Abstract
Insect eggs represent a threat for the plant as hatching larvae rapidly start with their feeding activity. Using a whole-genome microarray, we studied the expression profile of Arabidopsis (Arabidopsis thaliana) leaves after oviposition by two pierid butterflies. For Pieris brassicae, the deposition of egg batches changed the expression of hundreds of genes over a period of 3 d after oviposition. The transcript signature was similar to that observed during a hypersensitive response or in lesion-mimic mutants, including the induction of defense and stress-related genes and the repression of genes involved in growth and photosynthesis. Deposition of single eggs by Pieris rapae caused a similar although much weaker transcriptional response. Analysis of the jasmonic acid and salicylic acid mutants coi1-1 and sid2-1 indicated that the response to egg deposition is mostly independent of these signaling pathways. Histochemical analyses showed that egg deposition is causing a localized cell death, accompanied by the accumulation of callose, and the production of reactive oxygen species. In addition, activation of the pathogenesis-related1∷β-glucuronidase reporter gene correlated precisely with the site of egg deposition and was also triggered by crude egg extract. This study provides molecular evidence for the detection of egg deposition by Arabidopsis plants and suggests that oviposition causes a localized response with strong similarity to a hypersensitive response.
Plants under attack from herbivorous insects develop an array of defenses that are aimed at slowing the growth or development of the aggressor and specifically tailored to protect themselves from further attack. Numerous studies have revealed that, in addition to constitutive defenses represented by trichomes, thick secondary cell walls, or poisonous compounds, plants are equipped with inducible defenses that can be grouped into indirect (production of volatile odor blends to attract natural enemies of the attacker encountered) and direct responses (production of antidigestive proteins, toxic secondary compounds, and enzymes that affect insect growth and development; Karban and Baldwin, 1997; Walling, 2000; Dicke et al., 2003). In Arabidopsis (Arabidopsis thaliana), challenge with chewing herbivores has been shown to provoke substantial transcriptional changes largely controlled by the plant hormone jasmonic acid (JA; Reymond et al., 2004; De Vos et al., 2005).
One important aspect of plant/insect interaction that has received less attention is the potential recognition of insect egg deposition. Eggs represent a future threat for the plant and the anticipation of damage by a preactivation of defenses could provide an advantage to the host. Both direct and indirect responses to oviposition have been observed in plants (Hilker and Meiners, 2006). In pea (Pisum sativum) pods, growth of undifferentiated cells is triggered upon oviposition by the pea weevil, which results in elevating the egg from the surface, increasing the risk of desiccation, predation, or falling off the pod (Doss et al., 2000). Upon oviposition by the white-backed planthopper, rice (Oryza sativa) plants produce benzyl benzoate, an ovicidal substance that causes high egg mortality (Seino et al., 1996). The development of a necrotic zone at the site of egg deposition was observed in Brassica nigra, resulting in egg dessication and mortality (Shapiro and Devay, 1987). In one potato (Solanum tuberosum) hybrid clone, a necrotic zone around the oviposition site provoked the detachment and the fall of the eggs on the soil (Balbyshev and Lorenzen, 1997).
Indirect defenses induced by egg deposition comprise the release of volatiles or the modification of the surface chemistry by egg-laden leaves resulting in the attraction of egg parasitoids. Twigs of Scots pine (Pinus sylvestris) emit volatiles after oviposition by the pine sawfly Diprion pini, attracting the egg parasitoid Chrysonotomyia ruforum. This response was shown to be both local and systemic and could be mimicked by JA treatment (Hilker et al., 2002). Similar tritrophic interactions have been observed in elm (Ulmus spp.; Meiners and Hilker, 2000), bean (Phaseolus vulgaris; Colazza et al., 2004), and brussels sprout (Brassica oleracea gemmifera; Fatouros et al., 2005). The nature of the elicitor of oviposition-induced volatile release is not known but has been associated with oviduct secretions coating the egg (Meiners and Hilker, 2000; Hilker et al., 2005).
There is thus evidence that plants are capable of detecting the presence of insect eggs and that they respond by activating direct and indirect defenses. However, there is no information on the molecular changes that take place in the host following oviposition. In this study, we investigated the response of Arabidopsis to oviposition by pierid butterflies and analyzed whole-genome transcript profiles in egg-laden leaves for a period of 3 d. The combined results of transcriptome analysis and visualization of defense-associated markers reveal that oviposition triggers a local response similar to a programmed cell death (PCD) accompanied by the activation of many defense-related genes.
RESULTS
Expression Changes in Response to Pieris brassicae Eggs
After mating, female Pieris brassicae butterflies lay eggs on the underside of leaves of Brassicaceae family plants. Approximately 10 to 40 eggs are gently deposited close to each other without visible physical damage. From 4 to 6 d after oviposition, eggs start to hatch, young larvae eat the egg chorion, and then start to eat the leaf around the oviposition site. We wanted to know whether molecular changes take place in Arabidopsis during the first days after oviposition, before egg hatching and thus before larvae start to feed on the plant. Using near full-genome microarrays, we investigated transcriptional changes 24, 48, and 72 h after oviposition by P. brassicae. On average, butterflies laid one to two egg batches per plant and we collected five leaf discs of approximately 5 mm in diameter at the appropriate times. RNA was extracted, amplified, labeled, and hybridized to Complete Arabidopsis Transcriptome MicroArray (CATMA) microarrays containing 22,473 gene-specific tags (Allemeersch et al., 2005). As controls, an equal number of leaf discs from a distal egg-free leaf were sampled at each time point, and the RNA extracted. This ensured that any observed changes were due to the presence of eggs and not to signals or contacts generated by flying butterflies. Each experiment was repeated three times independently, and each sample was labeled either with Cy3-dCTP or Cy5-dCTP (dye-swap design), giving a total of six measurements per time point. Average expression values were computed and genes whose expression ratio was ≥2 or ≤0.5 and P value < 0.05 (both criteria had to be met in at least one time point) were considered as differentially regulated by egg deposition.
Eggs laid by P. brassicae triggered large changes in gene expression. A total of 303 genes were induced 24 h after oviposition, 416 after 48 h, and 671 after 72 h (Supplemental Table S1). In addition, oviposition caused a down-regulation of 53, 123, and 426 genes after 24, 48, and 72 h, respectively (Supplemental Table S1). This represents a maximal induction of approximately 4% and a repression of approximately 2% of the transcriptome. Analysis of the time course showed that the 56% of the genes that are induced at 72 h are already up-regulated at 24 or 48 h and that the expression of most genes gradually increased or stayed steady over the 3 d of measurements, indicating that the response is more likely due to the continuous presence of the eggs and not to a touch response to egg deposition. The majority of down-regulated genes (68%) was only repressed 72 h after oviposition (Supplemental Figs. S1–S3). It is possible that we have missed some early changes in gene expression but we observe that the response to oviposition corresponds to consistent and long-lasting transcriptional changes. In addition, we might have underestimated the number of differentially regulated genes after oviposition by using distal leaves as control tissue. As mentioned above, the rationale was to use leaves that had been in contact with butterflies as a control to eliminate the possibility of identifying genes responding to touch or butterfly derived chemicals. The limitation of this experimental design is that genes induced after oviposition at similar levels in oviposited leaves and in distal leaves will not be detected. However, one hybridization using leaf samples from plants that did not contain eggs indicated that the number of genes induced systemically is low (Supplemental Fig. S4).
We examined the potential function of differentially expressed genes and classified them according to gene ontology (GO) terms using the tools for GO annotations at The Arabidopsis Information Resource (http://www.arabidopsis.org; Berardini et al., 2004). The distribution of gene annotations in different functional classes revealed a relatively high abundance of defense- and stress-related genes (Table I). Since eggs represent a future threat for the plant as they generate chewing herbivores that will feed on the leaves, we were curious to compare the transcript profile after oviposition to that of plants experiencing herbivory. We performed transcriptional analyses of Arabidopsis plants challenged with third-instar larvae of the specialist Pieris rapae using CATMA microarrays. We have previously shown using a dedicated microarray containing defense-related genes that both P. brassicae and P. rapae larvae trigger highly similar expression changes (Reymond et al., 2000). Surprisingly, there was very little overlap between oviposition- and herbivory-induced genes; only 10% (75/766) of the genes induced by P. brassicae eggs were also induced by P. rapae herbivory. These genes included members of the phenylpropanoid pathway (prephenate dehydratase, At3g44720; PAL1, At2g37040; C4H, At2g30490; TAT3, At2g24850; flavonol synthase, At5g05600) implicated in the synthesis of defense compounds; several genes involved in the synthesis of Trp (ASA1, At5g05730; TRP1, At5g17990; TSB1, At5g54810; TSB2, At4g27070); genes involved in redox balance or oxidative stress response (thioredoxin, At1g45145; glutaredoxin, At1g28480; two peroxidases, At5g64120 and At5g05340); and stress-responsive transcription factors (HSF4, At4g36990; ZAT10, At1g27730; ZAT12, At5g59820). They also included genes potentially involved in the synthesis and response to JA, salicylic (SA), and ethylene defense signals (LOX4, At1g72520; OPR3, At2g06050; ACX1, At4g16760; JR1, At3g16470; EDS5, At4g39030; 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase, At1g05010; ATERF1, At4g17500; ERF2, At5g47220; Supplemental Table S1).
Table I.
Functional classification of genes regulated by P. brassicae oviposition using GO categories
| Biological Process | No. of Genes
|
|
|---|---|---|
| Up-Regulated | Down-Regulated | |
| Response to abiotic or biotic stimulus | 125 | 48 |
| Response to stress | 86 | 31 |
| Protein metabolism | 113 | 65 |
| Transport | 67 | 18 |
| Transcription | 45 | 18 |
| Developmental processes | 29 | 21 |
| Electron transport or energy pathways | 37 | 19 |
| Cell organization and biogenesis | 33 | 29 |
| Signal transduction | 34 | 24 |
| DNA or RNA metabolism | 2 | 1 |
| Other processes | 318 | 162 |
| Biological process unknown | 201 | 145 |
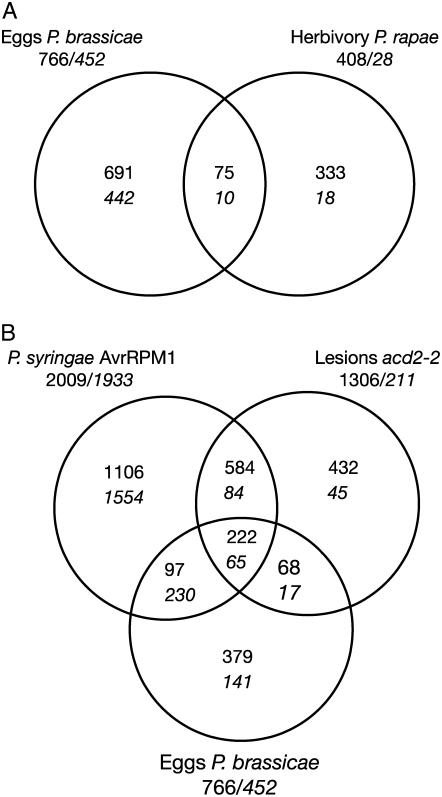
The overlap between oviposition- and herbivory-repressed genes was even smaller (Fig. 1A; Supplemental Table S1). Of the 10 genes repressed by both treatments, seven are annotated as expressed proteins with unknown function. The other three include an expansin (ATEXPA8, At2g40610), a germin (At5g20630), and a putative zinc-finger protein (At2g25900).
Figure 1.
A, Comparison of transcript profiles between oviposition by P. brassicae (24–72 h, three replicates each) and herbivory by P. rapae (challenge for 5 h by 3rd-instar larvae, seven replicates). B, Comparison of transcript profiles between oviposition, infection by P. syringae AvrRPM1 for 24 h (three replicates, data from AtGenExpress), or during lesion formation in the mutant acd2-2 (three replicates). Diagrams represent the numbers of induced (normal) or repressed (italic) genes in each category. All selected genes have an expression ratio ≥ 2 or ≤ 0.5 and a P value < 0.05. The total number of differentially expressed genes in each experiment is indicated outside the diagram. Genes induced after oviposition and any of the other treatments are listed in Supplemental Tables S1, S4, and S5.
Looking into the list of induced genes in more details we identified several markers of hypersensitive response (HR), which is a particular case of PCD. HR is an induced response triggered by the specific recognition of bacterial pathogens, viruses, fungi, and nematodes and is characterized by a localized cell death at the site of infection that prevents the progression of the disease (Kombrink and Somssich, 1995). We thus compared the expression profile after oviposition by P. brassicae with the profile observed after an HR caused by the bacterial pathogen Pseudomonas syringae AvrRPM1 (data from AtGenExpress; http://www.weigelworld.org/resources/microarray), and with the expression changes triggered by lesion formation in acd2-2. This mutant has spontaneous spreading cell death lesions (Mach et al., 2001). We observed an important overlap between the three transcriptome signatures; 50% of the egg-induced genes, respectively, 69% of the repressed genes, were also differentially regulated during either a bacterial HR or in leaves developing spontaneous lesions, indicating that eggs trigger a transcriptional response sharing a lot in common with those observed during the programmed death of plant cells (Fig. 1B; Supplemental Table S1). Interestingly, we observed that most of the 75 genes induced both by oviposition and insect herbivory (Fig. 1A) were also induced in acd2-2 mutant (62/75) and in P. syringae infected plants (64/75), indicating that these genes are likely markers of a general stress response. Similarly, the majority of the 10 genes repressed both by oviposition and insect herbivory were also down-regulated in acd2-2 mutants (8/10) and by P. syringae treatment (8/10; Supplemental Table S1).
Egg deposition by P. brassicae caused the up-regulation of many defense-related genes (Table II). Classical marker genes that accumulate during HR were induced, including pathogenesis-related (PR) genes PR2, PR3, PR4, and PR5, and regulators of innate immunity, like EDS1, PAD4, and SAG101. Some avirulence- or HR-responsive genes of unknown function were also up-regulated. Genes related to PCD included the antiapoptosis BAX-INHIBITOR-1 (Matsumura et al., 2003); two genes that repress cell death and promote cell growth, BONZAI1 (BON1) and BON1-associated protein (BAP1; Hua et al., 2001; Yang et al., 2006); two proteases belonging to the group of metacaspases that regulate plant PCD (Sanmartin et al., 2005); and a zinc-finger (MYND type) family protein predicted to be associated with PCD. Cells that undergo HR synthesize callose and phenolic compounds. A gene encoding a callose synthase (CALS1) and several members of the phenylpropanoid pathways were induced by oviposition. Cellular protection against an oxidative burst as well as defense responses are also activated in adjacent cells surrounding the site of HR. Several genes involved in the regulation of oxidative stress, like a superoxide dismutase, an l-ascorbate oxidase, and a glutathione S-transferase were induced. Up-regulated genes encoding defense proteins included chitinases, lectins, and proteinase inhibitors. The Trp pathway provides precursors for several defense compounds like camalexin and indole glucosinolates in Arabidopsis (Wittstock and Halkier, 2002; Glawischnig et al., 2004). Several genes involved in the biosynthesis of indole glucosinolates were induced by oviposition. Among other interesting genes potentially involved in defense or signaling, we identified 17 WRKY transcription factors that are typically involved in the regulation of defense- or senescence-induced programs (Eulgem et al., 2000), several MYB, NAC, ERF/AP2, and zinc-finger transcription factors, 29 protein kinase genes, 13 genes associated with calcium signaling, 21 genes encoding various transporters, and seven genes involved in redox metabolism (Supplemental Table S1).
Table II.
List of selected genes regulated after P. brassicae oviposition
Expression changes were analyzed 24, 48, and 72 h after egg deposition. Average expression ratios are calculated from three independent biological replicates. See also Supplemental Table S1. AGI, Arabidopsis Genome Initiative gene code.
| Gene Description | Gene Symbol | AGI | 24 h
|
48 h
|
72 h
|
|||
|---|---|---|---|---|---|---|---|---|
| Ratio | P Value | Ratio | P Value | Ratio | P Value | |||
| Induced genes | ||||||||
| PR proteins | ||||||||
| Callose synthase | CALS1 | At1g05570 | 2.14 | 0.0030 | 2.64 | 0.0061 | 3.01 | 0.0018 |
| Glucan endo-1,3-β-glucosidase | BGL2/PR2 | At3g57260 | 9.71 | 0.0006 | 11.40 | 0.0000 | 7.60 | 0.0015 |
| Chitinase | CHIB/PR3 | At3g12500 | 1.08 | 0.2720 | 1.36 | 0.1136 | 2.92 | 0.0427 |
| Chitinase | At2g43570 | 9.67 | 0.0011 | 37.01 | 0.0000 | 35.93 | 0.0009 | |
| Hevein-like protein | PR4/HEL | At3g04720 | 1.53 | 0.0580 | 4.81 | 0.0000 | 4.68 | 0.0002 |
| PR protein 5 | PR5 | At1g75040 | 4.10 | 0.0000 | 12.41 | 0.0000 | 15.80 | 0.0004 |
| Lectin | At5g18470 | 3.76 | 0.0002 | 6.80 | 0.0000 | 7.93 | 0.0000 | |
| Trypsin inhibitor | ATTI1 | At2g43510 | 2.70 | 0.0004 | 6.09 | 0.0014 | 5.38 | 0.0000 |
| Trypsin and protease inhibitor | At1g73260 | 2.24 | 0.3632 | 2.87 | 0.0270 | 42.77 | 0.0043 | |
| Regulators of innate immunity | ||||||||
| Disease resistance protein/lipase | EDS1 | At3g48090 | 2.27 | 0.0005 | 2.64 | 0.0000 | 2.22 | 0.0018 |
| Phytoalexin-deficient 4 protein/lipase | PAD4 | At3g52430 | 3.52 | 0.0000 | 4.51 | 0.0001 | 3.78 | 0.0023 |
| Leaf senescence-associated protein/lipase | SAG101 | At5g14930 | 1.64 | 0.0178 | 1.94 | 0.0039 | 2.19 | 0.0002 |
| Flavin-containing monooxygenase | FMO1 | At1g19250 | 1.17 | 0.3632 | 1.93 | 0.1514 | 2.23 | 0.0201 |
| MutT/nudix family protein | NUDT7 | At4g12720 | 3.16 | 0.0001 | 2.71 | 0.0003 | 2.88 | 0.0045 |
| Multidrug and toxin extrusion transporter | EDS5/SID1 | At4g39030 | 4.80 | 0.0058 | 4.35 | 0.0000 | 7.09 | 0.0000 |
| NPR1/NIM1-interacting protein 2 | NIMIN-2 | At3g25882 | 5.15 | 0.0000 | 7.74 | 0.0000 | 5.30 | 0.0000 |
| Biotic stress response | ||||||||
| Mildew resistance protein, RPW8 homolog | HR4 | At3g50480 | 3.41 | 0.0000 | 4.72 | 0.0003 | 3.21 | 0.0098 |
| Disease resistance protein (CC-NBS-LRR class) | ADR1 | At1g33560 | 2.14 | 0.0024 | 2.26 | 0.0003 | 2.32 | 0.0043 |
| Disease resistance protein (TIR class) | At1g66090 | 2.85 | 0.0100 | 3.20 | 0.0269 | 2.84 | 0.0003 | |
| Avirulence-induced gene | AIG1 | At1g33960 | 10.55 | 0.0006 | 20.83 | 0.0000 | 29.40 | 0.0012 |
| AvrRpt2-induced protein | AIG2 | At3g28930 | 1.49 | 0.0094 | 2.75 | 0.0002 | 3.20 | 0.0008 |
| HR-induced protein | At3g01290 | 2.90 | 0.0004 | 3.83 | 0.0012 | 2.32 | 0.0002 | |
| Harpin-induced protein | YLS9 | At2g35980 | 5.19 | 0.0119 | 5.17 | 0.0053 | 23.66 | 0.0002 |
| Pathogen-responsive α-dioxygenase | At1g73680 | 1.66 | 0.0347 | 1.71 | 0.0368 | 3.12 | 0.0017 | |
| Cell death | ||||||||
| Bax inhibitor-1 family | AtBI-1 | At5g47130 | 1.66 | 0.0868 | 2.51 | 0.0426 | 5.56 | 0.0005 |
| BON1-associated protein 1 | BAP1 | At3g61190 | 3.02 | 0.0053 | 4.46 | 0.0001 | 2.88 | 0.0016 |
| Copine BONZAI1 | BON1 | At5g61900 | 2.23 | 0.0010 | 2.45 | 0.0004 | 3.04 | 0.0015 |
| Zinc-finger (MYND type) family protein | At4g02220 | 1.95 | 0.0003 | 2.15 | 0.0020 | 2.15 | 0.0006 | |
| Caspase | AtMCP1c | At4g25110 | 2.71 | 0.0001 | 3.36 | 0.0000 | 1.96 | 0.0008 |
| Lesion inducing protein related | At4g14420 | 1.66 | 0.0074 | 2.00 | 0.0014 | 2.34 | 0.0019 | |
| Short-chain alcohol dehydrogenase | SAG13 | At2g29350 | 11.26 | 0.0001 | 33.92 | 0.0000 | 45.04 | 0.0001 |
| Proteolysis | ||||||||
| Cys proteinase | At3g49340 | 4.51 | 0.0108 | 16.88 | 0.0001 | 16.32 | 0.0000 | |
| Cathepsin B-like Cys protease | At4g01610 | 1.76 | 0.0014 | 2.67 | 0.0019 | 2.58 | 0.0000 | |
| Metalloproteinase | At1g24140 | 1.97 | 0.0061 | 2.13 | 0.0258 | 2.16 | 0.0088 | |
| Aspartyl protease | At5g10760 | 3.57 | 0.0002 | 7.76 | 0.0000 | 5.02 | 0.0000 | |
| Antioxidant defense system | ||||||||
| Superoxide dismutase | CSD1 | At1g08830 | 1.80 | 0.0068 | 2.16 | 0.0059 | 2.43 | 0.0000 |
| Monodehydroascorbate reductase | At5g03630 | 1.47 | 0.0018 | 2.27 | 0.0024 | 1.88 | 0.0007 | |
| l-Ascorbate oxidase | At4g39830 | 2.21 | 0.0511 | 2.27 | 0.0003 | 4.29 | 0.0001 | |
| Glutathione S-transferase | GSTU8 | At3g09270 | 1.00 | 0.3632 | 1.37 | 0.1791 | 3.23 | 0.0124 |
| Phenylpropanoid pathway | ||||||||
| Prephenate dehydratase | At3g44720 | 2.26 | 0.0046 | 1.67 | 0.0015 | 3.03 | 0.0052 | |
| Aminotransferase | TAT3 | At2g24850 | 10.68 | 0.0081 | 11.54 | 0.0000 | 8.24 | 0.0015 |
| Phenyl-alanine ammonia-lyase 1 | PAL1 | At2g37040 | 1.38 | 0.1247 | 0.94 | 0.4925 | 4.10 | 0.0027 |
| Cinnamic acid 4-hydroxylase | C4H | At2g30490 | 1.44 | 0.0010 | 1.50 | 0.0001 | 2.59 | 0.0030 |
| Cinnamyl-alcohol dehydrogenase | At1g72680 | 3.56 | 0.0087 | 2.43 | 0.0070 | 3.56 | 0.0002 | |
| Tryptophan pathway | ||||||||
| Chorismate synthase | At1g48850 | 1.71 | 0.0002 | 1.51 | 0.0023 | 2.76 | 0.0006 | |
| Anthranilate synthase, α-subunit | ASA1 | At5g05730 | 1.40 | 0.2477 | 1.91 | 0.1753 | 2.03 | 0.0026 |
| Anthranilate phosphoribosyltransferase | TRP1/PAT1 | At5g17990 | 2.44 | 0.0026 | 2.58 | 0.0001 | 3.47 | 0.0001 |
| Indole-3-glycerol phosphate synthase | IGPS | At2g04400 | 2.03 | 0.0109 | 2.34 | 0.0421 | 2.82 | 0.0003 |
| Tryptophan synthase, α-subunit | TSA1 | At3g54640 | 2.54 | 0.0162 | 3.57 | 0.0001 | 7.34 | 0.0000 |
| Tryptophan synthase, β-subunit 1 | TSB1 | At5g54810 | 1.23 | 0.0005 | 1.27 | 0.0267 | 2.37 | 0.0005 |
| Tryptophan synthase, β-subunit 2 | TSB2 | At4g27070 | 1.97 | 0.3494 | 2.34 | 0.0017 | 3.78 | 0.0023 |
| Cytochrome P450 | CYP79B2 | At4g39950 | 2.99 | 0.0015 | 3.75 | 0.0000 | 8.94 | 0.0000 |
| Cytochrome P450 | CYP83B1 | At4g31500 | 1.73 | 0.0021 | 2.36 | 0.0003 | 2.10 | 0.0009 |
| Aminotransferase | SUR1 | At2g20610 | 1.57 | 0.0025 | 1.59 | 0.0019 | 2.28 | 0.0030 |
| Terpenoid synthesis | ||||||||
| Triterpenoid cyclase | At1g66960 | 2.15 | 0.0582 | 4.70 | 0.0000 | 6.77 | 0.0000 | |
| Terpene synthase/cyclase | At1g61120 | 2.37 | 0.3632 | 3.64 | 0.0071 | 2.70 | 0.0787 | |
| Hormone biosynthesis | ||||||||
| Lipoxygenase | LOX4 | At1g72520 | 2.72 | 0.0134 | 3.33 | 0.0017 | 3.86 | 0.0302 |
| 12-Oxophytodienoate reductase | OPR3 | At2g06050 | 2.07 | 0.0000 | 2.39 | 0.0223 | 1.32 | 0.5373 |
| S-adenosyl-methionine synthetase 1 | SAM1 | At1g02500 | 1.47 | 0.0045 | 1.42 | 0.0009 | 2.07 | 0.0034 |
| ACC oxidase | EFE | At1g05010 | 2.17 | 0.0012 | 2.01 | 0.0015 | 2.71 | 0.0006 |
| ACC synthase | ACS6 | At4g11280 | 1.40 | 0.1027 | 1.17 | 0.1626 | 2.47 | 0.0048 |
| Isochorismate synthase 1 | ICS1/SID2 | At1g74710 | 6.06 | 0.0006 | 5.24 | 0.0044 | 8.56 | 0.0005 |
| Repressed genes | ||||||||
| Cell wall metabolism | ||||||||
| β-Expansin | ATEXPB1 | At2g20750 | 0.51 | 0.0348 | 0.30 | 0.0009 | 0.25 | 0.0003 |
| Expansin | ATEXPA11 | At1g20190 | 0.43 | 0.0042 | 0.41 | 0.0002 | 0.27 | 0.0020 |
| Expansin | ATEXPA5 | At3g29030 | 0.47 | 0.0042 | 0.49 | 0.0052 | 0.20 | 0.0004 |
| Arabinogalactan protein | AGP17 | At2g23130 | 0.78 | 0.1793 | 0.44 | 0.0181 | 0.42 | 0.0326 |
| Fasciclin-like arabinogalactan protein (FLA2) | FLA2 | At4g12730 | 0.56 | 0.0005 | 0.46 | 0.0032 | 0.21 | 0.0008 |
| α-(1,4)-Fucosyltransferase | FUT13 | At1g71990 | 0.69 | 0.0210 | 0.70 | 0.0813 | 0.42 | 0.0017 |
| Glycine-rich cell wall protein | At4g18280 | 0.81 | 0.1149 | 0.74 | 0.0274 | 0.38 | 0.0024 | |
| Cellulose synthase | CSLA09 | At5g03760 | 0.81 | 0.1271 | 0.51 | 0.0024 | 0.33 | 0.0001 |
| Pectate lyase | At5g48900 | 0.57 | 0.0304 | 0.54 | 0.0247 | 0.30 | 0.0014 | |
| Pectinesterase | At4g33220 | 0.62 | 0.0003 | 0.71 | 0.0026 | 0.48 | 0.0012 | |
| Xyloglucan endotransglycosylase | EXGT A1 | At2g06850 | 0.97 | 0.4332 | 0.63 | 0.0719 | 0.31 | 0.0024 |
| Xyloglucan endotransglycosylase | XTR8 | At3g44990 | 0.34 | 0.0000 | 0.39 | 0.0039 | 0.60 | 0.0523 |
| Cutin and wax biosynthesis | ||||||||
| Aldehyde decarbonylase | CER1 | At1g02205 | 1.01 | 0.7803 | 0.28 | 0.0006 | 0.37 | 0.0002 |
| Eceriferum protein | CER2 | At4g24510 | 0.73 | 0.0257 | 0.40 | 0.0001 | 0.23 | 0.0000 |
| Fatty acid elongase 3-ketoacyl-CoA synthase 1 | KCS1 | At1g01120 | 1.09 | 0.8883 | 0.35 | 0.0000 | 0.26 | 0.0008 |
| Long-chain fatty acid—CoA ligase | LACS2 | At1g49430 | 0.66 | 0.0104 | 0.42 | 0.0001 | 0.31 | 0.0004 |
| Very-long-chain fatty acid condensing enzyme | At2g16280 | 0.67 | 0.0105 | 0.75 | 0.0442 | 0.49 | 0.0100 | |
| β-Ketoacyl-CoA synthase (FIDDLEHEAD) | FDH | At2g26250 | 0.77 | 0.0641 | 0.54 | 0.0008 | 0.45 | 0.0000 |
| Fatty acid metabolism | ||||||||
| Enoyl-ACP reductase/fatty acid synthase | MOD1 | At2g05990 | 0.71 | 0.0564 | 0.55 | 0.0049 | 0.44 | 0.0008 |
| Omega-3 fatty acid desaturase | FAD7 | At3g11170 | 0.74 | 0.0258 | 0.83 | 0.1757 | 0.44 | 0.0005 |
| Omega-3 fatty acid desaturase | FAD3 | At2g29980 | 0.99 | 0.4369 | 0.63 | 0.0079 | 0.45 | 0.0045 |
| Photosynthetic activity | ||||||||
| PSI reaction center subunit VI | At3g16140 | 0.68 | 0.0435 | 0.73 | 0.0795 | 0.45 | 0.0070 | |
| PSII 5-kD protein | At1g51400 | 0.62 | 0.0155 | 0.61 | 0.0049 | 0.38 | 0.0019 | |
| PSII core complex psbY | PSBY | At1g67740 | 0.47 | 0.0004 | 0.58 | 0.0051 | 0.33 | 0.0004 |
| PSII reaction center PsbP | At2g39470 | 0.58 | 0.0025 | 0.65 | 0.0006 | 0.45 | 0.0006 | |
| PSII reaction center PsbW | At4g28660 | 0.73 | 0.0094 | 0.68 | 0.0150 | 0.50 | 0.0102 | |
| PSII assembly factor HCF136 | HCF136 | At5g23120 | 0.73 | 0.0050 | 0.65 | 0.0044 | 0.43 | 0.0001 |
| Mg-protoporphyrin IX chelatase | CHLI2 | At5g45930 | 0.49 | 0.0007 | 0.50 | 0.0007 | 0.38 | 0.0008 |
| Divinyl protochlorophyllide a 8-vinyl reductase | PCB2 | At5g18660 | 0.51 | 0.0068 | 0.37 | 0.0012 | 0.25 | 0.0014 |
| ATP synthase protein I | At2g31040 | 0.56 | 0.0033 | 0.70 | 0.0252 | 0.45 | 0.0074 | |
| Chlorophyll a/b-binding protein | LHCB3 | At5g54270 | 0.58 | 0.0102 | 0.51 | 0.0011 | 0.31 | 0.0013 |
| NAD(P)H:plastoquinone dehydrogenase | NDH O | At1g74880 | 0.56 | 0.0037 | 0.69 | 0.0266 | 0.41 | 0.0020 |
| Oxygen evolving enhancer 3 (PsbQ) | At1g14150 | 0.49 | 0.0075 | 0.67 | 0.0261 | 0.34 | 0.0049 | |
| Cytochrome c biogenesis protein | At1g49380 | 0.61 | 0.0001 | 0.65 | 0.0004 | 0.42 | 0.0013 | |
| Ferredoxin | At3g16250 | 0.39 | 0.0012 | 0.55 | 0.0214 | 0.24 | 0.0008 | |
| Ferredoxin-thioredoxin reductase | At5g08410 | 0.68 | 0.0103 | 0.66 | 0.0029 | 0.43 | 0.0002 | |
| Calvin cycle | ||||||||
| Glyceraldehyde-3-P dehydrogenase | At1g12900 | 0.52 | 0.0001 | 0.56 | 0.0001 | 0.39 | 0.0006 | |
| Fructose-1,6-bisphosphatase | At3g54050 | 0.68 | 0.0068 | 0.68 | 0.0023 | 0.45 | 0.0000 | |
| Ribose-5-P isomerase | At3g04790 | 0.57 | 0.0101 | 0.77 | 0.0734 | 0.43 | 0.0079 | |
| Starch biosynthesis | ||||||||
| ADP-Glc pyrophosphorylase | ADG2 | At5g19220 | 0.59 | 0.0002 | 0.60 | 0.0029 | 0.40 | 0.0001 |
| Starch synthase | At5g24300 | 0.75 | 0.0370 | 0.57 | 0.0009 | 0.46 | 0.0006 | |
In addition, we observed that 41 receptor-like kinases (RLKs) were induced by eggs from 24 to 72 h after oviposition. RLKs are transmembrane proteins containing an intracellular kinase domain and a variable extracellular domain thought to interact with different extracellular ligands. There are more than 600 RLKs in Arabidopsis (Shiu and Bleecker, 2001) and the role of only a few of them has been characterized, notably in development, growth, symbiosis, or defense (Zipfel and Felix, 2005). RLKs are known to be involved in the perception of pathogen-derived elicitors that activate defense responses. Recent studies with bacterial elicitors have shown that they induce more than 100 RLKs (Zipfel et al., 2004, 2006). By comparing our data with those of Zipfel et al. (2006) we found that 22 of the oviposition-induced RLKs were also induced after treatment with the bacterial elicitors flg22 or elf26 (Table III).
Table III.
RLK genes induced by P. brassicae oviposition
Expression ratios are averaged from three independent experiments. RLKs with a ratio ≥ 2 and a P value < 0.05 in at least one time point are shown.
| AGI | ECDa | Subfamilya | Gene Symbol | P. brassicae 24 h Ratio | P. brassicae 48 h Ratio | P. brassicae 72 h Ratio | flg22b | elf26b |
|---|---|---|---|---|---|---|---|---|
| At5g47850 | CR4L | CR4L | 1.65 | 1.50 | 2.68 | + | + | |
| At4g23200 | DUF26 | DUF26 | 1.70 | 1.73 | 2.53 | |||
| At4g23220 | TKT | DUF26 | 1.92 | 2.02 | 2.21 | |||
| At4g23190 | DUF26 | DUF26 | RLK3 | 1.61 | 2.08 | 2.67 | + | + |
| At4g23150 | DUF26 | DUF26 | 3.48 | 5.55 | 6.19 | |||
| At3g45860 | DUF26 | DUF26 | 2.71 | 2.61 | 2.25 | |||
| At4g23260 | DUF26 | DUF26 | 1.09 | 2.06 | 2.42 | |||
| At4g23320 | DUF26 | DUF26 | 1.51 | 1.17 | 2.13 | + | ||
| At4g23210 | DUF26 | DUF26 | 1.98 | 2.18 | 2.64 | + | + | |
| At4g04490 | DUF26 | DUF26 | 4.46 | 4.61 | 5.35 | |||
| At4g04500 | DUF26 | DUF26 | 3.65 | 4.32 | 4.85 | |||
| At4g11890 | K | DUF26 | 4.28 | 7.40 | 3.98 | + | ||
| At2g37710 | LEC | L-Lectin | LRK1 | 2.30 | 2.56 | 2.92 | ||
| At3g53810 | LEC | L-Lectin | 2.24 | 1.87 | 2.24 | + | + | |
| At5g01540 | LEC | L-Lectin | 3.47 | 4.60 | 5.80 | + | + | |
| At5g60280 | LEC | L-Lectin | 2.10 | 2.68 | 5.72 | |||
| At2g32800 | K | L-Lectin | 1.83 | 2.04 | 1.82 | + | + | |
| At1g66880 | LRKL | LRK10L-1 | 2.61 | 3.61 | 3.95 | + | + | |
| At5g15730 | K | LRR I | 1.82 | 1.74 | 2.26 | + | + | |
| At1g51890 | LRR3 | LRR I | 1.87 | 1.92 | 3.93 | + | + | |
| At5g45800 | LRR7 | LRR VII | 1.42 | 2.26 | 3.63 | |||
| At3g14840 | LRR9 | LRR VIII-2 | 1.50 | 1.43 | 2.03 | |||
| At3g09010 | K | LRR VIII-2 | 2.17 | 2.95 | 2.72 | + | ||
| At1g16670 | K | LRR VIII-2 | 1.74 | 1.59 | 2.02 | + | + | |
| At5g48380 | LRR4 | LRR X | 2.17 | 2.88 | 2.47 | + | + | |
| At5g42440 | K | LRR X | 2.07 | 2.34 | 2.04 | |||
| At1g74360 | LRR20 | LRR X | 1.78 | 1.87 | 2.43 | + | + | |
| At4g28490 | LRR21 | LRR XI | HAESA | 2.93 | 2.76 | 2.47 | ||
| At1g09970 | LRR19 | LRR XI | 2.65 | 2.98 | 2.86 | |||
| At5g25930 | LRR22 | LRR XI | 2.99 | 2.16 | 3.32 | + | + | |
| At3g47090 | LRR22 | LRR XII | 1.92 | 1.90 | 2.12 | |||
| At1g35710 | LRR27 | LRR XII | 2.19 | 2.39 | 1.77 | |||
| At4g39270 | LRR10 | N.A. | 2.14 | 1.85 | 2.89 | |||
| At5g58540 | K | RLCK I | 1.30 | 2.28 | 3.18 | |||
| At2g39660 | K | RLCK VII | 1.90 | 1.24 | 2.06 | + | + | |
| At1g14370 | K | RLCK VII | APK2a | 2.54 | 2.68 | 2.74 | + | + |
| At4g35600 | K | RLCK VII | 2.10 | 2.04 | 2.12 | + | + | |
| At5g47070 | K | RLCK VII | 1.89 | 2.22 | 1.72 | + | + | |
| At5g51770 | SK | RLCK XI | 2.04 | 1.59 | 1.93 | |||
| At1g61380 | SD | SD-1 | 1.69 | 1.41 | 2.11 | + | ||
| At4g18250 | THN | Thaumatin | 3.50 | 4.58 | 4.20 | + | + |
Extracellular domains (ECD) and subfamily annotations are taken from Shiu and Bleecker (2001).
Expression after treatment with the bacterial elicitors flg22 and elf26 are from Zipfel et al. (2006); +, Ratio > 2.
Genes down-regulated by P. brassicae eggs consisted of genes involved in cell wall metabolism, cuticle biosynthesis, and fatty acid synthesis. This was accompanied by the repression of many genes that participate in photosynthetic activities (Table II).
PR1 Is Induced by Oviposition
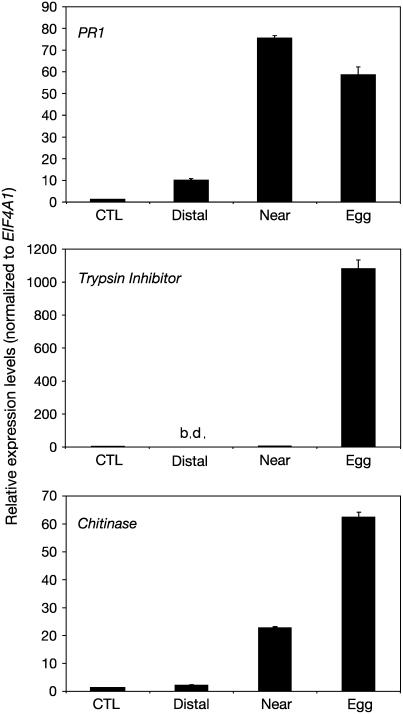
The up-regulation of two representative genes by P. brassicae oviposition was verified by quantitative real-time PCR after 72 h. A chitinase gene (At2g43570) was highly induced at and near the oviposition site, but only very weakly in the distal leaf (Fig. 2). The expression of a trypsin inhibitor gene (At1g73260) showed a highly localized response to P. brassicae eggs, as the gene was more than 1,000-fold more expressed at the egg site than in the near, distal, or control samples. We also analyzed the expression of PR1 since it is a known defense marker gene involved in innate immunity. A probe for this gene was lacking in the current CATMA microarray. We found that PR1 is highly induced (60- to 70-fold compared to control plants) 72 h after oviposition, both at the site of oviposition and in a region surrounding the egg batch (Fig. 2). There was also a small induction in the distal leaf.
Figure 2.
Real-time quantitative PCR analysis of selected egg-inducible genes. Relative expression levels of PR1 gene (PR1, At2g14610), a trypsin inhibitor gene (At1g73260), and a chitinase gene (At2g43570) were analyzed 72 h after oviposition by P. brassicae. Leaf samples were collected at the site of oviposition (egg), next to the egg batch (near), in egg-free distal leaves (distal), and in leaves from plants that were not in contact with butterflies (CTL). Expression levels were normalized with respect to the housekeeping gene EIF4A1 (At3g13920) and displayed relative to the expression control samples (expression value set at 1). Data bars represent the mean levels of transcripts (±se, n = 3). b.d., Below detection.
Role of JA and SA Pathways in Egg Responses
We next determined the contribution of known signaling pathways involved in defense in Arabidopsis. Given the importance of JA and SA in plant defense responses (Reymond and Farmer, 1998), we selected the jasmonate-insensitive mutant coi1-1 (Xie et al., 1998), and the SA-deficient mutant sid2-1 (Nawrath and Metraux, 1999). Changes in gene expression 72 h after oviposition by P. brassicae were measured with CATMA microarrays in three independent experiments. Transcript profiles between wild-type and mutant plants were very similar. We found almost no genes whose induction depended strictly on the JA or the SA pathway, i.e. genes that were induced in wild-type plants and not in the mutants after oviposition (Fig. 3). On the contrary, some of the induced genes were slightly more induced or more repressed in coi1-1 than in wild type, illustrating a potential negative regulation of these genes by JA in wild-type plants; 29% of the genes induced by P. brassicae oviposition were significantly more induced in coi1-1 than in wild-type plants (Fig. 3A; Supplemental Table S2). This bias was less pronounced with sid2-1 plants where only 16% of egg-regulated genes showed a statistical difference between mutant and wild-type plants, some genes being more induced in sid2-1 and some more in wild-type plants (Fig. 3B; Supplemental Table S2).
Figure 3.
Role of signaling mutants in response to P. brassicae eggs. Expression changes were monitored with CATMA microarrays 72 h after oviposition in the jasmonate-insensitive Arabidopsis mutant coi1-1 (A) and in the SA-deficient mutant sid2-1 (B). Expression ratios of mutant plants are plotted against expression ratios of wild-type plants. Pink dots represent genes that are differentially regulated (P < 0.05) between mutant and wild-type plants. Values are the mean of three independent replicates. Genes induced by oviposition in wild-type or mutant plants are listed in Supplemental Table S2.
We did not identify enrichment of particular functional categories among the genes that were differentially regulated by oviposition between coi1-1 and wild-type plants or between sid2-1 and wild-type plants. Moreover, most of the genes differentially regulated between mutant and wild-type plants were not the same in coi1-1 and in sid2-1 plants (Supplemental Table S2). An interesting exception consisted of two genes encoding NPR1-interacting proteins (NIMIN-1, At1g02450; NIMIN-2, At3g25882) that were significantly more induced by oviposition in coi1-1 and less induced in sid2-1. These proteins have been shown to modulate PR gene expression by negatively regulating distinct functions of NPR1 (Weigel et al., 2005). NPR1 plays a crucial role in the pathway cross talk between JA and SA (Spoel et al., 2003). We identified three germins in the list of genes induced 2-fold more in coi1-1 plants. Germins have oxalate oxidase activity leading to hydrogen peroxide (H2O2) production. Genes involved in cell wall remodeling exhibited also a similar expression bias, including a pectin methyl esterase (At3g47380), two endoxyloglucan transferases (At2g14620 and At4g18990), and a cellulase (At1g65610). Proteases and protease inhibitors were also enriched in this category, including two aspartyl proteases (At3g12700 and At5g37540), a Cys proteinase (At5g43060), a protease inhibitor (At3g22620), and a serpin (At2g26390; Supplemental Table S2).
Two known PR genes (PR2, At3g57260; HEL, At3g04720) and an avirulence-responsive gene (AIG2, At3g28930) were induced 2-fold more by oviposition in sid2-1 than in wild-type plants. On the opposite, five WRKY transcription factors were not or less induced by oviposition in sid2-1 plants (Supplemental Table S2). Accordingly, WRKY transcriptions factors have been implicated in the control of SA-dependent defense gene induction (Ulker and Somssich, 2004). In summary, although we do not see a major contribution of SA and JA in the control of oviposition-induced gene expression changes, more work will be necessary to assess the relative importance of these two pathways in the fine modulation of plant response to egg deposition.
Expression Changes after Oviposition by P. rapae
To gain more insight in the specificity of egg-induced expression changes, we extended our analysis to a species with a different oviposition behavior. In contrast to P. brassicae, P. rapae butterflies lay only one egg per site. We analyzed transcript profiles of Arabidopsis leaves 24 to 72 h after oviposition by P. rapae. Each Arabidopsis leaf contained approximately one to two single eggs and we collected leaf discs of approximately 5 mm at the site of oviposition. Samples from three independent experiments for each time point were hybridized to the microarrays. We observed that single egg deposition by P. rapae triggered a much weaker response than that by P. brassicae; only 23, 45, and 59 genes were up-regulated after 24, 48, and 72 h, respectively. Of those, 25 genes were induced in at least two time points, indicating that although the effect was small these changes were not due to random experimental variability. Furthermore, 15 of these 25 genes were also induced after oviposition by P. brassicae (Supplemental Table S3). In general, P. brassicae eggs caused a larger induction than P. rapae eggs but we also identified some genes specifically induced by P. rapae eggs, including a PR Bet v1 allergen protein (At1g70830) and others showing a quantitatively similar response to each treatment, including the hevein-like gene HEL/PR4 (Fig. 4). Only four genes were down-regulated 72 h after oviposition. However, three of these genes, including one germin-like protein (At5g20630) and two Gly-rich proteins (At1g04800 and At4g29020) were also down-regulated after oviposition by P. brassicae (Supplemental Table S3).
Figure 4.
Time course of expression changes after oviposition by P. brassicae (solid line) and P. rapae (dotted line). Microarray data from six representative genes are shown: short-chain alcohol dehydrogenase (SAG13, At2g29350), trypsin inhibitor (At1g73260), chitinase (At2g43570), Cys proteinase (At3g49340), PR Bet v1 allergen protein (At1g70830), and hevein-like protein (HEL/PR4, At3g04720). Values are the mean (±se) of three independent experiments.
Oviposition Induces Cellular Changes Associated with Cell Death
A careful examination of egg-laden Arabidopsis plants did not reveal any physical damage. Furthermore, we could not see obvious phenotypical changes, nor any sign of infection at the site of oviposition during the days between oviposition and hatching. Scanning electron microscope images of leaves containing P. brassicae and P. rapae eggs showed that eggs are fixed to the plant surface with a glue forming a visible meniscus surrounding the base of the egg, without apparent modification of the leaf epidermis (Fig. 5, A–C). Since the analyses of expression changes after oviposition identified many genes associated with PCD, we carried out histochemical experiments to analyze known cellular changes associated with these responses. First, trypan blue was used to stain dead cells in leaves 72 h after oviposition by P. brassicae. We observed a strong staining corresponding closely to the site where the egg batch was deposited (Fig. 5, D and E). As trypan blue is a dye that enters permeable dead or damaged cells, a control experiment was done where leaves were stained just after oviposition. In this case, we only saw a faint circular staining corresponding to the contact point between each egg and the leaf surface (data not shown), indicating that careful detachment of the eggs before the staining procedure was not damaging the plant surface and hence was not causing the strong cell death response observed after 3 d.
Figure 5.
sem analyses of eggs of P. brassicae (A and B) and P. rapae (C) showing the meniscus (arrows) formed by the glue that connects the egg to the plant surface. d to I, Cellular changes in response to P. brassicae eggs 3 d after oviposition. E, Leaves were stained with trypan blue to visualize cell death. G, Callose deposition (arrow) was detected by staining leaves with aniline blue and viewed under a fluorescent microscope. H, Close-up view of callose deposition at the site of egg batch deposition; some eggs were left on the leaf to better localize the site of oviposition. I, Accumulation of H2O2 (arrow) at the site of oviposition was revealed by DAB staining. Images taken before removing the eggs are presented to show the correspondence between egg batch location and cellular responses (D for E and F for G). Magnification bars = 1 mm in A, and D to I; 100 μm in B and C.
Second, we examined callose deposition in egg-laden leaves. The accumulation of autofluorescent phenolic compounds and deposition of callose are markers associated with lesion formation in response to pathogen invasion (Koga et al., 1988) and lesions found in lesion-mimic mutants (Dietrich et al., 1994). Leaves were stained with aniline blue 72 h after P. brassicae oviposition and viewed under a fluorescent microscope. We observed a strong accumulation of callose closely associated with the site of egg batch deposition (Fig. 5, F–H).
Third, PCD is often preceded by an early production of reactive oxygen species (Levine et al., 1994). To visualize the production of H2O2 after oviposition, leaves were submerged in a 3,3′-diaminobenzidine (DAB) solution, a stain that polymerizes with H2O2 and that produces a reddish-brown precipitate. In accordance with the detection of dead cells and callose deposition, the leaves stained with DAB 72 h after oviposition showed an accumulation of H2O2 at the site of egg deposition (Fig. 5I).
Fourth, having observed that the defense gene PR1 was highly induced by P. brassicae eggs (Fig. 2) we selected this gene as a marker to further study the response to egg deposition. We obtained a transgenic Arabidopsis line containing the PR1 promoter coupled to the β-glucuronidase (GUS) reporter gene and analyzed the activation of the promoter after egg deposition or application of egg extracts. PR1∷GUS activation was observed 48 and 72 h after oviposition by P. brassicae and corresponded precisely to the site of egg batch deposition (Fig. 6, A–D). However, we could never detect PR1∷GUS activation only 24 h after oviposition (data not shown). No PR1∷GUS activity was obtained when eggs were removed immediately after oviposition and leaves were stained after 24, 48, and 72 h (data not shown), ruling out a touch effect caused by egg deposition on the leaf surface. Eggs were also moved from the original site of oviposition to a new leaf. After 72 h, the leaves carrying the repositioned eggs were stained for the presence of PR1∷GUS without positive results, suggesting that the close contact of the egg and the epidermis in the process of oviposition contributes substantially to the expression of PR1 in the leaf (data not shown). Application of lanolin paste or sticking Parafilm egg-like structures to leaves for 72 h did not activate the PR1∷GUS reporter gene either (data not shown). In another test, eggs were extracted with MeOH or CHCl3, the extract was dried and resuspended in 0.05% Tween 20. The solution was applied to leaves for 72 h but no PR1∷GUS activity could be detected (data not shown). Finally, we observed PR1∷GUS activation after oviposition of single eggs by P. rapae (Fig. 6, E and F), although staining was only seen in less than 50% of the cases.
Figure 6.
Activation of the defense gene PR1 by egg deposition. Arabidopsis transgenic lines containing the PR1∷GUS reporter gene were visualized for GUS staining assay. Oviposition by P. brassicae for 48 h (A and B), and 72 h (C and D), and by P. rapae for 72 h (E and F, single egg indicated by an arrow). Application of P. brassicae crude egg extract activated PR1∷GUS expression (G and H, left side), but not when the extract was boiled (G and H, right side). Treatment with 1 μL (I, top) and 2 μL (I, bottom) of crude egg extract stored at −20°C was also effective. Application of egg masses dissected from P. brassicae activated PR1∷GUS expression (J). GUS staining in response to application of solid (K, top arrow) or liquid (K, bottom arrow) P. brassicae crude accessory gland extract was only observed for the liquid phase (L, bottom arrow). Images of leaves before staining are presented to show colocalization of egg deposition (A, C, and E) or crude extract application (G and K) and GUS staining. Magnification bars = 5 mm.
To carry out an initial study on the nature of the elicitor(s) responsible for activating PR1∷GUS expression, we collected P. brassicae eggs deposited on Arabidopsis leaves, gently crushed them in an Eppendorf tube and painted a few microliters of crude extract on a reporter plant. After 72 h, plants were stained for GUS activity and clearly showed activation of the PR1 promoter at the site of application (Fig. 6, G and H, left side). When the extract was boiled for 3 min before application, no activity could be detected (Fig. 6, G and H, right side). We then found that the supernatant of centrifuged crude egg extract (10,000g, 1 min) kept a strong activity after storage at −20°C (Fig. 6I). This provides a useful information for the future isolation of active fractions. To confirm that eggs, or compounds associated with eggs, are responsible for the induction of PR1, we dissected female butterflies and removed eggs from the ovaries. A portion of the eggs (equivalent to a normal batch or about 30 eggs) was applied to a leaf. After 72 h, a strong GUS staining was observed at the site of application (Fig. 6J). Female accessory glands produce a cement that allows the deposited eggs to be glued to the plant surface. We obtained P. brassicae accessory glands (Dr. J.J.A. van Loon, Wageningen University) and tested whether gland extracts can activate PR1∷GUS. The supernatant from accessory glands that were crushed and centrifuged caused GUS staining 72 h after application while the solid pellet did not (Fig. 6, K and L). The staining was, however, less intense than with crushed eggs or with dissected ovaries. All experiments were repeated several times with similar results.
DISCUSSION
Egg-Induced Responses
In this study, we show that Arabidopsis plants are able to detect the presence of eggs of the pierid butterflies P. brassicae and P. rapae, and trigger a response that has strong similarities with an HR. In the case of pathogenesis, HR is elicited after the recognition of specific pathogen-derived molecules by plant-specific resistance genes and triggers a localized response that is characterized by a restricted necrosis at the site of infection, the production of reactive oxygen species, the accumulation of secondary metabolites, and the induction of PR genes. Cells directly below the oviposition site stained strongly with trypan blue, indicating that these cells were negatively affected by the presence of the eggs and were undergoing cell death, though not to the extreme result of developing a necrotic zone around the eggs. Indeed, there was no physical difference between the leaf surface where eggs were recently removed and other parts of the leaf. However, the plant was responding strongly as was further evidenced by the very specific accumulation of callose at the oviposition site, along with the induction of a callose synthase gene. Callose has been implicated in plant defense, for example in the wound response where it acts as a plug to seal the wound site. It is also believed that callose is rapidly deposited as a physical barrier to impede microbial and fungal attack (Stone and Clarke, 1992). However, it has also been shown that an Arabidopsis mutant lacking a functional callose synthase is more resistant to powdery mildew (Nishimura et al., 2003), indicating that the role of callose in resistance has to be carefully evaluated in each biological interaction.
Another supporting factor of a defensive response by the plant was the production of H2O2 at the oviposition site. H2O2 is often produced as a result of external biotic and abiotic stimuli and has been shown to play a role in the control of HR (Levine et al., 1994) and in the induction of PR proteins in tobacco (Nicotiana tabacum; Chen et al., 1993; Chamnongpol et al., 1998). Moreover, oviposition provoked gene expression changes that were very similar to those observed during bacteria-induced HR or in lesion-mimic mutants. Well-known PCD and defense-associated genes were induced by oviposition. One of the strongly induced genes was SAG13, a short-chain alcohol dehydrogenase that has been identified as a marker of PCD in the lesion-mimic mutant acd11 (Brodersen et al., 2006b). The PR1∷GUS reporter line showed that the induction of the defense marker gene PR1 was specifically associated with the oviposition locations of both P. brassicae and P. rapae eggs. Interestingly, egg deposition induced EDS1, PAD4, and SAG101, three lipase-related proteins that are thought to play a fundamental role in transducing redox signals upon both biotic and abiotic stresses (Wiermer et al., 2005). These genes were initially identified as key defense regulators necessary for the resistance response evoked by TIR-NB-LRR resistance genes but recent studies have revealed additional activities associated with cell death (Wiermer et al., 2005). Moreover, the EDS1 pathway was shown to be controlled positively by the flavin-dependent monooxygenase FMO1 and negatively by the Nudix hydrolase NUDT7, two genes that were also induced by oviposition. Experiments with sid2-1 mutants have indicated that this control of EDS1 in disease resistance and cell death is independent of SA (Bartsch et al., 2006). Interestingly, we found that the transcriptional response of Arabidopsis to egg deposition does not require the accumulation of SA since sid2-1 and wild-type plants showed a strikingly similar expression profile after oviposition. Another recent study has identified PAD4/EDS1 as central regulators in the positive control of the SA pathway and in the negative control of the JA pathway (Brodersen et al., 2006a). The activation of these genes by egg deposition might thus play a dual role. First, PAD4/EDS1 might participate in the control of egg-induced cell death leading to the activation of defenses. Second, egg-derived elicitors might also suppress the JA pathway, highlighting a strategy of the insect to manipulate this signaling pathway. Indeed, a suppression of the JA pathway would impair the proper activation of JA-dependent responses that are required when hatching larvae start to feed. However, a functional JA perception pathway is not necessary for the induction of egg-inducible genes as demonstrated with our experiment with the jasmonate-insensitive coi1-1 mutant.
Until now, only two studies have reported that insect eggs can trigger an HR-like response in a plant. Eggs of P. brassicae and Pieris napi caused the apparition of a necrotic zone on B. nigra plants, increasing egg mortality (Shapiro and Devay, 1987). This response was only observed in some members of the plant population, illustrating a genetic basis for this mechanism. A hybrid clone of potato plant responded in a similar manner to oviposition by the Colorado beetle Leptinotarsa decemlineata. A necrotic zone containing the egg mass developed and detached from the leaf indirectly, causing a reduction in larval survival rate (Balbyshev and Lorenzen, 1997). Although we could not see a macroscopic effect of oviposition on Arabidopsis leaves, we identified a response that has the molecular and cytological features of an HR. It was recently reported that P. brassicae eggs cause a browning of the leaf surface at the site of oviposition on Brassica oleraceae plants (Hilker and Meiners, 2006). Interestingly, we also occasionally noticed this effect, suggesting that this species might react more strongly than Arabidopsis. A phenotypical analysis of different Arabidopsis ecotypes might reveal some genetically based variation in the response to egg deposition.
In addition, we observed a down-regulation of several expansins, a cellulose synthase, pectin-modifying enzymes, and genes involved in cuticle biosynthesis by P. brassicae eggs, indicating that there might be cell wall remodeling at the site of oviposition. Many photosynthesis-related genes were also repressed, corroborating the previous observation of a net decrease of photosynthetic activity in Scots pine after egg deposition by D. pini (Schroder et al., 2005). Since defense production has been shown to be costly (Baldwin, 1998), plants have likely to reallocate resources for this metabolic process at the expense of growth and photosynthesis.
Nature of the Elicitor
We showed that a compound present in the supernatant of crude egg extracts activated PR1∷GUS expression, indicating that the response to oviposition was not triggered by a physical damage or by simple contact of the butterfly during egg deposition. Extracts from accessory glands that produce the egg cement had, however, a weaker eliciting activity. Eggshell is constituted of several layers of proteinaceous structures and of a lipidic wax layer while the chemical nature of the egg cement is unknown (Trougakos and Margaritis, 2002). The only characterized elicitors of a plant response to egg deposition are the long-chain fatty acid derivatives called bruchins. These elicitors induce the formation of a tumor-like growth on pea pods and have been isolated from bruchid beetles (Doss et al., 2000). In the case of indirect response to oviposition, the elicitors responsible for attracting egg parasitoids in pine and elm have been associated with secretion coating the eggs (Meiners and Hilker, 2000; Hilker et al., 2005). When applied to artificial wounds, oviduct secretion could mimic the attraction of egg parasitoids by the pine sawfly and the eliciting activity was found to be sensitive to proteinase treatment (Hilker et al., 2005).
Insect eggs also contain defensive compounds that protect them from predation (Blum and Hilker, 2002). These toxins are either of insect origin or are acquired by feeding on a plant throughout the lifecycle of the female and further sequestered in the egg. For instance, breakdown products of glucosinolates have been detected in P. brassicae eggs (Aplin et al., 1975). Moreover, JA has been identified in eggs and neonates of several families of Lepidoptera (Tooker and De Moraes, 2005) with concentrations up to hundreds of times greater than the concentrations found in Arabidopsis. Interestingly, JA treatment was also found to trigger the emission of volatiles that attract egg parasitoids, mimicking the response of pine twigs to oviposition (Hilker et al., 2002). It has been hypothesized that JA could act as an egg-derived signal, although it is not known whether it is transferred from the chorion to the plant surface. We measured JA levels in P. brassicae eggs and found 25 ± 6 ng JA/g egg tissue (n = 3), a value comparable to resting levels of unchallenged Arabidopsis leaves (Reymond et al., 2004). This amount is similar to levels measured in Manduca sexta, a species where egg JA content was considered to be low and substantially lower than the 400 μg/g measured in Spodoptera exigua eggs (Tooker and De Moraes, 2005). Given the fact that JA levels in P. brassicae eggs are low and that the induction of most egg-responsive genes are independent on a functional JA pathway, it is unlikely that JA plays a role as an egg-derived elicitor.
Eggs may contain symbionts that are transmitted by the female and are also exposed to pathogens (Kellner, 2002). The response to oviposition could thus be due to microorganisms associated with the eggs. However, in both reports of HR-like responses to egg deposition, the authors could not isolate plant pathogens associated with the eggs and concluded that the induction of the necrotic response by bacteria was unlikely (Shapiro and Devay, 1987; Balbyshev and Lorenzen, 1997). In our experiments, we could not detect obvious signs of bacterial disease while we observed the induction of a transcript pattern similar to that caused by the infiltration of high doses of a plant pathogen. Finally, we showed that two species of insects with different oviposition behavior provoked a similar response. It seems unlikely that eggs from different species carry the same plant pathogens. There is thus little evidence that microorganisms are the cause of egg-derived elicitors but further experiments will be necessary to identify the chemical nature and precise origin of these elicitors.
The perception of chemical signals at the cell surface is mediated by receptors containing an extracellular ligand-binding domain and a cytoplasmic kinase domain. Arabidopsis contains more than 600 RLKs of which only a few have a defined role (Shiu and Bleecker, 2001). We observed that 41 RLKs were induced by egg deposition. These egg-induced RLKs belong to different classes defined by a specific extracellular domain, suggesting that they might respond to different egg-derived elicitors. Some RLKs have been shown to play an important role in the detection of pathogen-associated molecular patterns. For example, FLS2 is a receptor for flagellin, a protein subunit of the flagellar filament, and EFR1 mediates the perception of the bacterial protein EF-Tu (Chinchilla et al., 2006; Zipfel et al., 2006). Upon activation, both receptors induce a similar set of defense genes, pointing to a general surveillance mechanism at the cell surface responding to multiple pathogen-associated molecular patterns and converging to a common transcriptional response (Zipfel et al., 2006). Since about half of the egg-induced RLKs were also found to respond to flagellin and EF-Tu (Zipfel et al., 2006), we cannot exclude that some of them were induced by egg-associated microorganisms. Further studies on the nature of the egg elicitors will help in elucidating the molecular basis of RLK activation.
Role of Egg-Induced Genes in Plant Resistance
Since the egg shell is composed of proteins and wax, our finding that proteases and lipases are induced by egg deposition could indicate that these enzymes act as a direct defense against the egg. In addition, several chitinase genes were also induced by P. brassicae eggs. Chitin is a major component of insects where it plays a scaffolding role supporting the cuticle as well as peritrophic matrices in the gut epithelium, but it does not seem to be present in the eggshell (Merzendorfer and Zimoch, 2003). Chitinases might thus rather play a defensive role against newly hatched larvae and their early induction might anticipate the feeding damage. Similarly we also observed the induction of lectins and proteases inhibitors that are known to have some antiinsect properties. Moreover, we observed that two terpene cyclases were up-regulated by egg deposition. Oviposition might thus also trigger the release of volatiles to attract parasitoid wasps, as it has been documented in other plant/insect interactions. Although this phenomenon has not yet been observed in Arabidopsis, these two genes are potential candidates for the synthesis of indirect defense compounds. In summary, our data suggest that the transcriptional response to oviposition constitutes a direct defense against the egg and that it can also contribute to defense in the early stages after egg hatching. At present we do not know whether the strong and localized HR-like response is really effective against egg deposition by interfering with the egg development and/or hatching rate, or whether it slows down the initial larval development. Interestingly, we noticed that hatching P. brassicae larvae always start to feed at the oviposition site, supporting a role for a localized plant defense activation. Further studies using Arabidopsis mutants impaired in the induction of egg-responsive genes might help to answer this question.
In conclusion, we provide here a molecular evidence for the detection of egg deposition by Arabidopsis plants and show that oviposition causes a strong and localized defense response that resembles an HR. It remains to be shown whether this response constitutes a direct defense against the eggs or whether plants have evolved a mechanism to anticipate the threat posed by future feeding larvae.
MATERIALS AND METHODS
Plant and Insect Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 and mutants were grown as described previously (Reymond et al., 2000). Mutant seeds of acd2-2 were obtained from the Nottingham Arabidopsis Stock Center. Seeds of coi1-1 (nonglabrous) were a gift from Dr. Jane Glazebrook (University of Minnesota, St. Paul), sid2-1 a gift from Dr Christiane Nawrath (University of Lausanne), and PR1∷GUS from Dr Allan D. Shapiro (Florida Gulf Coast University). Homozygous coi1-1 mutants were selected on plates containing 50 μm JA as described previously (Xie et al., 1998) and were tranferred to soil after 10 d of growth. Rearing and handling of Pieris rapae (small cabbage white butterfly) and Pieris brassicae (large white butterfly) were described previously (Reymond et al., 2000). Plants were grown under a regime of 10/14 h photoperiod (100 μmol m−2 s−1, 22°C, and 65% relative humidity).
Plant Treatments
Plants were 6 weeks old at the time of treatment. For insect oviposition, 24 to 30 plants (8–10 plants per time point) were transferred to the greenhouse for 2 to 4 h during which butterflies were allow to lay eggs. On average P. brassicae butterflies were laying one or two batches of 10 to 40 eggs per plant, and P. rapae were laying approximately 20 single isolated eggs per plant. Plants were then placed in a phytotron (10/14 h photoperiod, 100 μmol m−2 s−1, 22°C, 65% relative humidity) for a period of 24, 48, or 72 h before harvesting. Leaf discs of approximately 5 mm in diameter containing all egg batches or single eggs were collected at the appropriate times, the eggs were immediately removed carefully avoiding damage to the surface of the leaf disc, and the leaf samples were frozen in liquid nitrogen for later RNA extraction. As controls, an equal number of leaf discs from a distal egg-free leaf were sampled at each time point. Each experiment was repeated at least three times independently at intervals of several weeks.
Microarray Experiments and Data Analysis
CATMA microarrays (Hilson et al., 2004) were obtained from Paul Van Hummelen (Microarray facility, Flanders Interuniversity Institute for Biotechnology) as part of the Compendium of Arabidopsis Gene Expression (CAGE) project (http://www.cagecompendium.org). Total RNA was extracted from leaf discs using TRIzol (Invitrogen) followed by a purification step (RNeasy clean up, Qiagen). RNA was then amplified using the MessageAMPII aRNA kit (Ambion). Fluorescent probes for hybridization were prepared using 5 μg of a RNA combined with 2 μg of 9 n primers in a total volume of 9 μL. The mix was heated to 70°C for 10 min, chilled on ice, and then reverse transcribed for 2 h at 42°C using 300 units of SuperscriptII (Invitrogen) in the presence of 10 mm dithiothreitol, dNTPS (0.5 mm dATP, dGTP, dTTP; 0.1 mm dCTP), 0.5 units of RNase Inhibitor (Invitrogen), and 2 nmol of Cy3-dCTP or Cy5-dCTP fluorescent dye (Amersham) in a final volume of 20 μL. The reaction was treated with 2 μL of 1 m NaOH and heated for 10 min at 65°C to degrade the RNA, then acidified with 2 μL of 1 m HCl and 2 μL of 3 m NaAc, pH 5.2 for effective purification using the MinElute DNA purification kit (Qiagen). Labeled samples were eluted in 20 μL of Tris-EDTA buffer. Samples labeled with Cy3 or Cy5 were treated separately throughout the procedure and quantified using a Nanodrop spectrophotometer (Witec). Each of three biological samples were labeled either with Cy3-dCTP or Cy5-dCTP (dye-swap design), giving a total of six measurements per time point.
For hybridization, 40 pmol of the Cy3-labeled sample and 40 pmol of the Cy5-labeled sample were combined with 3× SSC, 0.4% SDS, 18 μg yeast (Saccharomyces cerevisiae) tRNA, and 50% formamide in a final volume of 90 μL. The solution was boiled for 1 min, centrifuged for 1 min, and applied to the microarray under a lifterslip (Erie Scientific). The microarray was sealed in a hybridization cassette (Telechem) under an atmosphere of 3× SSC and submerged in a waterbath at 42°C overnight. The microarray was then washed once for 10 min in 1× SSC/0.1% SDS at 50°C, followed by two washes for 10 min in 0.1× SSC/0.1% SDS at 50°C, one wash for 1 min in 0.1× SSC at 37°C, and dipped several times in 0.1× SSC at room temperature. The microarray was immediately dried by centrifugation for 2 min at 2,600 rpm and scanned with a ScanArray 4000 (Packard BioScience SA). The average fluorescence intensity for each fluor and for each gene was determined using the ImaGene program (BioDiscovery). Median background fluorescence signal around each gene spot was subtracted from each spot and to normalize signal intensities between the Cy3 and Cy5 channels a normalization factor was computed using the Loess method. Signal values < 500 (2–3 times the average background sd) were raised to 500 to avoid extreme expression ratios.
Normalized signal intensities were used to calculate expression ratios. We previously observed that a threshold of 2-fold and a P value < 0.05 strongly increase the chance of identifying differentially expressed genes (Reymond et al., 2004) and therefore used both criteria to identify genes differentially expressed by oviposition. We performed a Student's t test with log2-transformed expression ratios (one sample hypothesis, H0: log2 ratio = 0) and only considered genes with an expression ratio ≥ 2 in at least one time point and a P value < 0.05. To address the issue of multiple comparisons, we calculated a false discovery rate using the method of Storey and Tibshirani (Storey and Tibshirani, 2003). This method computes a q value for each gene using the distribution of P values of all measurements. For oviposition experiments with P. brassicae, the false discovery rate at P = 0.05 was 14%, 8%, and 5% for 24, 48, and 72 h time points, respectively. For the comparison of expression changes after oviposition in wild-type and signaling mutants, log2-transformed expression ratios were analyzed by a Student's t test (two-sample hypothesis, H0: log2 ratioWT = log2 ratiomutant) and genes with a P value < 0.05 were considered as differentially regulated.
For the comparison of expression changes between oviposition and infection with Pseudomonas syringae AvrRPM1, ATH1 Affymetrix data from three independent experiments were obtained from the AtGenExpress project at http://www.weigelworld.org/resources/microarray. Expression ratios were calculated from gcRMA-normalized values of control (MgCl2 infiltration) and infected (P. syringae infiltration 24 h) samples. For P. rapae herbivory, CATMA microarray data from seven independent experiments were used. Expression ratios were calculated from normalized values (loess) of unchallenged leaves and leaves challenged for 5 h with third- to fourth-instar P. rapae larvae. For experiments with acd2-2 mutant, CATMA microarray data from three independent experiments were used. Expression ratios were calculated from normalized values (loess) of leaves with no visible lesions and leaves where approximately 10% to 20% of the surface presented necroses. For all experiments, a P value was computed on log ratios with a Student's t test and only genes with an expression ratio ≥ 2 or ≤ 0.5 and a P value < 0.05 were kept for the comparative analysis.
All microarray data have been deposited in ArrayExpress under accession numbers E-CAGE-189 (P. rapae oviposition), E-CAGE-190 (P. brassicae oviposition), E-MEXP-911 (P. rapae herbivory), and E-MEXP-912 (acd2-2 mutant).
Real-Time PCR Analysis
Leaf discs for quantitative real-time PCR reactions were harvested 72 h after oviposition. All samples were harvested using a scalpel to enable cutting the precise area of the egg batch within 1 mm of the eggs (egg sample). The near sample was the 1 to 3 mm section of leaf surrounding previously cut egg disc. The distal leaf disc sample was cut to a similar size as the egg batch from a leaf without eggs but on the same plant as the eggs. A control sample was also harvested consisting of leaf discs of similar size to the egg leaf discs but on a plant that was not exposed to the butterflies. The eggs were immediately and carefully removed, avoiding damage to the surface of the leaf disc, and the leaf sample was frozen in liquid nitrogen for later RNA extraction.
RNA samples were first reverse transcribed using Superscript II (Invitrogen) based on the manufacturer's instructions with 5 μg total RNA (purified with an RNeasy Qiagen kit followed by a DNaseI treatment) in a final volume of 100 μL. The resulting cDNA samples were stored at −80°C. Real-time PCR was performed in triplicate using FullVelocity SYBR green kit (Stratagene). In a 25 μL reaction volume, 1 μL of the cDNA sample was combined with 3.75 μL of 6-carboxy-X-rhodamine (1/5,000 dilution), 12.5 μL of 2X SYBR, and 2.5 μL of primer mix where both forward and reverse primers are at a 1 μm concentration. The reaction was placed in a MX300 real-time PCR machine (Stratagene) with a temperature profile of 95°C for 5 min, then 40 cycles of 95°C for 10 s and 55°C for 30 s. The following gene-specific primers were used: EIF4A1, At3g13920 forward (5′-CCAGAAGGCACACAGTTTGATGCA-3′), At3g13920 reverse (5′-AGACTGAGCCTGTTGAATCACATC-3′); PR1, At2g14610 forward (5′GTGGGTTAGCGAGAAGGCTA-3′), At2g14610 reverse (5′-ACTTTGGCACATCCGAGTCT-3′); chitinase, At2g43570 forward (5′-GGAGAGTACTGCGACACAGAGAAA-3′), At2g43570 reverse (5′-GGCAGGAACCTGGTCTTGAGCAA-3′); trypsin inhibitor, At1g73260 forward (5′-CCTCGTGGTTGCTGGTCCAAA-3′), At1g73260 reverse (5′-CCTCTCACATAGTCTTGGACGAAA-3′). Relative mRNA abundance was calculated using the comparative ΔΔCT method. The values were normalized to the constitutively expressed gene EIF4A1 (At3g13920) and to the control sample (plants without oviposition).
Histochemical Staining
For visualization of cell death, eggs were very carefully removed from leaves 72 h after oviposition avoiding leaf damage and leaves were submerged in trypan blue solution (5 mL lactic acid, 10 mL 50% glycerol, 1 mg trypan blue, and 5 mL phenol) at 30°C for 2 to 3 h. Leaves were then destained in 70% chloral hydrate followed by washes in 70% to 95% ethanol.
Callose deposition was detected 72 h after oviposition. Eggs were removed, leaving a few on the border of the egg batch for localization purposes. Ethanol-destained leaves were placed in 0.15 m phosphate buffer pH 8.5 containing 0.01% aniline blue for 1.5 to 3 h. Leaves were examined under a Leica MZ16FA epifluorescence microscope (excitation 340–380 nm, emission 420 nm filter) and images were acquired using a Leica DC300F camera.
H2O2 accumulation was measured 72 h after oviposition. After the eggs were removed, the leaves were submerged in a 1 mg/mL solution of DAB and placed in a phytotron (100 μmol m−2 s−1, 20°C and 65% relative humidity) for 8 h. The leaves were then destained by boiling in 95% ethanol for 10 min, and stored at room temperature in 95% ethanol until photographs could be taken.
GUS staining for PR1 expression analyses was performed on leaves 24, 48, and 72 h after oviposition. Eggs were removed and leaves were treated with 90% acetone at room temperature for 30 to 60 min, rinsed in a 50 mm phosphate buffered mixture containing 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.05% Triton X-100. Leaves were then submerged in the same solution with the addition of 0.5 μg/μL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, vacuum infiltrated for 20 min, and left at 37°C overnight. Chlorophyll was further destained in 70% ethanol and photographs were taken.
For PR1∷GUS activity after application of P. brassicae egg extract, egg batches were collected on Arabidopsis plants 2 to 3 h after oviposition and crushed with a conical pestle in an Eppendorf tube. A few microliters of crude extract were directly applied on an Arabidopsis leaf with a pipette tip and the plant was left for 72 h before GUS staining. An aliquot of the extract was also boiled for 3 min and painted on a leaf. In another experiment, the crude egg extract was centrifuged (10,000g, 1 min) and the supernatant was stored at −20°C before the application of 1 or 2 μL to a leaf, equivalent to one-third or two-thirds of an egg batch, respectively. For the treatment with female's ovaries, the P. brassicae reproductive system was removed with a scalpel, homogenized, and applied to a leaf. Isolated accessory glands from P. brassicae female butterflies were obtained from J.J.A. van Loon (Wageningen University). Six glands (stored at −80°C for 3 d) were crushed in an Eppendorf tube, and centrifuged (10,000g, 1 min). Leaves were spotted with 2 μL of supernatant (equivalent to one accessory gland) and the plant was left for 72 h before staining. Aliquots of the solid pellet equivalent to one accessory gland were also spotted on the leaves.
JA Measurements
JA measurements were done according to previously published protocols (Weber et al., 1997).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Clustering of genes induced after oviposition by P. brassicae.
Supplemental Figure S2. Clustering of genes repressed after oviposition by P. brassicae.
Supplemental Figure S3. Venn diagram showing the distribution of P. brassicae egg-regulated genes between 24, 48, and 72 h time points.
Supplemental Figure S4. Comparison of expression changes after oviposition by P. brassicae. Expression ratios of oviposited versus egg-free leaves are plotted against expression ratios of oviposited versus control leaves from nonoviposited plants.
Supplemental Table S1. List of genes regulated by P. brassicae eggs, and comparison with infection with P. syringae AvrRPM1, and lesion formation in acd2-2.
Supplemental Table S2. List of genes regulated by P. brassicae eggs in wild type and in the mutant plants coi1-1 and sid2-1.
Supplemental Table S3. List of genes regulated after oviposition by P. rapae.
Supplemental Table S4. List of genes regulated after feeding by P. rapae.
Supplemental Table S5. List of genes regulated during lesion formation in acd2-2.
Supplementary Material
Acknowledgments
We gratefully acknowledge Edward E. Farmer for providing financial support for C.G.-D. and for critical reading of the manuscript. We wish to thank Boris Kunstner for maintenance of the plants, Aurore Chételat for help with JA measurements, Dr. Gustavo Bonaventure for help with real-time PCR, Dr. Robin Liechti for establishing the Nomad database, and Natacha Bodenhausen for helpful comments. We also thank Dr. Jane Glazebrook for the coi1-1 nonglabrous mutant, Dr. Christiane Nawrath for the sid2-1 mutant, and Dr. Allan Shapiro for the PR1∷GUS plants. Dr. Joop J.A. van Loon kindly provided P. brassicae accessory glands.
This work was supported by a genomics grant from the University of Lausanne, by a Secrétariat d'etat à 1'Education et à la Recherche grant (SER 02.0346), and by the Compendium of Arabidopsis Gene Expression project that is funded by the European Commission within its Fifth Framework Programme (grant no, QLK3–CT–2002–02035).
The author responsible for distribution of materials integral to the findings presented in this article in accord with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Philippe Reymond (philippe.reymond@unil.ch).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allemeersch J, Durinck S, Vanderhaeghen R, Alard P, Maes R, Seeuws K, Bogaert T, Coddens K, Deschouwer K, Van Hummelen P, et al (2005) Benchmarking the CATMA microarray: a novel tool for Arabidopsis transcriptome analysis. Plant Physiol 137 588–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin RT, d'Arcy Ward L, Rothschild M (1975) Examination of large white and small white butterflies (Pieris spp.) for presence of mustard oils and mustard oil glycosides. J Entomol Ser A 50 73–78 [Google Scholar]
- Balbyshev NF, Lorenzen JH (1997) Hypersensitivity and egg drop: a novel mechanism of host plant resistance to Colorado potato beetle (Coleoptera: Chrysomelidae). J Econ Entomol 90 652–657 [Google Scholar]
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang PF, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum MS, Hilker M (2002) Chemical protection of insect eggs. In M Hilker, T Meiners, eds, Chemoecology of Insect Eggs and Egg Deposition. Blackwell Publishing, Malden, MA, pp 61–90
- Brodersen P, Petersen M, Nielsen HB, Zhu SJ, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006. a) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47 532–546 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Ødum N, Jørgensen LB, Brown RE, Mundy J (2006. b) Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev 16 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamnongpol S, Willekens H, Moeder W, Langebartels C, Sandermann H, Van Montagu M, Inze D, Van Camp W (1998) Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc Natl Acad Sci USA 95 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Silva H, Klessig DF (1993) Active oxygen species in the induction of plant systemic acquired-resistance by salicylic-acid. Science 262 1883–1886 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colazza S, McElfresh JS, Millar JG (2004) Identification of volatile synomones, induced by Nezara viridula feeding and oviposition on bean spp., that attract the egg parasitoid Trissolcus basalis. J Chem Ecol 30 945–964 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18 923–937 [DOI] [PubMed] [Google Scholar]
- Dicke M, van Poecke RMP, de Boer JG (2003) Inducible indirect defence of plants: from mechanisms to ecological functions. Basic Appl Ecol 4 27–42 [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77 565–577 [DOI] [PubMed] [Google Scholar]
- Doss RP, Oliver JE, Proebsting WM, Potter SW, Kuy SR, Clement SL, Williamson RT, Carney JR, DeVilbiss ED (2000) Bruchins: insect-derived plant regulators that stimulate neoplasm formation. Proc Natl Acad Sci USA 97 6218–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Fatouros NE, Bukovinszkine'Kiss G, Kalkers LA, Gamborena RS, Dicke M, Hilker M (2005) Oviposition-induced plant cues: do they arrest Trichogramma wasps during host location? Entomol Exp Appl 115 207–215 [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker M, Kobs C, Varma M, Schrank K (2002) Insect egg deposition induces Pinus sylvestris to attract egg parasitoids. J Exp Biol 205 455–461 [DOI] [PubMed] [Google Scholar]
- Hilker M, Meiners T (2006) Early herbivore alert: insect eggs induce plant defense. J Chem Ecol 32 1379–1397 [DOI] [PubMed] [Google Scholar]
- Hilker M, Stein C, Schroder R, Varama M, Mumm R (2005) Insect egg deposition induces defence responses in Pinus sylvestris: characterisation of the elicitor. J Exp Biol 208 1849–1854 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (1997) Induced Responses to Herbivory. University of Chicago Press, Chicago
- Kellner RL (2002) The role of microorganisms for eggs and progeny. In M Hilker, T Meiners, eds, Chemoecology of Insect Eggs and Egg Deposition. Blackwell Publishing, Malden, MA, pp 149–167
- Koga H, Zeyen RJ, Bushnell WR, Ahlstrand GG (1988) Hypersensitive cell-death, autofluorescence, and insoluble silicon accumulation in barley leaf epidermal-cells under attack by Erysiphe graminis F Sp Hordei. Physiol Mol Plant Pathol 32 395–409 [Google Scholar]
- Kombrink E, Somssich IE (1995) Defense responses of plants to pathogens. In JH Andrews, IC Tommerup, eds, Advances in Botanical Research, Vol 21. Academic Press, London, pp 1–34
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 583–593 [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J 33 425–434 [DOI] [PubMed] [Google Scholar]
- Meiners T, Hilker M (2000) Induction of plant synomones by oviposition of a phytophagous insect. J Chem Ecol 26 221–232 [Google Scholar]
- Merzendorfer H, Zimoch L (2003) Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206 4393–4412 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972 [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RM, Krishnamurthy V, Dicke M, Farmer EE (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1 404–411 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartin M, Jaroszewski L, Raikhel NV, Rojo E (2005) Caspases: regulating death since the origin of life. Plant Physiol 137 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder R, Forstreuter M, Hilker M (2005) A plant notices insect egg deposition and changes its rate of photosynthesis. Plant Physiol 138 470–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino Y, Suzuki Y, Sogawa K (1996) An ovicidal substance produced by rice plants in response to oviposition by the whitebacked planthopper, Sogatella furcifera (HORVATH) (Homoptera: Delphacidae). Appl Entomol Zool (Jpn) 31 467–473 [Google Scholar]
- Shapiro AM, Devay JE (1987) Hypersensitivity reaction of Brassica nigra L (Cruciferae) kills eggs of Pieris butterflies (Lepidoptera, Pieridae). Oecologia 71 631–632 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BA, Clarke AE (1992) Chemistry and Biology of (1-3)β-d-Glucans. La Trobe University Press, Victoria, Australia
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Genome Biol 100 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooker JF, De Moraes CM (2005) Jasmonate in lepidopteran eggs and neonates. J Chem Ecol 31 2753–2759 [DOI] [PubMed] [Google Scholar]
- Trougakos IP, Margaritis LH (2002) Novel morphological and physiological aspects of insect eggs. In M Hilker, T Meiners, eds, Chemoecology of Insect Eggs and Egg Deposition. Blackwell Publishing, Malden, MA, pp 3–31
- Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19 195–216 [DOI] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8 383–389 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7 263–270 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yang SH, Yang HJ, Grisafi P, Sanchatjate S, Fink GR, Sun Q, Hua J (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45 166–179 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8 353–360 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.