Abstract
The biosynthesis of jasmonic acid (JA) in plant peroxisomes requires the action of acyl-coenzyme A oxidase (ACX). Among the five expressed members (ACX1–5) of the ACX gene family in Arabidopsis (Arabidopsis thaliana), only ACX1 is known to serve a role in JA production. Here, we used transgenic promoter-reporter lines to show that ACX1 is highly expressed in mature and germinating pollen, stem epidermal cells, and other tissues in which jasmonate-signaled processes occur. Wound-induced JA accumulation was reduced in a mutant that is defective in ACX1 and was abolished in a mutant that is impaired in both ACX1 and its closely related paralog, ACX5. The severe JA deficiency in acx1/5 double mutants was accompanied by decreased resistance to the leaf-eating insect Trichoplusia ni. The double mutant also showed reduced pollen viability and fecundity. Treatment of acx1/5 plants with JA restored both protection against T. ni larvae and normal seed set. Unexpectedly, acx1/5 plants accumulated JA in response to infection by the necrotrophic fungal pathogen Alternaria brassicicola. In contrast to mutants that are impaired in jasmonate perception or early steps of the JA biosynthetic pathway, acx1/5 plants maintained resistance to A. brassicicola infection. These results indicate that ACX1/5-mediated JA synthesis is essential for resistance to chewing insects and male reproductive function and further suggest that other ACX isozymes contribute to JA production in response to A. brassicicola challenge. Thus, different types of biotic stress may induce JA synthesis via distinct enzymatic routes.
Plant responses to biotic stress are coordinated by a network of signal transduction pathways that control a wide range of physiological processes. Jasmonic acid (JA) and related members of the jasmonate family of signaling compounds (collectively called JAs) play a central role in orchestrating these responses (Gfeller and Farmer, 2004; Browse, 2005; Devoto and Turner, 2005). Although JAs are often regarded as signals for plant defense (Howe, 2004; Glazebrook, 2005; Halitschke and Baldwin, 2005), it is now clear that they also regulate a variety of developmental processes. Included among these are carbon/nitrogen partitioning (Creelman and Mullet, 1997), tendril coiling (Weiler et al., 1993), glandular trichome development (Li et al., 2004), root growth (Staswick et al., 1992), and various aspects of male and female reproductive function (Feys et al., 1994; McConn and Browse, 1996; Li et al., 2004). A current challenge in the field of jasmonate signaling is to understand the molecular mechanisms by which individual bioactive JAs regulate specific target processes.
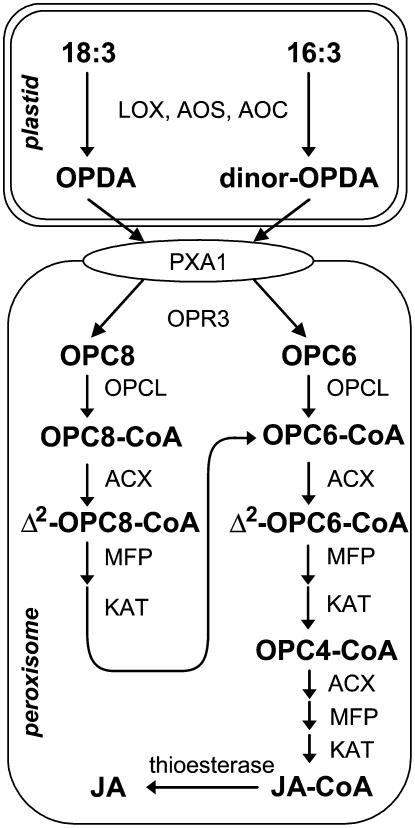
The octadecanoid pathway for JA biosynthesis is initiated in the chloroplast and terminated in peroxisomes (Fig. 1). Many of the enzymes and corresponding genes involved in the pathway have been identified (Schaller et al., 2005). A chloroplastic lipoxygenase initiates JA synthesis by adding molecular oxygen to linolenic acid (18:3). The resulting 13-hydroperoxy fatty acid is converted to 12-oxo-phytodienoic acid (OPDA) by the sequential action of allene oxide synthase (AOS) and allene oxide cyclase. Very little is known about the mechanism of plastid-to-peroxisome transport of octadecanoids. Recently, however, an ATP-binding cassette transporter was implicated in the import of ODPA into peroxisomes (Theodoulou et al., 2005). OPDA reductase (OPR3) transforms OPDA to 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid (OPC-8:0) within the peroxisome. OPC-8:0 is then converted to its corresponding CoA derivative by OPC-8:0-CoA ligase1, which is the starting substrate for three rounds of β-oxidation that ultimately yield JA (Koo et al., 2006). A parallel JA biosynthetic pathway starting from chloroplastic pools of hexadecatrienoic acid (16:3) has also been described (Weber et al., 1997; Fig. 1).
Figure 1.
The JA biosynthetic pathway. Trienoic fatty acids (18:3 and 16:3) are converted within the chloroplast to OPDA and dinor-OPDA, respectively. These cyclopentenone intermediates are transported to the peroxisome by an ATP-binding cassette transporter (PXA1; also known as CTS or PED3) or by a PXA1-independent pathway (not shown) and then reduced by OPR3. The resulting cyclopentanone compounds (OPC8 and OPC6) are converted to their corresponding CoA derivatives by OPC-8:0-CoA ligase. Successive rounds of β-oxidation yield JA. The three core enzymes in the β-oxidation cascade are ACX, MFP, and KAT. LOX, Lipoxygenase; AOC, allene oxide cyclase; OPC6, 3-oxo-2-(2′-pentenyl)-cyclopentane-1-hexanoic acid; OPC4, 3-oxo-2-(2′-pentenyl)-cyclopentane-1-butyric acid; JA, (+)-7-iso-JA.
Peroxisomal β-oxidation has been studied extensively for its role in the metabolism of storage lipids and a variety of other plant compounds (Baker et al., 2006; Poirier et al., 2006). The three core enzymes involved in this pathway are acyl-CoA oxidase (ACX), a multifunctional protein (MFP) possessing 2-trans-enoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase activities, and 3-keto-acyl-CoA thiolase (KAT; Graham and Eastmond, 2002; Fig. 1). Seminal work by Vick and Zimmerman (1984) provided the first evidence that JA biosynthesis involves β-oxidation. Only recently, however, have specific enzymes been implicated in this stage of the pathway. The ACX1A gene product in tomato (Lycopersicon esculentum) was shown to metabolize OPC-8:0-CoA and to contribute to the vast majority of JA production in wounded leaves (Li et al., 2005). Genetic evidence also indicates that the ACX1 and KAT2 (also known as PED1) genes in Arabidopsis (Arabidopsis thaliana) have a role in wound-induced JA synthesis (Cruz Castillo et al., 2004; Afitlhile et al., 2005; Pinfield-Wells et al., 2005). The persistence of significant levels of JA in acx1 and kat2 mutants, however, suggests that additional members of these gene families contribute to JA production in Arabidopsis.
Increasing evidence indicates that different JAs regulate distinct and overlapping physiological responses (Mithofer et al., 2005; Schaller et al., 2005; Schilmiller and Howe, 2005). Genetic studies have shown that JA is strictly required for male fertility in Arabidopsis (McConn and Browse, 1996; Stintzi and Browse, 2000; Browse, 2005) and induced resistance of tomato to lepidopteran insects (Li et al., 2005). Analysis of mutants that are defective in the production of jasmonoyl-Ile further indicates that this conjugated form of JA is an important signal for plant protection against pathogens (Staswick et al., 1998) and insects (Kang et al., 2006). These findings raise the possibility that defense responses previously attributed to JA and methyl-JA (MeJA) are in fact mediated by JA-amino acid conjugates. OPDA rather than JA is thought to be the active signal for the tendril coiling response of Bryonia (Weiler et al., 1993). Studies of the Arabidopsis opr3 mutant suggest that OPDA can promote jasmonate-based resistance to Bradysia impatiens and Alternaria brassicicola in the absence of JA (Stintzi et al., 2001). Thus, metabolism of OPDA to JA may be required for some but not all jasmonate-signaled responses. DNA microarray studies indicate that OPDA-induced gene expression occurs via at least two distinct signaling pathways (Stintzi et al., 2001; Taki et al., 2005). One pathway depends on the Coronatine-Insensitive1 (COI1) F-box protein that is essential for most, if not all, JA-mediated processes. A second set of OPDA-responsive genes is activated in a COI1-independent manner. Whereas the physiological significance of the COI1 pathway in plant growth and development is well established, much less is known about wound- and OPDA-induced responses that occur independently of COI1 (Howe, 2004; Devoto and Turner, 2005).
A better understanding of the physiological roles of JA (and its derivatives) and C18 jasmonate precursors (e.g. OPDA) would be facilitated by the identification of Arabidopsis mutants that are impaired in the β-oxidation stage of JA synthesis. Although mutants affected in this part of the pathway have been reported (Cruz Castillo et al., 2004; Afitlhile et al., 2005; Pinfield-Wells et al., 2005), none have been shown to exhibit physiological hallmarks of JA deficiency. Here, we report that simultaneous disruption of two ACX genes (ACX1 and ACX5) depletes JA to a level that compromises both male fertility and resistance to leaf-eating insects. Unexpectedly, acx1/5 plants accumulated JA in response to the fungal pathogen A. brassicicola. These findings indicate that ACX1/5-mediated JA synthesis is essential for both reproductive and antiinsect defensive processes in Arabidopsis and that a different metabolic route is involved in JA production in A. brassicicola-infected leaves.
RESULTS
ACX1 Is Broadly Expressed during Arabidopsis Development
The Arabidopsis ACX1 gene plays a role in fatty acid degradation during seedling establishment and in JA synthesis in leaves of mature plants (Cruz Castillo et al., 2004; Adham et al., 2005; Pinfield-Wells et al., 2005). These observations prompted us to investigate the spatial and temporal expression pattern of ACX1 through the use of promoter∷reporter gene fusions. A 1.3-kb fragment containing the ACX1 promoter was fused to the β-glucuronidase (GUS)-encoding uidA or yellow fluorescent protein (YFP) reporter genes (Fig. 2A). The YFP reporter was engineered with a type 1 peroxisomal targeting sequence (PTS1) that directs ACX1 to peroxisomes. Transgenic plants expressing these fusions were analyzed by 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) staining (for GUS activity) or laser scanning confocal microscopy (for YFP fluorescence). Although histochemical (GUS) and fluorescent (YFP) reporter proteins offer different advantages for analysis of gene expression, both approaches yielded very similar results with respect to ACX1 expression. For example, both reporter lines showed robust reporter ACX1 expression in 3- to 5-d-old germinating seedlings, consistent with the role of ACX1 in catabolism of lipid stores during seedling establishment (Adham et al., 2005; Pinfield-Wells et al., 2005; Fig. 2, B and C). X-gluc staining of whole plants further showed that ACX1 was uniformly expressed in leaves and roots of 16-d-old plants, whereas expression in flowering plants was mostly restricted to young rosette leaves, the upper part of the stem, and flower buds (Fig. 2, D–F). In general, the expression pattern of ACX1 as determined by these fusion constructs was consistent with DNA microarray data (Schmid et al., 2005), as well as the spatial distribution of JA in whole plants (Muller et al., 2002).
Figure 2.
Developmental and tissue-specific expression of ACX1. A, Schematic drawing of ACX1-promoter∷YFP-PTS1 and ACX1-promoter∷GUS constructs. B, X-gluc staining of a 4-d-old germinating seedling expressing ACX1∷GUS. C, Confocal image of a 4-d-old germinating seedling expressing ACX1∷YFP-PTS1. Bar = 200 mm. D and E, X-gluc staining of 16-d-old plate-grown (D) and 35-d-old soil-grown (E) plants expressing ACX1∷GUS. Bars = 5 mm. F, G, and I, X-gluc staining of the inflorescence at various stages: emerging inflorescence (F), developing floral buds before stage 13 (G), and floral stage 12 (I). Arrows indicate areas of high GUS expression. X-gluc-stained dehiscent pollen grains are shown in the inset of I. Bar = 50 mm. H, Cross section of inflorescence stem from a plant expressing ACX1∷YFP-PTS1. The arrow indicates the epidermal layer where YFP fluorescence (green in the image) was detected. Bar = 50 mm. J, Confocal image of a single optical section across an anther (stage 13 flower) from a plant expressing ACX1∷YFP-PTS1. Bar = 100 mm. K, Confocal image of a pollinated pistil on an ACX1∷YFP-PTS1-expressing plant. Bar = 200 mm. L, Germinating pollen grain from an ACX1∷YFP-PTS1 plant. Bar = 10 mm. The red color in the confocal images results from autofluorescence of chlorophyll.
The expression of ACX1 in floral tissues was studied in greater detail. Analysis of cross sections of the inflorescence stem showed that the ACX1 promoter was active mainly in the epidermal cell layer (Fig. 2H). Relatively high expression in young buds of the flower cluster was also observed (Fig. 2G). Prior to dehiscence (i.e. stage 12 flowers), ACX1 promoter activity was observed in ovaries and pollen grains within the anther (Fig. 2I). These patterns of expression are similar to that reported for the Arabidopsis AOS gene (Kubigsteltig et al., 1999). YFP reporter lines confirmed that ACX1 expression in anthers of stage 12 flower buds was largely restricted to developing pollen grains and that expression remained high after dehiscence and pollination of the stigma (Fig. 2, J and K). Confocal imaging of YFP fluorescence in single in vitro-germinated pollen grains revealed numerous fluorescing particles that were distributed uniformly within the pollen grain and the elongating tube. The size of these particles (approximately 700 nm) is consistent with their identity as peroxisomes (Olsen, 1998), which is the expected destination of ACX1 and the YFP-PTS1 reporter. These results are consistent with a role for ACX1 in pollen development and germination.
ACX1 and ACX5 Catalyze Wound-Induced JA Biosynthesis in Arabidopsis Leaves
The persistence of significant levels of JA in acx1 mutants of Arabidopsis (Cruz Castillo et al., 2004; Pinfield-Wells et al., 2005) suggests that additional ACX isozymes contribute to production of the hormone. A good candidate for such an enzyme is ACX5, which is closely related to both ACX1 and the ACX1A isozyme that catalyzes JA biosynthesis in tomato leaves (Fig. 3A). Our approach for testing this hypothesis was to isolate and characterize T-DNA knockout mutants that are defective in ACX1 (acx1 plants), ACX5 (acx5 plants), or both ACX1 and ACX5 (acx1/5 plants; see “Materials and Methods”). Reverse transcription (RT)-PCR and RNA-blot experiments (see below) showed that the acx1 and acx5 mutants identified fail to express functional ACX1 and ACX5 transcripts, respectively, indicating that we were assessing the null phenotypes.
Figure 3.
The Arabidopsis ACX1 and tomato ACX1A isozymes are immunologically related. A, Phylogenetic analysis of the Arabidopsis ACX family. A neighbor-joining phylogeny was constructed, as previously described (Li et al., 2005) in PAUP4.0* from the deduced amino acid sequences of the Arabidopsis ACX family members (AtACX1–AtACX6). Also included in the phylogeny is tomato ACX1A, which has an established role in JA biosynthesis. Numbers indicate percent bootstrap support for each branch of the phylogeny. B, Western-blot analysis of ACX protein levels in wild-type and acx knockout plants. Protein from 3-week-old rosette leaves of the indicated genotype was blotted and probed with a polyclonal antibody against tomato ACX1A (LeACX1A; left) or an equivalent amount of preimmune serum (right). The arrow indicates an Arabidopsis ACX protein that specifically cross reacts with the immune serum. Molecular mass standards (kilodalton) are indicated on the left side of each blot.
To determine whether the various acx mutants are affected in the expression of ACX isozymes that have a putative role in JA biosynthesis, we performed immunoblot assays with a polyclonal antibody raised against tomato ACX1A (LeACX1A). The antiserum reacted strongly and specifically with a protein in wild-type but not acx1 leaves (Fig. 3B). The apparent Mr of this polypeptide was in good agreement with the calculated Mr of ACX1/5 (approximately 74,300). The identity of this protein as ACX1 is consistent with the observation that acx5 leaves accumulate normal levels of the LeACX1A-related protein, whereas acx1/5 leaves do not (Fig. 3B). These results indicate that AtACX1 and LeACX1A are immunologically related and that ACX1 does not accumulate in either acx1 or acx1/5 leaves.
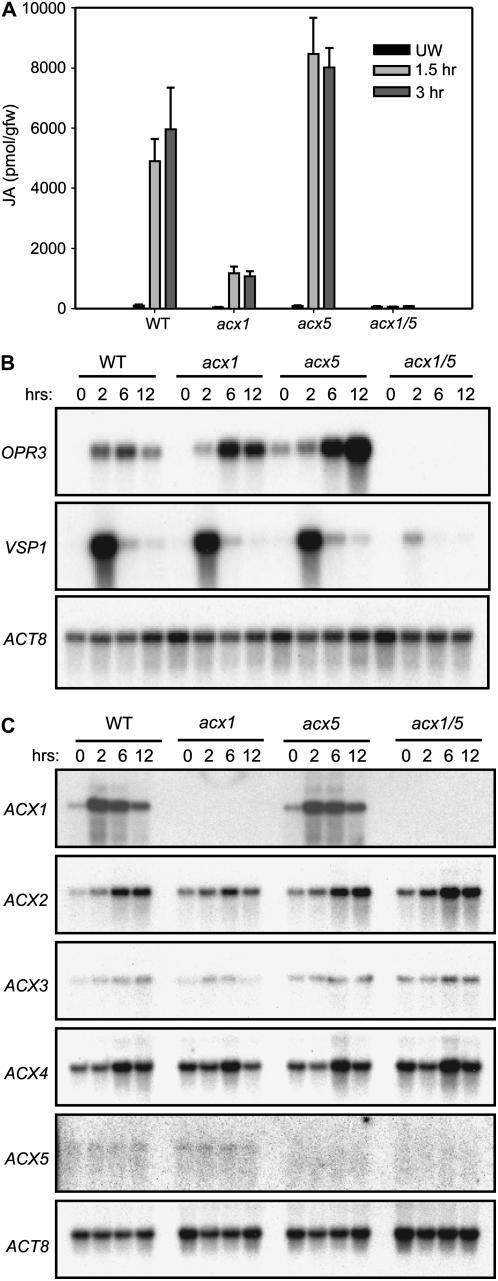
To determine the relative contribution of ACX1 and ACX5 to JA synthesis, we used gas chromatography-mass spectrometry (GC-MS) to measure the endogenous levels of JA in unwounded (control) and mechanically wounded leaves (Fig. 4A). The basal level of JA in unwounded acx1 and acx1/5 plants (39 ± 11 and 50 ± 37 pmol/g fresh weight, respectively) was reduced approximately 2-fold in comparison to JA levels in wild-type and acx5 plants (95 ± 31 and 83 ± 23 pmol/g fresh weight, respectively). Wound-induced JA levels in acx1 leaves were reduced to about 20% of wild-type levels, which is consistent with the results of previous studies (Cruz Castillo et al., 2004; Pinfield-Wells et al., 2005). JA accumulation in wounded acx5 plants slightly exceeded (by 1.5-fold) that in wild-type plants. In contrast to the relatively large (>25-fold) wound-induced increase in JA levels in wild-type, acx1, and acx5 plants, JA levels in the acx1/5 mutant did not increase significantly in response to wounding. We estimated that the total amount of JA in wounded acx1/5 leaves was approximately 1% of wild-type levels.
Figure 4.
Effect of acx mutations on wound-induced JA accumulation and gene expression. A, JA levels in acx knockouts in response to mechanical wounding. Leaves on 4-week-old plants of the indicated genotype were wounded twice across the midvein. At various times after wounding (1.5 or 3 h), tissue was collected for JA extraction and quantification by GC-MS. Unwounded (UW) leaf tissue was collected as a control. Values represent the mean and sd for three independent JA extractions per genotype. B and C, Time course expression of wound-induced genes. Leaves were wounded, as described above. At the indicated times after wounding, leaf tissue was harvested for RNA extraction. Leaf tissue from unwounded control plants was used for the 0 h time point. B, RNA blots were hybridized to 32P-labeled cDNA probes for OPR3 and VSP1. C, RNA blots were hybridized to gene-specific probes for five members (ACX1–ACX5) of the ACX gene family. Blots were hybridized to a probe for Actin-8 (ACT-8) as a loading control. For purposes of comparing the expression level of ACX1 and ACX5, note that ACX1- and ACX5-probed blots were exposed to autoradiographic film for 2 and 11 d, respectively.
RNA-blot analysis was used to determine the effect of the acx mutations on two genes, OPR3 and VSP1, whose wound-induced expression is regulated by the jasmonate-signaling pathway (Berger et al., 1995; Mussig et al., 2000). Mechanical wounding activated the expression of both genes in wild-type, acx1, and acx5 leaves (Fig. 4B). Wound-induced expression of VSP1 and OPR3 was severely diminished in acx1/5 leaves; expression of these genes in the acx1/5 background was detected only upon prolonged exposure of autoradiographs. As expected, expression of ACX1 and ACX5 was not detected in the corresponding single mutants or in the acx1/5 double mutant (Fig. 4C). These results demonstrate that both ACX1 and ACX5 are involved in the production of JA pools that activate the expression of wound-responsive genes.
Detection of ACX5 transcripts in RNA isolated from unwounded plants required autoradiograph exposure times that were 5 to 10 times longer than that needed to produce a comparable signal on ACX1-probed blots. Because the specific activity of the gene-specific probes was similar, we conclude that ACX1 expression in undamaged leaves is considerably higher (at least 7-fold) than that of ACX5. Mechanical wounding strongly activated the expression of ACX1 but not ACX5 (Fig. 4C). Thus, the level of ACX1 mRNA in wounded leaves greatly exceeded that of ACX5. ACX2, ACX3, and ACX4 expression was either not altered by wounding or increased modestly at later points in the time course. Because the wound-induced pattern of ACX2, ACX3, and ACX4 expression was not affected in the acx1/5 mutant, we conclude that expression of these genes does not depend on JA synthesis (Fig. 4C).
acx1/5 Plants Are More Susceptible to Attack by Trichoplusia ni Larvae
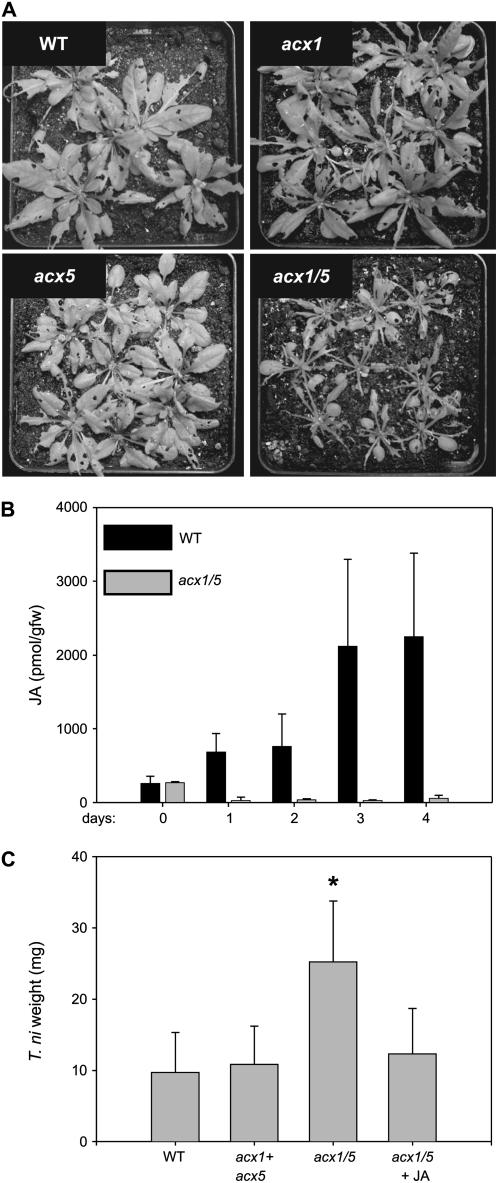
To determine the role of ACX1/5 in resistance of Arabidopsis to a leaf-chewing insect, wild-type and acx mutant plants were challenged with Trichoplusia ni larvae. In three independent feeding trials, acx1/5 plants reproducibly suffered more damage than wild-type, acx1, or acx5 plants (Fig. 5A). The increased susceptibility of acx1/5 plants to attack by T. ni larvae was associated with a lack of herbivore-induced JA accumulation in damaged leaves (Fig. 5B). Consistent with these observations, the average weight of larvae reared on acx1/5 plants was greater than that of larvae grown on the other host genotypes (Fig. 5C). Treatment of acx1/5 plants with JA immediately before the start of the feeding trial was sufficient to restore protection of the mutant to T. ni attack (data not shown). Larval weight measurements showed the T. ni performance on JA-treated acx1/5 plants was not significantly different (P = 0.25) from that on untreated wild-type plants (Fig. 5C). These results demonstrate that ACX1/5 function is essential for jasmonate-based resistance of Arabidopsis to attack by T. ni.
Figure 5.
acx1/5 plants are susceptible to attack by T. ni. A, Four-week-old wild-type and acx mutant plants were challenged with newly hatched T. ni larvae (one first-instar larva per plant). Plants were photographed 10 d after the start of the feeding trial. B, JA levels in wild-type and acx1/5 damaged leaves were measured each day during the feeding trial. Data show the mean and sd of three independent samples per time point. C, The average weight of larvae reared on each host genotype was determined at the end of the feeding trial. In this experiment, larvae were allowed to move freely between acx1 and acx5 plants. Thus, larval weight data for caterpillars recovered from these two genotypes were combined. Additional experiments showed that larval performance on acx1 and acx5 plants was not significantly different (data not shown). Also shown is larval weight data for acx1/5 plants that were treated with JA prior to initiation of the feeding trial (acx1/5 + JA). Data show the mean and sd of the following number of larvae: wild type, 40; acx1 + acx5, 42; acx1/5, 39; acx1/5 + JA, 8. *, The weight of larvae grown on acx1/5 plants was significantly greater (P < 0.001; Mann-Whitney rank sum test) than that of larvae reared on wild-type plants.
acx1/5 Plants Maintain Jasmonate-Dependent Resistance to Infection by A. brassicicola
The fungal pathogen A. brassicicola activates jasmonate-dependent defense responses in Arabidopsis (Penninckx et al., 1996; Thomma et al., 1998; Stintzi et al., 2001). The ability of OPDA to promote these responses in the absence of JA (Stintzi et al., 2001) prompted us to determine whether acx1/5 plants are altered in their resistance to A. brassicicola. Wild-type and acx1/5 rosette leaves were inoculated with a suspension of A. brassicicola spores. The jasmonate-insensitive coi1-1 mutant, which is compromised in resistance to A. brassicicola (Thomma et al., 1998), was also inoculated as a control for the susceptibility phenotype. At 5 d postinoculation (dpi), both wild-type and acx1/5 plants developed a typical resistance response that was manifested by the formation of brown necrotic lesions at the site of spore inoculation (Fig. 6, A and B). In contrast, coi1 leaves were heavily colonized by the pathogen (Fig. 6, C and E). Susceptibility was also observed in a mutant that harbors a T-DNA insertion in the AOS gene that encodes a plastidic enzyme of the octadecanoid pathway (Figs. 1 and 6, D and F).
Figure 6.
acx1/5 plants maintain resistance and produce JA in response to A. brassicicola infection. A to F, Four-week-old plants of the indicated genotype were inoculated with A. brassicicola applied in a 5-μL drop of a spore suspension (5 × 105 spores/mL) on the leaf surface. Plants were photographed 5 d after inoculation. E and F, Close-up view of leaves depicted in C and D, respectively. G, JA levels were measured daily for 4 d in leaves of wild type and acx1/5 after either mock treatment with water or treatment with A. brassicicola. Data show the mean and sd of three independent samples per time point. *, Significant difference in JA levels (P < 0.05; Student's t test) between pathogen-treated wild-type and acx1/5 plants at the indicated time point. H, Effect of A. brassicicola infection on gene expression. Four-week-old wild-type and acx1/5 plants were inoculated on the leaf surface with either water (mock) or A. brassicicola applied as a 5-μL drop of a spore suspension (5 × 105 spores/mL). Inoculated leaf tissue was collected for RNA extraction at 1 and 2 dpi. Untreated plant tissue was collected for the 0 time point. RNA blots were hybridized to the indicated probes. For purposes of comparing the expression level of ACX1 and ACX5, note that ACX1- and ACX5-probed blots were exposed to autoradiographic film for 16 h and 12 d, respectively.
To test the hypothesis that a C18 JA precursor promotes resistance of acx1/5 plants to A. brassicicola in the absence of JA, we measured JA levels in wild-type and acx1/5 leaves at various times after pathogen inoculation. JA levels in pathogen-treated wild-type leaves increased sharply at 1 dpi and continued to rise during the remainder of the time course (Fig. 6G). Contrary to our expectation, pathogen-challenged acx1/5 leaves also produced high levels of JA. The time course of this response in the mutant was delayed by approximately 1 d in comparison to wild type. At later stages of symptom development (i.e. 4 dpi), JA levels in wild-type and acx1/5 leaves were not significantly different. Treatment of wild-type and mutant plants with a mock control did not affect JA levels during the time course (Fig. 6G). These results indicate that loss of ACX1/5 function is not sufficient to eliminate JA accumulation in A. brassicicola-infected leaves.
RNA-blot analysis was used to determine whether acx1/5 plants are affected in gene expression in response to A. brassicicola challenge. PDF1.2, a well-characterized pathogen-responsive gene, was expressed to similar levels in wild-type and mutant plants 1 dpi (Fig. 6H). At 2 dpi, PDF1.2 mRNA levels in the mutant waned relative to the wild type. We also conducted experiments to determine the effect of A. brassicicola treatment on the expression of ACX genes. Of the five family members examined, only ACX1 mRNA levels increased in response to pathogen challenge (Fig. 6H). These results are in agreement with DNA microarray studies showing that ACX1 expression is stimulated in response to A. brassicicola treatment (Schenk et al., 2003).
acx1/5 Plants Are Impaired in Male Fertility
Severe deficiencies in JA synthesis or perception in Arabidopsis cause a combination of defects in pollen viability, anther elongation, and anther dehiscence (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000; Devoto and Turner, 2005). acx1 and acx5 single knockout lines exhibited normal anther development and seed production (data not shown). Anther dehiscence and elongation in acx1/5 flowers also appeared normal. However, the double mutant produced fewer seed-containing siliques and fewer viable seeds than wild type (Table I). Treatment of acx1/5 flowers with JA restored silique development and viable seed production (Fig. 7A). The severity of the fertility defect appeared to vary with the environmental growth conditions. We observed that acx1/5 plants exhibiting stress symptoms (e.g. anthocyanin accumulation) produced more seed than nonstressed acx1/5 plants.
Table I.
Reduction of seed-containing siliques in acx1/5 plants
Wild-type and acx1/5 plants were grown side by side in the same growth chamber. The total number of siliques per plant and the number of seed-containing siliques per plant were determined for 14 wild-type and 32 acx1/5 plants that were fully senesced. The percentage of siliques per plant that contained at least one developed seed was also calculated. Values for wild type and acx1/5 represent the mean and sd. P values were calculated with the Student's t test.
| Wild Type | acx1/5 | P Value | |
|---|---|---|---|
| Siliques/plant | 115.1 ± 25.8 | 105 ± 34.3 | P = 0.399 |
| Seed-containing siliques/plant | 83.8 ± 26.1 | 14.6 ± 9.3 | P < 0.001 |
| Percent siliques with seed | 71.9% ± 10.1% | 13.1% ± 6.5% | P < 0.001 |
Figure 7.
acx1/5 plants are defective in JA-mediated pollen development. A, Photograph of an acx1/5 inflorescence. In the absence of JA treatment, the majority of siliques fail to develop and produce no viable seed. Treatment of stage 12 flowers with JA restored silique development (arrows) and seed production. B, Pollen from newly dehisced flowers was collected and stained with FDA and PI to determine percent viability. All measurements were performed in quadruplicate, with between 100 and 500 pollen grains per genotype. *, Pollen viability in acx1/5 plants was significantly less than that in wild-type plants (P = 0.002, Mann-Whitney rank sum test). C, JA levels are reduced in acx1/5 flowers. Data show the mean and sd of three independent experiments. The experiment involved at least three JA extractions from independent pools of similarly staged flowers.
Fluorescein diacetate (FDA)-propidium iodide (PI) staining was used to compare the viability of pollen from wild-type and acx plants. In four independent experiments, pollen collected from wild-type, acx1, and acx5 flowers exhibited approximately 80% viability (Fig. 7B). In contrast, the viability of pollen obtained from acx1/5 plants was approximately 30%. Decreased pollen viability in acx1/5 plants was correlated with a steep decline in JA levels in flower buds (P < 0.001, Student's t test; Fig. 7C). These results indicate that the JA deficiency caused by acx1/5 reduces plant fecundity, most likely as a result of a defect in pollen development. A role for ACX5 in male fertility is consistent with DNA microarray studies showing that this gene is expressed to its highest levels in stamens and pollen (Schmid et al., 2005).
DISCUSSION
Role of ACX1 and ACX5 in JA Biosynthesis
The goal of this study was to determine the role of the ACX gene family in various jasmonate-signaled processes in Arabidopsis. Analysis of an acx1 T-DNA knockout mutant showed that ACX1 is responsible for the majority (approximately 80% of wild-type levels) of JA production in wounded Arabidopsis leaves, in agreement with previous studies (Cruz Castillo et al., 2004; Pinfield-Wells et al., 2005). Despite the importance of this isozyme for JA synthesis, loss of ACX1 function did not significantly reduce the expression of JA-regulated genes in wounded leaves (Fig. 4B). This finding indicates that the amount of JA produced in acx1 plants is sufficient to activate the expression of wound-responsive genes and that other ACXs contribute to JA synthesis in the absence of ACX1. Indeed, introduction of an acx5 null mutation into the acx1 background further reduced the level of wound-induced JA to approximately 1% of wild-type levels. The severity of this deficiency was confirmed by experiments showing that expression of wound-responsive genes such as OPR3 and VSP1 is blocked in acx1/5 plants.
Our results indicate that ACX5 has the capacity to synthesize JA in an acx1 genetic background. However, because acx5 leaves are not JA deficient, it would appear that ACX5 contributes little, if any, to JA production in wild-type leaves. This interpretation is consistent with the fact that the expression level of ACX5 in leaves is much lower than that of ACX1. That the accumulation of JA in wounded acx5 leaves exceeded that in wild-type plants (Fig. 4A) raises the possibility that ACX5 may actually impede ACX1-catalyzed JA synthesis. For example, it is possible that the catalytic efficiency of ACX5 is low in comparison to ACX1. Because ACX1 is a biological dimer (Pedersen and Henriksen, 2005), ACX5 may suppress JA production in wild-type plants by forming ACX1/ACX5 heterodimers that are less active than ACX1 homodimers.
A function for ACX1 and ACX5 in JA biosynthesis is in agreement with previous studies showing that a homologous enzyme (ACX1A) from tomato plays an important role in JA synthesis (Li et al., 2005). Structural and functional conservation between Arabidopsis ACX1 and tomato ACX1A is supported by recent x-ray crystallography studies (Pedersen and Henriksen, 2005; Powers et al., 2006), as well as our finding that antibodies raised against ACX1A cross-react specifically with Arabidopsis ACX1 (Fig. 3B). Pedersen and Henriksen (2005) showed that the substrate binding pocket of Arabidopsis ACX1 is relatively wide in comparison to other ACXs for which structural information is available. The unique features of the binding pocket may account for the ability of this class of ACXs to metabolize C18 cyclopentanoid-CoA precursors of JA, as well as a variety of medium- and long-chain acyl-CoAs (Hooks et al., 1999; Li et al., 2005; Pedersen and Henriksen, 2005).
Several lines of evidence indicate that ACXs involved in JA synthesis may also participate in the breakdown of storage lipids during seedling establishment. For example, both ACX1 and ACX1A act on a broad range of medium- and long-chain fatty acyl-CoAs in vitro (Hooks et al., 1999; Li et al., 2005). It has also been shown that enzyme extracts from acx1 mutant seedlings are deficient in the metabolism of long-chain acyl-CoAs (Adham et al., 2005). The ability of acx1/5 seedlings to grow in the absence of exogenous Suc indicates that these isozymes are not absolutely required for normal seedling establishment (Adham et al., 2005). However, the Suc-dependent growth phenotype observed upon introduction of the acx1 mutation into a genetic background that lacks ACX2, which metabolizes long chain acyl-CoAs, indicates that ACX1 can participate in the breakdown of storage lipids under some conditions (Hooks et al., 1999; Adham et al., 2005; Pinfield-Wells et al., 2005). Our finding that ACX1 is highly expressed in germinating seedlings (Fig. 2) lends additional support to this idea. It thus seems likely that ACX1 and related enzymes serve different physiological functions at various stages of plant growth and development, including fatty acid catabolism during seedling development and JA biosynthesis in mature leaves and reproductive tissues. Our results thus support the view (Adham et al., 2005) that ACX isozymes have distinct and overlapping functions in vivo.
Involvement of ACX1 and ACX5 in Male Fertility
Our results demonstrate that the β-oxidation stage of JA synthesis is essential for male reproductive development in Arabidopsis. A role for ACX1 and ACX5 in pollen development is consistent with the tissue-specific expression pattern of these genes. Analysis of YFP and GUS reporter fusions showed that the ACX1 promoter is highly active in floral organs and pollen grains. Likewise, gene expression maps indicate that ACX5 is expressed to its highest levels in stamens and pollen (Schmid et al., 2005). These findings thus confirm and extend previous studies (Browse, 2005) showing that JA is strictly required for male fertility in Arabidopsis. The Arabidopsis aim1 mutant that is defective in the MFP-catalyzed step of β-oxidation also exhibits severely reduced fertility (Richmond and Bleecker, 1999). However, it has not yet been determined whether aim1-mediated sterility is caused by decreased JA production.
acx1/5 flowers do not exhibit obvious defects in anther elongation or pollen dehiscence that occur in other JA-deficient mutants such as fad3/7/8 and opr3 (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000). Rather, the acx1/5 metabolic block appears to impair male fertility by decreasing pollen viability. Support for this hypothesis comes from the finding that ACX1 expression in the stamen is much more prominent in the pollen than it is in the anther or filament (Fig. 2). Additional ACX family members that are expressed in stamens may produce enough JA to satisfy these other aspects of JA-dependent anther development. The absence of anther elongation and dehiscence phenotypes in acx1/5 flowers indicates that fertility defect is less severe than that caused by fad3/7/8 and opr3, which is consistent with the ability of the mutant to produce viable seeds under some growth conditions. The amount of JA in acx1/5 flowers (approximately 25% of wild-type levels) may thus be close to the threshold level that is needed for normal reproductive vigor. Although the reduced fertility phenotype was reproducible for plants grown under identical conditions, it is important to note that the severity of the seed-set phenotype varied considerably under different growth conditions. We speculate that subtle changes in environmental conditions such as humidity, light quality, and soil-borne microbes may affect the basal level of JA in acx1/5 plants, thereby influencing the seed-set phenotype.
Role of ACXs in Plant Defense
The severe JA deficiency in wounded acx1/5 leaves was associated with increased susceptibility to T. ni larvae. This finding, together with the ability of exogenous JA to restore protection against T. ni, demonstrates that JA is essential for defense against this lepidopteran herbivore. We also observed that acx1/5 plants have significantly reduced resistance to western flower thrip (Frankliniella occidentalis; Supplemental Fig. S2), a cell content-feeding herbivore that activates jasmonate-based defenses (Li et al., 2002; De Vos et al., 2005). The increased susceptibility of acx1/5 plants to folivores is consistent with studies in tomato showing that ACX1A is required for induced defense responses to Manduca sexta (Li et al., 2005), as well as more recent studies highlighting the importance of jasmonoyl-Ile in plant antiinsect defense (Kang et al., 2006). We thus conclude that conversion of C18 cyclopentanones to JA via peroxisomal β-oxidation is required for defense responses to multiple arthropod herbivores. This role of the jasmonate pathway distinguishes plants from their animal counterparts in which β-oxidation plays a primary role in the inactivation of oxylipin signals derived from arachidonic acid (Ramwell et al., 1980).
The jasmonate-signaling pathway is essential for resistance of Arabidopsis to A. brassicicola (Pozo et al., 2004; Glazebrook, 2005). Mutants that are defective in COI1 or early steps (e.g. AOS) in JA synthesis are highly susceptible to this pathogen. In contrast, mutants that are blocked in later steps in JA synthesis, including opr3 (Stintzi et al., 2001) and acx1/5 (this study), maintain resistance to A. brassicicola. opr3 and acx1/5 mutants are also similar in that they express PDF1.2 in response to pathogen treatment, albeit at levels that are reduced in comparison to wild-type plants. These observations are consistent with the proposal OPDA can promote resistance to A. brassicicola in the absence of JA (Stintzi et al., 2001). We were surprised to find, however, that A. brassicicola-treated acx1/5 plants accumulate relatively high levels of JA. Thus, we are unable to draw conclusions about the relative contribution of OPDA versus JA in promoting resistance to A. brassicicola. This observation notwithstanding, our results are consistent with previous studies indicating that the outcome of the Arabidopsis-A. brassicicola interaction is shaped by jasmonate signals other than, or in addition to, JA (Stintzi et al., 2001; De Vos et al., 2005, 2006).
It is remarkable that acx1/5 plants accumulate high levels of JA in response to infection by A. brassicicola yet fail to produce JA in response to mechanical wounding or T. ni attack. Because some plant pathogenic fungi are known to produce JA (Miersch et al., 1999), it is possible that the increased JA pool in Alternaria-treated leaves is derived from the fungus rather than the plant. The observation that aos (Fig. 6D) and fad3/7/8 (Stintzi et al., 2001) mutants are susceptible to infection indicates that if A. brassicicola produces JA during colonization of Arabidopsis, this pool of JA is not sufficient to promote host resistance. An alternative explanation for this phenomenon is that insect herbivory and A. brassicicola activate de novo JA synthesis via enzymatic routes that utilize different ACX isozymes. That is, ACXs in addition to ACX1/5 are involved in JA production in pathogen-infected leaves. Two good candidates for this function are ACX2 and ACX3. These isoforms act on long- and medium-chain acyl-CoAs, respectively, and are expressed in rosette leaves (Graham and Eastmond, 2002; Adham et al., 2005). The relatively broad substrate specificity of ACX2 and ACX3 indicates that they may be capable of metabolizing OPC-8:0-CoA or OPC-6:0-CoA, particularly if these intermediates accumulate to high levels in pathogen-infected leaves. The existence of alternative routes for JA synthesis that are activated by different types of environmental stress suggests that the cellular mechanisms underlying jasmonate homeostasis may be more complex than previously realized.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants (Arabidopsis [Arabidopsis thaliana] ecotype Columbia [Col]) were grown in soil in a growth chamber maintained at 21°C under 16 h of light (100 μE m−2 s−1) and 8 h of dark. T-DNA-tagged lines from the SALK collection (Alonso et al., 2003) were obtained from the Arabidopsis Biological Resource Center (Ohio State University). PCR assays were used to screen for plants that are homozygous for T-DNA insertions in ACX1 (SALK_041464) and ACX5 (SALK_009998; Supplemental Fig. S1). These assays employed the LBb1 T-DNA oligonucleotide (http://signal.salk.edu/tdna_FAQs.html) and appropriate gene-specific primers (acx1, 64 L 5′-GCAGACAGGAAGAATTTGTGAGAGTTTGG-3′ and 64R 5′-GTGGTGGACATGGATACTTGTGGTG-3′; acx5, 98 L 5′-CCGAGTCATTGAGTGGATCCT-3′ and 98R 5′-CTGGAAAGGCTCCTTCTGGGA-3′). The T-DNA insertion locations were confirmed by DNA sequencing of the amplified PCR products. The acx1/5 double mutant was generated by crossing the single mutants and screening the resulting F2 progeny for plants exhibiting reduced seed set. A total of 12 double mutant plants were recovered from 210 F2 progeny, in good agreement with the hypothesis that T-DNA insertions in ACX1 and ACX5 segregate as independent recessive mutations. The identity of the double knockouts was confirmed by PCR (Supplemental Fig. S1b). Fertility of acx1/5 plants was restored by treatment of flowers with MeJA, as previously described (Stintzi and Browse, 2000).
RT-PCR analysis with primers that hybridize to the 5′ untranslated region (5′-CACACTCGAGAATCTGAGACAATAG-3′) of ACX1 and the 3′ end of the ACX1 open reading frame (5′-GGGTCGACTCAGAGCCTAGCGGTACGAAG-3′) detected full-length ACX1 transcripts in RNA isolated from wild-type but not acx1 leaves (data not shown). Prolonged exposures of ACX1-probed northern blots showed that low levels of ACX1-related transcripts accumulate in acx1 leaves. RT-PCR experiments and DNA sequencing showed that these transcripts are derived from transcriptional read through of ACX1 into the T-DNA insertion (data not shown). The corresponding ACX1 proteins are predicted to lack amino acids encoded by at least one ACX1 exon and thus can be assumed to be nonfunctional. RT-PCR analysis of acx5 plants with primers that hybridize to the 5′ (5′-CACACTCGAGAATCTGAGACAATAG-3′) and 3′ (5′-GAGTTAGAGTTTGGCAGAGCGG-3′) untranslated regions of ACX5 detected ACX5 transcripts in RNA isolated from wild-type but not acx5 leaves (data not shown). PCR screening was used to identify a homozygous aos T-DNA insertion mutant (SALK_017756). Primers used for these reactions were 5′-TTCTCTCCTTCTTCTCCGACG-3′ and 5′-GATCCATCGGAGCCTAAACAC-3′.
Western-Blot Analysis
Recombinant His-tagged LeACX1A was affinity purified, as previously described (Li et al., 2005). Rabbit polyclonal antibodies against this antigen were produced by a commercial vendor (Cocalico Biologicals) according to their standard protocol. Western-blot analysis was performed with 30 μg of total protein extracted from 3-week-old rosette leaves. Proteins were separated on 10% SDS-polyacrylamide gels and transferred to Immobilon-P membranes (Millipore) according to standard procedures (Harlowe and Lane, 1988). Membranes were incubated at 24°C for 1 h with anti-LeACX1A antibodies that were diluted 1:1,000 in Tris-buffered saline with 0.1% Tween 20 containing 1% nonfat milk. As a control, duplicate protein blots were incubated with preimmune serum obtained from the same rabbit that was immunized with LeACX1A. Blots were washed three times with Tris-buffered saline with 0.1% Tween 20 and then incubated with a peroxidase-conjugated anti-rabbit secondary antibody (1:25,000 dilution; Sigma). ACX protein-antibody complexes were visualized with the SuperSignal West Pico Chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
JA Measurements
Four-week-old plants were wounded twice with a hemostat across the midvein of each rosette leaf. At various times after wounding, leaf tissue (200–300 mg) from at least three different plants of the same genotype was pooled and frozen in liquid nitrogen. Tissue was also collected from unwounded control plants grown in the same flat. JA was extracted according to the vapor-phase extraction procedure (Schmelz et al., 2004), and quantified by GC-MS, as previously described (Li et al., 2005). For JA measurements in flowers, unopened buds and the first two opened flowers within the flower cluster were pooled from at least five inflorescences of each genotype. A total of 50 to 60 mg of flower tissue was used for each JA extraction. Because this procedure involves methylation of the endogenous JA pool, data are expressed in MeJA equivalents.
RNA-Blot Analysis
Wounding and harvesting of leaf tissue was done as described above. RNA extraction and gel-blot analysis were performed as previously described (Li et al., 2002). Probes prepared using clones obtained from the Arabidopsis Biological Resource Center were as follows: full-length cDNA clones were used for VSP1 (stock no. 114D3), OPR3 (U13428), ACX3 (U24900), and ACX4 (U21944). A partial-length cDNA clone (approximately 2 kb) was used for ACX2 (G12C9). Gene-specific probes corresponding to the 3′ end of ACX1 and ACX5 were generated by PCR amplification of genomic DNA, using the following primers: ACX1, 5′-CTAATGCGGTTGCACTTGTGGA-3′ (F) and 5′-TATCCCACATATCCTTCTCAGAGTAG-3′ (R); ACX5, 5′-AACGCTTCCGCTCTGCCAAACTC-3′ (F) and 5′-GAAGGTAAAGCAAAAGGGCA-3′ (R). RNA quality and equal loading was confirmed by staining duplicate gels with ethidium bromide, as well as by hybridization of blots to a cDNA probe for Actin-8. The Actin-8 cDNA was obtained by RT-PCR with the following primers: 5′-GARAARATGACNCARATNATGTTYGARACNTT-3′ and 5′-TCYTTNCTNATRTCNACRTCRCAYTTCATDAT-3′.
Insect Feeding Trials and Fungal Pathogenicity Assays
Trichoplusia ni eggs were obtained from Benzon Research and hatched at 30°C. Within 8 h of hatching, a single larva was transferred to 4-week-old plants. Feeding trials were conducted over a period of 10 d in a growth chamber maintained at 21°C under 12 h of light (100 μE m−2 s−1) and 12 h of dark. At the end of the trial, individual larvae were weighed, and the plants were photographed. For analysis of JA levels, triplicate samples of T. ni-damaged leaf tissue were collected from wild-type and acx1/5 plants at different times after initiation of the feeding trial. In trials involving JA-treated plants, 20 μL of a solution containing 1 mm JA (Sigma) and 0.1% Tween 20 was applied to each rosette leaf immediately before insect challenge.
Western flower thrips (Frankliniella occidentalis) were obtained from a colony reared on marigolds (Tagetes patula). Adult and larval stages of thrips were collected onto filter paper by shaking infested marigold flowers. Thrips were then transferred to 4-week-old plants in a growth chamber and allowed to feed for 3 weeks. Plants of each of the four genotypes were spatially randomized within the growth chamber.
A. brassicicola (strain MUCL20297) was grown on potato (Solanum tuberosum) dextrose agar at 25°C for 10 d, at which time conidia/conidiospores were collected in water. Four-week-old soil-grown Arabidopsis plants were inoculated on the leaf surface with a 5-μL drop of a suspension containing 5 × 105 spores mL−1. Flats containing the inoculated plants were covered with transparent plastic to maintain high humidity. Plants were maintained at 25°C for 5 d prior to assessing the disease phenotype. Gene expression and JA analysis were performed on samples collected daily from inoculated wild-type and acx1/5 leaves. As a control, mock-inoculated (5 μL water) samples were collected at each of the time points.
Pollen Viability Measurements
Pollen viability was measured by double staining of pollen grains with FDA and PI. The procedure was essentially as described by McConn and Browse (1996), with minor modifications. A total of 2 mg mL−1 FDA in acetone was added drop wise to a 20% (w/v) Suc solution. Pollen from newly dehisced anthers was transferred to a glass slide. Following the addition of equal volumes of FDA and PI solution (1 μg mL−1) to the pollen, the sample was covered with a coverslip and incubated in the dark for approximately 10 min. Pollen was visualized under UV illumination with an epifluorescence microscope (Zeiss Axiophot) equipped with a 4′,6-diamino-phenylindole filter set (excitation at 365 nm; emission at 450 nm longpass). FDA is deesterified within living cells to fluorescein, which emits a green fluorescence signal under UV excitation. Only nonviable cells incorporate PI, which fluoresces red orange under UV light. In vitro pollen germination was performed as previously described (Thorsness et al., 1993).
Construction of Plasmids and Transgenic Plants
For localization of ACX1 expression in various tissues, transgenic plants expressing GUS or YFP under the control of the ACX1 promoter were generated. A 1.3-kb promoter fragment upstream of the ACX1 translation initiation site was amplified by PCR from Arabidopsis (Col-0) genomic DNA. Primers used for the amplification of the promoter fragment that was incorporated into the ACX1∷GUS construct were as follows: 5′-AATTATCGATGCTAACATACGCCACTTCCTAGTC-3′ (underlined, ClaI site); 5′-TTAATCTAGAAATTCCTTCCATGATTCGTAATTC-3′ (underlined, XbaI site). This promoter was subcloned into the ClaI and XbaI sites of pBI121 (replacing the cauliflower mosaic virus 35S promoter) to generate the ACX1∷GUS construct (Fig. 2A). A modified form of pBI121 was used to facilitate the construction of ACX1∷YFP-PTS1. Briefly, two pairs of complementary primers (pair 1, 5′-GATCCACTAGTCTCGAG-3′ and 5′-ACGTGCTCGAGACTAGTG-3′; pair 2, 5′-CACGTGGTCGACGGTACCGAGCT-3′ and 5′-CGGTACCGTCGACC-3′) were annealed and ligated to a pBI121 vector that was digested with BamHI and SacI. This manipulation removed the GUS gene (also known as uidA) and introduced a new set of restriction enzyme sites for cloning. The EYFP open reading frame and the PTS1 (SKL) were amplified from EYFP-Peroxi (CLONTECH) with primers 5′-CGCGGATCCATGGTGAGCAAGGGCGAG-3′ (underlined, BamHI site) and 5′-CGGCTCGAGCTACAGCTTGGACTTGTAC-3′ (underlined, XhoI site). The EYFP-PTS1 PCR product was ligated into the BamHI and XhoI sites of the modified pBI121 vector to create a 35S promoter-driven EYFP-PTS1 construct. The ACX1 promoter used for the ACX1∷GUS construct was then subcloned into the ClaI and XbaI sites of the 35S∷EYFP-PTS1 vector to generate the ACX1∷YFP-PTS1 construct (Fig. 2A).
The ACX1∷YFP-PTS1 and ACX1∷GUS constructs were transformed into Agrobacterium tumefaciens strain C58C1. The resulting strains were used to transform Arabidopsis ecotype Col-0 with the floral dip method (Clough and Bent, 1998). T1 seeds were screened for the presence of the transgene on Murashige and Skoog media containing 50 μg mL−1 kanamycin. Multiple lines harboring each construct were analyzed for expression of the reporter, as described below.
Histochemical Detection of GUS Activity
Whole transgenic plants, or tissues obtained from these plants, were collected in ice-cold 90% acetone and then incubated for 20 min at ambient temperature in the same solution. These tissues were washed briefly with staining solution consisting of 50 mm sodium phosphate, pH 7.2, 0.01% Triton X-100 (v/v), 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, and 0.5 mg mL−1 X-gluc (Gold Bio Technology) and then incubated in the same solution overnight at 37°C. Stained tissues were cleared with a series of ethanol washes (30%–70%). Images of X-gluc-stained tissues were taken with either a Wild Heerbrugg dissecting microscope or a Zeiss Axiophot epifluorescence microscope equipped with a Nikon Coolpix 4500 camera.
Confocal Microscopy
Tissues of transgenic Arabidopsis expressing YFP under the control of the ACX1 promoter were hand sectioned with razor blades and mounted in distilled water between a slide and coverslip. For mounting of whole seedlings, seedlings grown on Murashige and Skoog media supplemented with 0.5% (w/v) Suc were placed on glass slides with distilled water and pressed gently. Confocal fluorescence images of the specimens were taken with a Zeiss LSM5 Pascal laser-scanning confocal microscope equipped with an argon laser. Chloroplast autofluorescence was excited with a 488-nm argon laser and was detected after passage through a long pass 650-nm emission filter. YFP fluorescence was excited with a 488-nm laser and was detected after passage through a band pass 505- to 530-nm emission filter.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of SALK lines harboring T-DNA insertions in ACX1 and ACX5.
Supplemental Figure S2. acx1/5 plants exhibit increased susceptibility to thrips.
Supplementary Material
Acknowledgments
We thank Gyu In Lee and Chuanyou Li for assistance in isolating T-DNA insertion lines, Guanghui Liu for help with PCR genotyping and RNA-blot analysis, John Scott-Craig for assistance with culturing of A. brassicicola, and Bart Thomma for providing the A. brassicicola strain.
This work was supported by Michigan State University (Graduate School Dissertation Completion Fellowship to A.L.S.), by the National Institutes of Health (grant no. GM57795), and by the U.S. Department of Energy (grant no. DE–FG02–91ER20021 to G.A.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gregg A. Howe (howeg@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J 41 859–874 [DOI] [PubMed] [Google Scholar]
- Afitlhile MM, Fukushige H, Nishimura M, Hildebrand DF (2005) A defect in glyoxysomal fatty acid β-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol Biochem 43 603–609 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodolou FL (2006) Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci 11 124–132 [DOI] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27 933–942 [DOI] [PubMed] [Google Scholar]
- Browse J (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72 431–456 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48 355–381 [DOI] [PubMed] [Google Scholar]
- Cruz Castillo M, Martinez C, Buchala A, Metraux JP, Leon J (2004) Gene-specific involvement of β-oxidation in wound-activated responses in Arabidopsis. Plant Physiol 135 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact 18 923–937 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Zaanen W, Koornneef A, Korzelius JP, Dicke M, Van Loon LC, Pieterse CMJ (2006) Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol 142 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Turner JG (2005) Jasmonate-regulated Arabidopsis stress signalling network. Physiol Plant 123 161–172 [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A, Farmer EE (2004) Keeping the leaves green above us. Science 306 1515–1516 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41 156–181 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT (2005) Jasmonates and related compounds in plant-insect interactions. J Plant Growth Regul 23 238–245 [Google Scholar]
- Harlowe E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Hooks MA, Kellas F, Graham IA (1999) Long-chain acyl-CoA oxidases of Arabidopsis. Plant J 20 1–13 [DOI] [PubMed] [Google Scholar]
- Howe GA (2004) Jasmonates as signals in the wound response. J Plant Growth Regul 23 223–237 [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Chung HS, Kobayashi Y, Howe GA (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281 33511–33520 [DOI] [PubMed] [Google Scholar]
- Kubigsteltig I, Laudert D, Weiler EW (1999) Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208 463–471 [DOI] [PubMed] [Google Scholar]
- Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, et al (2005) Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Williams MM, Loh YT, Lee GI, Howe GA (2002) Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol 130 494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O, Bohlmann H, Wasternack C (1999) Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 50 517–523 [Google Scholar]
- Mithofer A, Maitrejean M, Boland W (2005) Structural and biological diversity of cyclic octadecanoids, jasmonates, and mimetics. J Plant Growth Regul 23 170–178 [Google Scholar]
- Muller A, Duchting P, Weiler EW (2002) A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216 44–56 [DOI] [PubMed] [Google Scholar]
- Mussig C, Biesgen C, Lisso J, Uwer U, Weiler E, Altmann T (2000) A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a link between brassinosteriod action and jasmonic acid synthesis. J Plant Physiol 157 143–152 [Google Scholar]
- Olsen LJ (1998) The surprising complexity of peroxisome biogenesis. Plant Mol Biol 38 163–189 [PubMed] [Google Scholar]
- Pedersen L, Henriksen A (2005) Acyl-CoA oxidase 1 from Arabidopsis thaliana: structure of a key enzyme in plant lipid metabolism. J Mol Biol 345 487–500 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, Graham IA (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43 861–872 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK (2006) Peroxisomal β-oxidation-A metabolic pathway with multiple functions. Biochim Biophys Acta 1763 1413–1426 [DOI] [PubMed] [Google Scholar]
- Powers RA, Rife CL, Schilmiller AL, Howe GA, Garavito RM (2006) Structure determination and analysis of acyl-CoA oxidase (ACX1) from tomato. Acta Crystallogr D Biol Crystallogr 62 683–686 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Van Loon LC, Pieterse CMJ (2004) Jasmonates: signals in plant-microbe interactions. J Plant Growth Regul 23 211–222 [Google Scholar]
- Ramwell PW, Foegh M, Loeb R, Leovey EMK (1980) Synthesis and metabolism of prostaglandins, prostacyclin, and thromboxanes: arachidonic acid cascade. Semin Perinatol 4 3–12 [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB (1999) A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11 1911–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Schaller A, Stintzi A (2005) Biosynthesis and metabolism of jasmonates. J Plant Growth Regul 23 179–199 [Google Scholar]
- Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DJ (2003) Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol 132 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA (2005) Systemic signaling in the wound response. Curr Opin Plant Biol 8 369–377 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT (2004) The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J 39 790–808 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15 747–754 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al (2005) 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants: implications for transport of jasmonate precursors into peroxisomes. Plant Physiol 137 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness M, Kandasamy M, Nasrallah M, Nasrallah J (1993) Genetic ablation of floral cells in Arabidopsis. Plant Cell 5 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick B, Zimmerman D (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiol 75 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW, Albrecht T, Groth B, Xia ZQ, Luxem M, Liss H, Andert L, Spengler P (1993) Evidence for the involvement of jasmonates and their octadecanoid precursors in the tendril coiling response of Bryonia dioica. Phytochemistry 32 591–600 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.