in protection from oxidative stress, as well as induction of transcriptional regulators. Besides the two largest categories that are composed of genes with unknown function and those that did not fit into any of the given functional categories (others), two categories (secondary metabolism and biotic stress) comprise 14% and 16% of all genes induced at least 2-fold, respectively. These findings are in general agreement with previous studies (Suzuki et al., 2005) and confirm the responsiveness of the utilized M. truncatula suspension cell line on the transcriptome level.
Figure 2.
Classification of the 336 genes found to be at least 2-fold induced in M. truncatula suspension cells upon 4-h treatment with 50 μg mL−1 invertase. The genes classified had to meet the prerequisites −1 ≥ M ≥ 1, P ≤ 0.05, A ≥5.5, and n ≥ 6. A complete list of the classified genes is given in Supplemental Table S1. The number of genes allocated to each functional category is indicated, the functional categories are defined.
The Visualization Tool MapMan Allows Fast and Easy Evaluation of Transcriptome-Profiling Datasets on the Functional Level
Transcriptome-profiling studies can give valuable information on global cellular responses; however, evaluation and interpretation of the datasets are time consuming and require detailed knowledge of the existing literature. Bioinformatic tools can help to organize datasets according to preexisting biological knowledge and therefore enable comprehensive interpretation on the functional level. MapMan is a visualization tool developed to display plant genomic datasets onto pictorial diagrams (Thimm et al., 2004). The software organizes genes into blocks of cognate functions to allow even tentatively assigned genes and individual members of gene families to be displayed. This structure allows detection of general trends in the system's response and, in cases where the ontology is further developed, more precise resolution at the single pathway level. Recently, MapMan has been extended to enable statistical evaluation of differences in the responses of sets of genes assigned to different biological functions, allowing more validated interpretation of global cellular responses (Usadel et al., 2005). Because MapMan was initially designed to process datasets obtained from Arabidopsis (Arabidopsis thaliana) ATH1 microarrays, adaptation of this tool for employment with other microarray formats and plant species was an important step. At the beginning of the year, Urbanczyk-Wochniak et al. (2006) published the extension of MapMan for displaying Solanaceous datasets. The basis for this adaptation is the classification of the genes represented on the microarray according to the ontology used in MapMan.
To use MapMan as a tool to visualize transcriptomic datasets obtained from M. truncatula Mt16kOLI1 and Mt16kOLI1Plus microarrays, we assigned the 16,470 TCs represented on the Mt16kOLI1Plus array to MapMan BINS and subBINS based on a combination of automated BLAST searches against The Arabidopsis Information Resource 6.0 database and protein domain information. TCs lacking a good hit in Arabidopsis were assigned manually according to The Institute for Genomic Research (TIGR) annotation. It has to be mentioned that the ongoing sequencing efforts and annotation updates providing new information on the M. truncatula genome led to an urgent need to constantly update the classification of the set of TCs. Furthermore, M. truncatula as a legume plant poses a challenge in the need to assign genes with a function in nodulation, a class of genes that is absent in Arabidopsis. To account for this legume-specific class of genes, a new subBIN, named nodulins (subBIN 26.31), was defined. Even though this BIN was not displayed in the visualization of the datasets presented here, it is a crucial prerequisite for the evaluation of data obtained from symbiotically active Medicago plants. Because MapMan is designed to allow for the creation of user-defined diagrams and visualization maps, this tool is well suited for employment in a symbiotic context.
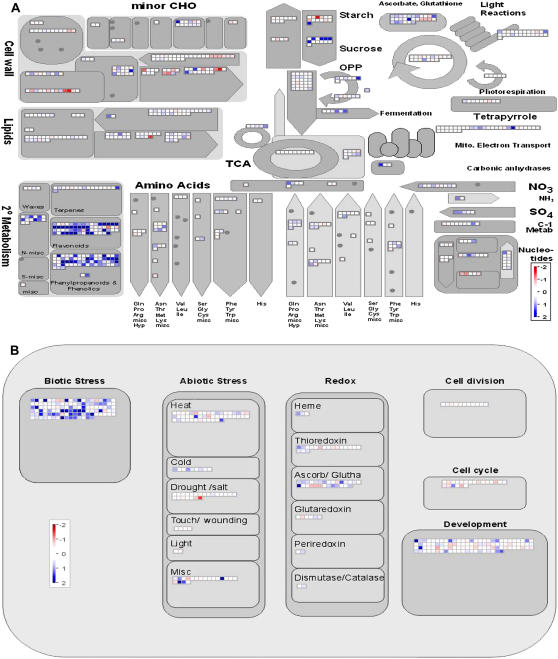
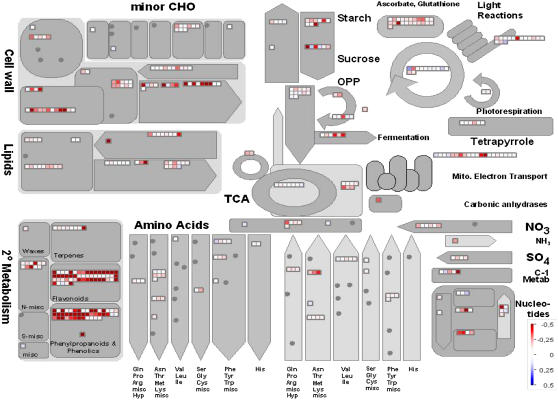
To demonstrate the value of the visualization of transcriptome data using MapMan, the dataset obtained from invertase-treated suspension cell cultures described above was used for a proof-of-concept study. In Figure 3, two different images are shown: one representing a general overview of the cell's basic metabolic pathways (Fig. 3A) and the other displaying different cellular responses (Fig. 3B). As discussed above, induction of secondary metabolism is one of the most noticeable responses in elicited M. truncatula cell cultures. Visualization of the cell's general metabolism confirms this cellular response (Fig. 3A). Furthermore, organization into hierarchical modules of biological relevance facilitates a more detailed resolution of global responses. For example, the three modules representing the secondary metabolism pathways flavonoids, phenylpropanoids, and phenolics, as well as N-misc, are clearly induced in this MapMan visualization. The strong induction of the pathways involved in lignification and phytoalexin synthesis was detected by standard evaluation methods as described above, whereas induction of the alkaloid metabolism (N-misc) had remained undetected. According to the M. truncatula TIGR gene index 7.0, most Medicago genes contained in this BIN cannot be assigned unambiguously to a specific cellular function, rendering detection of an induction in alkaloid biosynthesis via standard evaluation methods impossible. Nevertheless, automatic assignment of Medicago genes to the corresponding BIN as described in “Materials and Methods” leads to the identification of otherwise undetected response patterns, such as induction of the alkaloid pathway highlighting the applicability of the gene classification used to employ MapMan for the evaluation of Medicago datasets. Stimulation of alkaloid formation upon elicitation with fungal signal molecules has been shown before for different plant species (Blechert et al., 1995). Although production of alkaloids in Arabidopsis is poorly understood, induction of the berberine biosynthetic pathway upon wounding was described (Cheong et al., 2002). Therefore, induction of alkaloid metabolism upon elicitation can also be expected in M. truncatula. Another conspicuous response of invertase-treated cell cultures is the transcriptomic rearrangement of cell wall metabolism. In this context, the mixture of induced and repressed genes can be easily interpreted by closer examination of this module. Genes encoding for pectin methylesterases were induced upon elicitation with invertase. The transcriptional activation of pectin methylesterases upon herbivore attack has been shown before (von Dahl et al., 2006). In this context, demethylation of the cell wall pectin has been proposed to enable cross-linking via calcium ions, resulting in reinforcement of the cell wall. Other enzymes transcriptionally up-regulated upon invertase treatment are cellulose synthases and polygalacturonase inhibitor proteins, whereas proteins involved in cell wall degradation (i.e. cellulases, polygalacturonases, etc.) are repressed. Cell wall reinforcement is known to be a crucial response upon pathogen attack, but transcriptional initiation of this process besides the activation of lignifying enzymes or other secondary metabolic pathways has been rarely reported (Schenk et al., 2000). Another response that has been proposed to be induced following pathogen attack is the degradation of fatty acids by β-oxidation. Although the cluster of lipid metabolism did not show any marked changes, closer examination of the transcriptional changes of this cluster reveals a minor induction of the β-oxidation pathway, as well as of the glyoxylate cycle. This is in accordance with a previous study that monitored transcriptional adaptations upon attempted infection of Arabidopsis with Pseudomonas syringae pv tomato (Scheideler et al., 2002). Although it is tempting to speculate that mobilization of fatty acids might be utilized for generation of energy, it should be emphasized that, on the other hand, our data indicate a repression of starch degradation. This could hint to a role of β-oxidation in processes different from energy generation, like the provision of precursor molecules for biosynthetic pathways. Furthermore, our data show induction of the oxidative pentose phosphate pathway that has been shown to be essential in supplying NADPH for the activity of NADPH oxidase during oxidative burst (Pugin et al., 1997). Another response toward elicitation is induction of the cluster representing the synthesis of Asp family amino acids. This pattern is mainly caused by the induction of several S-adenosyl-methionine synthetase enzymes involved in the production of the phytohormone ethylene that has been identified as one of the key players in orchestrating plant disease resistance (Feys and Parker, 2000). Finally, a trend in the induction of transcripts involved in glutathione and ascorbate metabolism can be detected. Glutathione and ascorbic acid are indispensable for the protection of plant cells from oxidative stress. Even though the elicitor-induced oxidative burst is an important element in plant defense and defense-associated signaling, scavenging of reactive oxygen intermediates (ROI) is a crucial process to prevent oxidative damage of cellular compounds or induction of programmed cell death (Mittler, 2002; Apel and Hirt, 2004). Figure 3B visualizes transcriptomic datasets with respect to different cellular responses. In this representation, the most striking response observed was for a class of genes assigned as involved in biotic stress. In fact, only five of these genes appear to be suppressed upon invertase-mediated elicitation of cell cultures. Three of these are annotated as protease inhibitor proteins and could possibly be involved in mechanisms different from inhibition of proteases of pathogenic origin. Other patterns that can be observed are the induction of several genes involved in the response to different kinds of stress apart from drought and salt stress. Cross talk between the responses toward different kinds of stresses is a frequent observation, although stronger induction of wound-induced responses could have been expected (Desikan et al., 2001; Cheong et al., 2002). The class of miscellaneous genes involved in the response to abiotic stress contains four highly induced genes. All four are annotated as encoding germin-like proteins, which are well known to be elicitor-inducible proteins. The above-discussed induction of genes involved in protection from oxidative stress is visualized in more detail in Figure 3B, demonstrating that, apart from ascorbate and glutathione metabolism, little transcriptional reprogramming can be observed upon elicitation. Induction of antioxidative proteins like dismutases and catalases is missing despite the potential need for scavenging ROI produced during oxidative burst. This could hint to a role of ascorbate and glutathione cycle intermediates in addition to detoxification of ROI. It has been proposed that glutathione itself is involved in signaling events leading to the activation of plant defense responses (Dron et al., 1988; Gomez et al., 2004). On the other hand, a function as a superoxide dismutase has been described for a germin-like protein from the tobacco nectar, nectarin I. Although such a function remains purely speculative, the above-described induction of four germin-like proteins could be due to their function in the scavenging of ROI. Finally, some transcriptional rearrangements can be observed in the cluster of development-associated genes predominantly showing repression of this class. The transcriptomic adaptation of genes involved in cell maintenance and development has been described before (Schenk et al. 2000). However, no agreement on the single gene level could be made by comparing our results to the list of regulated genes described earlier.
Figure 3.
Changes in transcript levels in invertase-treated M. truncatula suspension cell cultures. Cells were elicited for 4 h using 50 μg mL−1 invertase. Displayed are genes associated with general metabolism (A) and cellular responses (B). Red and blue displayed signals represent a decrease and an increase in transcript abundance, respectively, relative to water-treated control cells. The scale used for coloration of the signals (log2 ratios) is presented. The image can be viewed and descriptions for each signal accessed under http://gabi.rzpd.de.
In summary, the clear patterns of plant response upon elicitation visualized using MapMan are in accordance with existing literature and validate the usefulness of this software tool for the evaluation of transcriptome datasets. Nevertheless, it has to be emphasized that interpretation of the results leads to identification of genes, which did not follow the general trend observed in the class to which they were assigned. This can be explained either by opposing regulation of transcripts marking isoenzymes with different cellular functions or by assigning such genes to a wrong BIN.
The LPS of the Microsymbiont S. meliloti Suppresses Elicitor-Induced Transcriptional Reprogramming
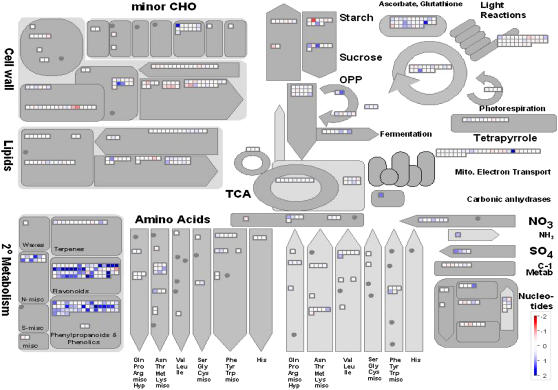
To investigate the effect of S. meliloti LPS on defense-associated gene expression in M. truncatula, we treated suspension cell cultures simultaneously with invertase and S. meliloti LPS with final concentrations of 50 μg mL−1 each. This concentration suppresses the invertase-induced oxidative burst in M. truncatula suspension-cultured cells to nearly 90% (Fig. 1). In contrast, concomitant treatment of cell cultures with invertase and LPS of the phytopathogen X. campestris pv campestris did result in oxidative burst with 91% maximal H2O2 production as compared to elicitation with invertase alone (Supplemental Fig. S1). These results indicate that the suppression effect is a specific feature of the M. truncatula-S. meliloti (legume-Rhizobia) interaction, which is further substantiated by elicitation of oxidative burst by S. meliloti LPS in tobacco cells (Albus et al., 2001). For transcriptional profiling analyses, cells were incubated with invertase and S. meliloti LPS for 4 h, separated from the medium, frozen, and RNA extracted and processed for microarray hybridizations. Transcriptomic reprogramming induced by this treatment combining pathogenic and symbiotic stimulus was compared to the transcriptome profiles obtained by elicitation with invertase alone. Comparison of the two datasets was made possible by using the same RNA of water-treated cells as a reference. A list of all genes in the dataset obtained from the combined treatment meeting the prerequisites P ≤ 0.05, A ≥ 5.5, and n ≥ 6 is included in Supplemental Table S1. Fourteen of the 20 most strongly induced genes are also among the 20 genes with highest induction rates after elicitation with invertase listed in Table I. Comparison of gene induction rates given in Table I shows that, for all 20 genes most strongly induced upon invertase treatment, there is also strong induction following combined treatment with invertase and S. meliloti LPS. Nevertheless, in each case, the induction rate is markedly reduced, giving a first hint to moderating activity of the rhizobial LPS. The same effect can be observed for 19 of the 20 genes most strongly repressed upon elicitation with invertase. To investigate general patterns of cellular adaptation induced by simultaneous subjection to invertase and S. meliloti LPS, the dataset was visualized using MapMan. Figure 4 shows a representation of the cell's basic metabolic pathways in response to the combined treatment. The overall response shows strong induction of secondary metabolism, some rearrangements in cell wall metabolism, and induction of several transcripts involved in ascorbate and glutathione metabolism. In general, the responses observed upon treatment with invertase and rhizobial LPS resembled transcriptional rearrangements followed by elicitation with invertase alone. Nevertheless, the described response patterns are induced to a lesser extent in the presence of S. meliloti LPS. To investigate a possible suppression effect of the rhizobial LPS on the global level, the differences between induction rates for each gene of both datasets were calculated. Genes for which rhizobial LPS had a moderating effect on transcriptional adjustment were characterized by negative values, whereas for genes that were even more strongly altered after simultaneous treatment with invertase and S. meliloti LPS compared to elicitation with invertase alone, the calculated values were positive. To analyze the global effects of S. meliloti LPS on defense-associated gene expression, the resulting values for differences in transcriptional response ratios were visualized by MapMan, using a representation of the cell's general metabolism (Fig. 5). It has to be emphasized that, unlike the diagrams discussed before, this representation does not visualize an unprocessed microarray dataset, but the calculated differences between two datasets that were compared. The comparison of the two datasets clearly showed that S. meliloti LPS has a moderating effect for nearly all genes that are displayed in Figure 5. This affects genes that are induced by invertase elicitation as well as genes that are down-regulated in response to the pathogenic stimulus. The visualization shows very few genes for which the rhizobial LPS seems to have a stimulating effect on defense-associated transcriptional rearrangement. A closer examination of these genes showed that, with one exception (TC80744, a gene annotated as unknown), the relative response ratios for these transcripts were comparably low in both datasets that were compared. Thus, the calculated positive values for the differences in response ratios could be due to the inaccuracy of the M values obtained in the microarray experiments. Of the 323 genes induced 2-fold or more after invertase treatment, which were compared to the dataset of cells treated with invertase and S. meliloti LPS, only 19 transcripts showed a positive value for the differences in relative response ratios. For all other genes, the calculated values indicated a moderating effect of rhizobial LPS on elicitor-induced transcription activity (i.e. values of differences in relative response ratios were negative). Nevertheless, the datasets obtained by the microarray experiment do not indicate a fixed degree for the suppression of transcriptional reorganization. Closer examination of nodule-specific genes did confirm the suppression effect of S. meliloti LPS on those nodulins that were induced upon invertase treatment (e.g. TC77131 encoding a nodulin 12A precursor). Nevertheless, significant transcriptional rearrangements in response to rhizobial LPS alone could not be observed.
Figure 4.
Changes in transcript levels in M. truncatula suspension cell cultures treated simultaneously with invertase and S. meliloti LPS relative to water-treated control cells. Cell cultures were subjected for 4 h to treatment with 50 μg mL−1 invertase and S. meliloti LPS each. Displayed are genes associated with general metabolism. Red and blue displayed signals represent a decrease and an increase in relative transcript abundance, respectively. The scale used for coloration of the signals (log2 ratios) is presented. The image can be viewed and descriptions for each signal accessed under http://gabi.rzpd.de.
Figure 5.
Visualization of the suppression effect of S. meliloti LPS on defense-associated gene transcription in M. truncatula cell cultures. Represented are the differences between the relative response ratios of invertase-treated (50 μg mL−1) suspension cells and cell cultures treated with 50 μg mL−1 invertase and 50 μg mL−1 S. meliloti LPS each. Red and blue displayed signals represent a moderating or increasing effect of the rhizobial LPS on elicitation-induced transcriptomic reorganization, respectively. The scale used for coloration of the signals (differences of the log2 ratios of the compared datasets) is presented. The image can be viewed and descriptions for each signal accessed under http://gabi.rzpd.de.
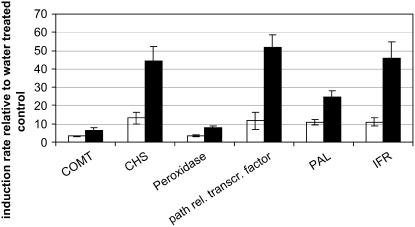
To confirm the moderating effect of rhizobial LPS on elicitor-induced transcriptional regulation, we monitored gene induction of eight genes using real-time reverse transcription (RT)-PCR. TCs selected for this approach included six genes induced upon invertase treatment and two genes that were repressed after elicitation and for which the microarray approach had shown a strong suppression effect mediated by S. meliloti LPS. It should be mentioned that, for the peroxidase gene (TC85174), no unique primers could be designed and melting curves indicated amplification of at least two different transcripts. Therefore, real-time RT-PCR monitored the abundance of a mixture of at least two different peroxidase genes. For all six TCs that were identified via microarray hybridization to be induced upon elicitation, induction could be confirmed (Fig. 6). Furthermore, induction of the genes was significantly suppressed in cells treated simultaneously with invertase and S. meliloti LPS (P ≤ 0.05; t test). Regarding the two TCs that were identified to be repressed upon invertase treatment only for TC81805 encoding a MYB-related transcription factor, repression of transcriptional activity could be confirmed. For both of these genes, rhizobial LPS did not show any significant moderating effect on transcriptional regulation (data not shown). Therefore, the suppression effect could only be confirmed for genes that are induced during elicitation, although the calculated differences in relative response ratios visualized in Figure 5 clearly indicate that S. meliloti LPS has a moderating effect on both induction and repression of genes.
Figure 6.
Relative suppression of invertase-induced gene regulation in M. truncatula mediated by S. meliloti LPS. Regulation of six genes (see “Materials and Methods”) that were confirmed via microarray hybridization to be regulated upon invertase treatment was monitored via real-time RT-PCR. Transcript abundance of cells treated with 50 μg mL−1 invertase or with 50 μg mL−1 invertase and 50 μg mL−1 S. meliloti LPS each were determined relative to water-treated control cells and normalized against mtTefα gene expression. Black and white columns represent relative induction rates of cell cultures treated with invertase or invertase and S. meliloti LPS, respectively. Experiments were performed in four biological and two technical replicates. Presented here are the arithmetic mean and the se of the data. COMT, Caffeic acid O-methyltransferase; path. rel. transcr. factor, pathogenesis-related transcription factor.
These results show two important characteristics of S. meliloti LPS-mediated suppression of defense-associated gene expression: (1) Global visualization of the differences in relative transcript levels of cells elicited with invertase and cells concomitantly treated with invertase and S. meliloti LPS shows that LPS has a moderating effect on the transcriptional adaptation of genes induced as well as on those repressed upon elicitation; (2) the suppression effect applies to all kinds of cellular processes, indicating early interference of LPS with signal transduction pathways.
However, by which means rhizobial LPS targets transmission of recognition of a pathogenic signal molecule into plant defense responses remains elusive. A different kind of suppressor molecule of rhizobial origin, the cyclic 1,3-1,6-β-glucans from the soybean (Glycine max) symbiont Bradyrhizobium japonicum have been shown to succeed in competition with the β-glucan elicitor of P. sojae for its binding sites (for review, see Mithöfer, 2002). This competitive binding not only inhibits the production of phytoalexins (Mithöfer et al., 1996), but also earlier defense-associated signaling events (i.e. an increase in cytosolic Ca2+ concentration [Mithöfer et al., 1999]). However, unlike S. meliloti LPS, the cyclic β-glucans from B. japonicum do not suppress, but rather induce, production of ROI in the host plant.
Another mechanism that could be responsible for S. meliloti LPS-mediated suppression of plant defense is the interference with basal elements of the signal transduction pathways responsible for the activation of defense responses. In the case of the phytopathogenic fungus Mycosphaerella pinodes, it has been shown that a glycopeptide termed supprescin is able to suppress plant defense at different levels, including induction of PAL transcription (Wada et al., 1995). The mechanism of suppression is likely to involve interference of the supprescin glycopeptide with a plasma membrane ATPase via direct interaction (Kato et al., 1993). Furthermore, the supprescin of M. pinodes does interfere with proteinase inhibitor metabolism in the plant plasma membrane that is essential for activation of defense responses (Shiraishi et al., 1994). It has been shown in studies on pea (Pisum sativum) plasma membranes and epicotyls that elicitation leads to a rapid and biphasic increase in levels of phosphatidylinositol-4,5-bisP and inositol 1,4,5-triP, whereas addition of the suppressor molecule inhibited this effect (Toyoda et al., 1992, 1993). These findings show that inhibition of compounds of very early signaling cascades is used by pathogens as a comprehensive mechanism of suppression. Another possibility as to how suppressors could operate in interfering with defense responses is active recognition of molecules via specific receptors. This idea has been proposed for the glycopeptide supprescin from M. pinodes by Shiraishi et al. (1994), although to date there is no proof for such a mode of action. However, specific recognition of suppressors via receptor proteins could mediate fine-tuned regulation of defense responses that are switched on or off upon concomitant recognition of an elicitor and a suppressor. In contrast, inhibition of early signaling processes necessarily blocks all downstream processes without any possibility of regulation. In the case of suppressors of pathogenic origin, the presence of a receptor protein without any other function appears unlikely because it would cause an immense evolutionary disadvantage, rendering the plant susceptible to pathogens producing the appropriate suppressor molecule. Nevertheless, in the case of symbiotic interactions, the presence of receptors specific for suppressors of symbiotic origin would enable different kinds of beneficial responses, including fine-tuned suppression of plant defense.
Surface polysaccharides act as bacterial signals in pathogenic and symbiotic interactions (Djordjevic et al., 1987). Although, to date, the role of the rhizobial LPS in the establishment and maintenance of symbiosis could not be elucidated in detail, there is evidence for its importance in the interaction. LPS mutants of Rhizobium leguminosarum as well as S. meliloti show severe defects in the establishment of symbiosis with their host plants (Vedam et al., 2004). In some cases, induced nodules are small, poorly infected, and show signs of plant defense (e.g. accumulation of cell wall material and phenolic compounds and autofluorescence [Perotto et al., 1994; Niehaus et al., 1998]). In both studies, bacteria were able to induce and colonize infection threads, but failed to differentiate after infecting nodule tissue. The death of rhizobial mutants during the infection process was thought to be due either to impairment of bacterial protection against unfavorable conditions or to inability to interfere with the host's defense response. Another mutant study showed that not only the presence, but also the correct modification, of LPS is needed for optimal nodulation activity (Keating et al., 2002). Furthermore, purified LPS of the microsymbiont has been shown to promote infection in the case of the R. leguminosarum bv trifolii—white clover (Trifolium repens) symbiosis—and to cause the production of novel proteins (Dazzo et al., 1991). The principle of the ability of rhizobial LPS to suppress plant defense responses in the host was achieved by demonstrating the suppression effect on elicitor-induced oxidative burst (Albus et al., 2001). Nevertheless, it is known that, during symbiotic interaction, prolonged production of H2O2 can be detected in the infection threads and the infection zone of the nodule, a process most likely involved in regulation of nodulation (Santos et al., 2001). The fact that oxidative burst during the infection process does not lead to abortion of the symbiotic interaction is due to the high activity of antioxidative proteins (Dalton et al., 1998). Therefore, it was a crucial step to demonstrate the suppression of plant defense responses different from oxidative burst. This study demonstrates that S. meliloti LPS has the capacity to interfere with the transcriptional response toward pathogenic signaling, further emphasizing the importance of LPS during successful symbiotic interaction.
MATERIALS AND METHODS
Plant Material
Medicago truncatula Jemalong cell suspension cultures were obtained and maintained as described by Scheidle et al. (2005).
Cultivation of Bacteria and Isolation of LPS
Sinorhizobium meliloti wild-type strain 2011 (Casse et al., 1979) was grown on tryptone yeast (Saccharomyces cerevisiae) agar plates supplemented with 0.4% Glc (w/v) at 28°C for 3 d. Bacteria were washed from the plates with 0.9% NaCl (w/v) and centrifuged at 5,000g and 4°C for 20 min. Cells were washed three times with cold water, resuspended in water, and LPS was extracted via the hot phenol-water method (Westphal and Jann, 1965). The water phase was dialyzed for 3 d against water. Proteins and nucleic acids were removed by treatment with 100 μg mL−1 DNase I (Serva), 15 μg mL−1 RNase (Serva), and 150 μg mL−1 proteinase K (Roche). Fractions were dialyzed against water for 2 d and lyophilized. The lyophilisate was resuspended in water and purified by ultracentrifugation at 100,000g and 4°C for 8 h. Centrifugation was repeated and the pellet resuspended in water and lyophilized. For the experiments, the LPS was dissolved in water and ultrasonicated on ice for 20 min.
Treatment of Plant Cell Cultures
For each biological replicate of microarray hybridizations, an appropriate number of M. truncatula suspension cell cultures was pooled. A sample of the pooled cells was sieved and 4 × 2 g of cells were removed for oxidative burst measurements. The remaining cell suspensions were realiquoted to sterile 100-mL flasks and incubated under standard cultivation conditions for 4 to 5 h. After determination of oxidative burst using the cells removed from the pooled cultures had confirmed responsiveness of the suspension cells, realiquoted cultures were treated with water (500 μL), 50 μg mL−1 yeast invertase (Sigma), 50 μg mL−1 S. meliloti LPS, or a combination of 50 μg mL−1 invertase and 50 μg mL−1 S. meliloti LPS. After treatment for 4 h, each cell culture was sieved to remove the cells from the medium and cells were rapidly frozen in liquid nitrogen. Four independent biological replicates were used for subsequent isolation of RNA.
Oxidative Burst Measurements
To confirm responsiveness of the cell suspensions used for microarray hybridization, 2 g of cells were resuspended in 8 mL of preincubation medium (3% Suc [w/v] in 4% Murashige and Skoog [v/v]; Murashige and Skoog, 1962). Cells were incubated under culture conditions for 3 h. Oxidative burst was determined using the H2O2-dependent chemiluminescence reaction according to Warm and Laties (1982). After cell cultures were subjected to one of the treatments used in these experiments, 200 μL of suspension-cultured cells were added to 700 μL 50 mm phosphate buffer (pH 7.9) and 100 μL 1.2 mm luminol (Sigma). The reaction was started by adding 100 μL of 14 mm potassium-hexacyanate (Fluka). Luminescence was monitored using a Sirius luminometer (Berthold).
RNA Extraction and Microarray Hybridization
Total RNA from suspension-cultured cells was extracted using TRIzol reagent (Sigma) and purified using Microcon-30 columns (Millipore). Twenty micrograms of RNA were used to synthesize Cy3- and Cy5-labeled cDNA as described by Hohnjec et al. (2005). Microarray analysis was performed using Mt16kOLI1Plus microarrays. The microarray is based on the Mt16kOLI1 microarray described by Hohnjec et al. (2005), but carries an additional set of 384 oligonucleotides. The array definition file of the microarray can be viewed at http://www.ebi.ac.uk/arrayexpress (accession no. A-MEXP-138). Shortly, the microarray is composed of 16,470 70-mer oligonucleotide probes representing all TCs of TIGR M. truncatula gene index, release 5.0, and several controls as well as 384 oligomers representing transcription factors and other regulators. Each probe, as well as the controls, is spotted in two replicates per microarray. Hybridization of targets, image acquisition, and analysis of image data were performed in accordance with Hohnjec et al. (2005). Data files were analyzed using EMMA 2.0 software (Dondrup et al., 2003). Lowess normalization was performed using a floor value of 20 and a t test was applied to identify differentially expressed genes.
Microarray analysis was performed using four independent biological replicates per experimental treatment hybridized to four independent microarrays, each facilitating two technical replicates. Complete transcriptome profile datasets can be viewed at http://www.ebi.ac.uk/arrayexpress (accession no. E–MEXP–924).
Adaptation of the Software Tool MapMan to the M. truncatula System
To adapt MapMan software (Thimm et al., 2004; Usadel et al., 2005; http://gabi.rzpd.de/projects/MapMan) to the M. truncatula Mt16kOLI1Plus microarray, the 34 major MapMan BINs and their subBINS used for the classification of Arabidopsis (Arabidopsis thaliana) genes were transferred to the M. truncatula system. Additionally, the subBIN 26.31, misc.nodulins, was added for the nodulins not appearing in Arabidopsis. The predicted proteins encoded by the 16,470 TC sequences present on the M. truncatula microarray were compared to Arabidopsis sequences (The Arabidopsis Information Resource release 6.0) using an initial arbitrary E value cutoff of 1 e−10. In addition, Interpro domains were identified and used for functional classification.
The classification based on the similarity to Arabidopsis proteins was then automatically checked against domains that have been assigned to a MapMan category. Furthermore, Medicago TCs that did not meet the prerequisites for an automatic draft assignment were classified manually according to their respective TIGR annotation (TIGR release 7.0). Finally, all draft assignments were corrected manually for potential mistakes.
RT-PCR
To validate results obtained by microarray hybridizations, RT-PCR was performed for eight genes using gene-specific primers (Supplemental Table S2). Peroxidase-specific primers (TC 85182) were not unique and led to the amplification of at least two different transcripts. Primers had a calculated melting temperature of 53°C ± 0.4°C and amplifications were not longer than 300 bp. RT-PCR was performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen). Fifty nanograms of DNaseI-treated total RNA (Serva) were used in a total volume of 25 μL. Amplification was monitored using the Opticon real-time PCR cycler (MJ Research) and quantified via Opticon Monitor analysis software, version 1.05. The program used for amplification and the method for calculation of relative gene expression are described in Hohnjec et al. (2003). For normalization of gene expression, the constitutively expressed gene encoding an elongation factor-1α (TC85181; TIGR M. truncatula gene index 7.0) was used as a reference.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of LPS from S. meliloti and X. campestris pv campestris on the invertase-induced oxidative burst in M. truncatula suspension-cultured cells.
Supplemental Table S1. Compilation of relative log2 expression ratios for M. truncatula suspension cell cultures treated with either 50 μg mL−1 invertase or 50 μg mL−1 S. meliloti LPS or a combination of 50 μg mL−1 invertase and 50 μg mL−1 S. meliloti LPS in comparison to water-treated control cells.
Supplemental Table S2. Primers used for real-time RT-PCR experiments.
Supplementary Material
Acknowledgments
Manuela Meyer is gratefully acknowledged for her assistance in the synthesis of microarray targets and in microarray hybridization. Axel Nagel is acknowledged for his support in maintenance and updating of MapMan software and databases. Thomas Bekel and Alex Goesmann are acknowledged for providing information on protein sequences and domains via the SAMS system. Moreover, we thank Thomas Patschkowski for a critical reading of the manuscript.
This work was supported by the German Research Council “Bioinformatics and genome research,” by the European Community's Human Potential Programme (under contract HPRN–CT–2002–00251), and by the German Ministry for Education and Research (grant nos. GABI 313112 and 313110).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Karsten Niehaus (karsten.niehaus@genetik.uni-bielefeld.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albus U, Baier R, Holst O, Pühler A, Niehaus K (2001) Suppression of an elicitor-induced oxidative burst reaction in Medicago sativa cell cultures by Sinorhizobium meliloti lipopolysaccharides. New Phytol 151 597–606 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia S (1968) Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol 22 87–108 [DOI] [PubMed] [Google Scholar]
- Basse CW, Bock K, Boller T (1992) Elicitors and suppressors of the defense response in tomato cells—purification and characterization of glycopeptide elicitors and glycan suppressors generated by enzymatic cleavage of yeast invertase. J Biol Chem 267 10258–10265 [PubMed] [Google Scholar]
- Benhamou N (1996) Elicitor-induced plant defence pathways. Trends Plant Sci 1 233–240 [Google Scholar]
- Blechert S, Brodschelm W, Hölder S, Kammerer L, Kutchan TM, Mueller MJ, Xia ZQ, Zenk MH (1995) The octadecanoic pathway: signal molecules for the regulation of secondary pathways. Proc Natl Acad Sci USA 92 4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Meyer A, Holst O, Pühler A, Niehaus K (2005) Characterization of the Xanthomonas campestris pv campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol Plant-MicrobeInteract 18 674–681 [DOI] [PubMed] [Google Scholar]
- Campbell GRO, Reuhs BL, Walker GC (2002) Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci USA 99 3938–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson RW, Kalembasa S, Turowski D, Pachori P, Noel KD (1987) Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J Bacteriol 169 4923–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2000) Tobacco nectarin I—purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem 275 36726–36733 [DOI] [PubMed] [Google Scholar]
- Casse F, Boucher C, Julliot JS, Michel M, Dénarié J (1979) Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Gen Microbiol 113 229–242 [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkin K, Edwards R, Edington B, Dixon RA (1990) Stress responses in alfalfa (Medicago sativa L.). I. Induction of phenylpropanoid biosynthesis and hydrolytic enzymes in elicitor-treated cell suspension cultures. Plant Physiol 92 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton DA, Joyner SL, Becana M, Iturbe-Ormaetxe I, Chatfield JM (1998) Antioxidant defenses in the peripheral cell layers of legume root nodules. Plant Physiol 116 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazzo FB, Truchet GL, Hollingsworth RI, Hrabak EM, Pankratz HS, Philip-Hollingsworth S, Salzwedel JL, Chapman K, Appenzeller L, Squartini A, et al (1991) Rhizobium lipopolysaccharide modulates infection thread development in white clover root hairs. J Bacteriol 173 5371–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3 371–390 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ (1994) Early events in the activation of plant defense responses. Annu Rev Phytopathol 32 479–501 [Google Scholar]
- Djordjevic MA, Gabriel DW, Rolfe BG (1987) Rhizobium—the refined parasite of legumes. Annu Rev Phytopathol 25 145–168 [Google Scholar]
- Dondrup M, Goesmann A, Bartels D, Kalinowski J, Krause L, Linke B, Rupp O, Sczyrba A, Puhler A, Meyer F (2003) EMMA: a platform for consistent storage and efficient analysis of microarray data. J Biotechnol 106 135–146 [DOI] [PubMed] [Google Scholar]
- Dron M, Clouse SD, Dixon RA, Lawton MA, Lamb CJ (1988) Glutathione and fungal elicitor regulation of a plant defense gene promoter in electroporated protoplasts. Proc Natl Acad Sci USA 85 6738–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel J, Mithöfer A (1998) Early events in the elicitation of plant defence. Planta 206 335–348 [Google Scholar]
- Erbs G, Newman MA (2003) The role of lipopolysaccharides in induction of plant defence responses. Mol Plant Pathol 4 421–425 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18 265–276 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE (2000) Interplay of signalling pathways in plant disease resistance. Trends Genet 16 449–455 [DOI] [PubMed] [Google Scholar]
- Gerber IB, Laukens K, Witters E, Dubery IA (2006) Lipopolysaccharide-responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol Biochem 44 369–379 [DOI] [PubMed] [Google Scholar]
- Gerber IB, Zeidler D, Durner J, Dubery IA (2004) Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta 218 647–657 [DOI] [PubMed] [Google Scholar]
- Gomez LD, Noctor G, Knight MR, Foyer CH (2004) Regulation of calcium signalling and gene expression by glutathione. J Exp Bot 55 1851–1859 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7 251–256 [DOI] [PubMed] [Google Scholar]
- Graham TL, Sequeira L, Huang TSR (1977) Bacterial lipopolysaccharides as inducers of disease resistance in tobacco. Appl Environ Microbiol 34 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC (2000) Nonhost resistance and non-specific plant defenses. Curr Opin Plant Biol 3 315–319 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant-Microbe Interact 16 903–915 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Shiraishi T, Toyoda K, Saitoh K, Satoh Y, Tahara M, Yamada T, Oku H (1993) Inhibition of ATPase activity in pea plasma membranes by fungal suppressors from Mycosphaerella pinodes and their peptide moieties. Plant Cell Physiol 34 439–445 [PubMed] [Google Scholar]
- Keating DH, Willits MG, Long SR (2002) A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J Bacteriol 184 6681–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DN, Chappell J, Boudet A, Hahlbrock K (1984) Induction of phenylalanine ammonia-lyase and 4-coumarate:CoA ligase mRNAs in cultured plant cells by UV light or fungal elicitor. Proc Natl Acad Sci USA 81 1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagares A, Caetano-Anollés G, Niehaus K, Lorenzen J, Ljunggren HD, Pühler A, Favelukes G (1992) A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J Bacteriol 174 5941–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Pühler A, Niehaus K (2001) The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta 213 214–222 [DOI] [PubMed] [Google Scholar]
- Mithöfer A (2002) Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci 7 440–444 [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Bhagwat AA, Feger M, Ebel J (1996) Suppression of fungal β-glucan-induced plant defence in soybean (Glycine max L.) by cyclic 1,3-1,6-β-glucans from the symbiont Bradyrhizobium japonicum. Planta 199 270–275 [Google Scholar]
- Mithöfer A, Ebel J, Bhagwat AA, Boller T, Neuhaus-Url G (1999) Transgenic aequorin monitors cytosolic calcium transients in soybean cells challenged with β-glucan or chitin elicitors. Planta 207 566–574 [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Newman MA, Daniels MJ, Dow JM (1997) The activity of lipid A and core components of bacterial lipopolysaccharides in the prevention of the hypersensitive response in pepper. Mol Plant Microbe Interact 10 926–928 [DOI] [PubMed] [Google Scholar]
- Newman MA, von Poepenack-Lahaye E, Parr A, Daniels MJ, Dow JM (2002) Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J 29 487–495 [DOI] [PubMed] [Google Scholar]
- Ni W, Fahrendorf T, Balance GM, Lamb CJ, Dixon RA (1996) Stress responses in alfalfa (Medicago sativa L.) XX. Transcriptional activation of phenylpropanoid pathway genes in elicitor-induced cell suspension cultures. Plant Mol Biol 30 427–438 [DOI] [PubMed] [Google Scholar]
- Niehaus K, Lagares A, Pühler A (1998) A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol Plant Microbe Interact 11 906–914 [Google Scholar]
- Nürnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198 249–266 [DOI] [PubMed] [Google Scholar]
- Perotto S, Brewin NJ, Kannenberg EL (1994) Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol Plant Microbe Interact 7 99–112 [Google Scholar]
- Poulton JE (1990) Cyanogenesis in plants. Plant Physiol 94 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin A, Franchisse JM, Tavernier E, Bligny R, Gout E, Douce R, Guern J (1997) Early events induced by the elicitor cryptogein in tobacco cells: involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate pathway. Plant Cell 9 2077–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YY, West CA (1992) Elicitation of diterpene biosynthesis in rice (Oryza sativa L.) by chitin. Plant Physiol 99 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Hérouart D, Sigaud S, Touati D, Puppo A (2001) Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol Plant Microbe Interact 14 86–89 [DOI] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, Hoheisel JD (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J Biol Chem 277 10555–10561 [DOI] [PubMed] [Google Scholar]
- Scheidle H, Groß A, Niehaus K (2005) The LipidA substructure of the Sinorhizobium meliloti lipopolysaccharide is sufficient to suppress the oxidative burst in host plants. New Phytol 165 559–566 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JK, McNeil M, Albersheim P (1984. a) The primary structures of one elicitor-active and seven elicitor-inactive hexa(β-d-glucopyranosyl)-d-glucitols isolated from the mycelial walls of Phytophtora megasperma f. sp. glycinea. J Biol Chem 259 11321–11336 [PubMed] [Google Scholar]
- Sharp JK, Valent B, Albersheim P (1984. b) Purification and partial characterization of a β-glucan fragment that elicits phytoalexin accumulation in soybean. J Biol Chem 259 11312–11320 [PubMed] [Google Scholar]
- Shiraishi T, Yamada T, Saitoh K, Kato T, Toyoda K, Yoshioka H, Kim HM, Ichinose Y, Tahara M, Oku H (1994) Suppressors: determinants of specificity produced by plant pathogens. Plant Cell Physiol 35 1107–1119 [Google Scholar]
- Silipo A, Molinaro A, Sturiale L, Dow JM, Erbs G, Lanzetta R, Newman MA, Parrilli M (2005) The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J Biol Chem 280 33660–33668 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Reddy MSS, Naoumkina M, Aziz N, May GD, Huhman DV, Sumner LW, Blount JW, Mendes P, Dixon RA (2005) Methyl jasmonate and yeast elicitor induce differential transcriptional and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220 696–707 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Toyoda K, Shiraishi T, Yamada T, Ichinose Y, Oku H (1993) Rapid changes in phophoinositide metabolism in pea in response to fungal signals. Plant Cell Physiol 34 729–735 [Google Scholar]
- Toyoda K, Shiraishi T, Yoshioka H, Yamada T, Ichinose Y, Oku H (1992) Regulation of phosphoinositide metabolism in pea plasma membranes by elicitor and suppressor from a pea pathogen, Mycosphaerella pinodes. Plant Cell Physiol 33 445–452 [Google Scholar]
- Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Bläsing O, Kowalczyk M, Weicht D, Polinceusz A, et al (2006) Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60 773–792 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Conceição Piques M, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedam V, Haynes JG, Kannenberg EL, Carlson RW, Sherrier DJ (2004) A Rhizobium leguminosarum lipopolysaccharide lipid-A mutant induces nitrogen-fixing nodules with delayed and defective bacteroid formation. Mol Plant Microbe Interact 17 283–291 [DOI] [PubMed] [Google Scholar]
- von Dahl CC, Hävecker M, Schlögl R, Baldwin IT (2006) Caterpillar-elicited methanol emission: a new signal in plant-herbivore interactions? Plant J 46 948–960 [DOI] [PubMed] [Google Scholar]
- Wada M, Kato H, Malik K, Sriprasertsak P, Ichinose Y, Shiraishi T, Yamada T (1995) A supprescin from a phytopathogenic fungus deactivates transcription of a plant defense gene encoding phenylalanine ammonia-lyase. J Mol Biol 249 513–519 [DOI] [PubMed] [Google Scholar]
- Warm E, Laties GG (1982) Quantification of hydrogen peroxide in plant extracts by the chemiluminscence reaction with luminol. Phytochemistry 21 827–831 [Google Scholar]
- Westphal O, Jann K (1965) Bacterial lipopolysaccharides. Extraction with phenol-water and further application of the procedure. In RL Whistler, JN BeMiller, Wolfrom ML, eds, Methods in Carbohydrate Chemistry, Vol 5. Academic Press, New York, pp 83–91
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.