Abstract
Viviparous1 (Vp1) encodes a B3 domain-containing transcription factor that is a key regulator of seed maturation in maize (Zea mays). However, the mechanisms of Vp1 regulation are not well understood. To examine physiological factors that may regulate Vp1 expression, transcript levels were monitored in maturing embryos placed in culture under different conditions. Expression of Vp1 decreased after culture in hormone-free medium, but was induced by salinity or osmotic stress. Application of exogenous abscisic acid (ABA) also induced transcript levels within 1 h in a dose-dependent manner. The Vp1 promoter fused to β-glucuronidase or green fluorescent protein reproduced the endogenous Vp1 expression patterns in transgenic maize plants and also revealed previously unknown expression domains of Vp1. The Vp1 promoter is active in the embryo and aleurone cells of developing seeds and, upon drought stress, was also found in phloem cells of vegetative tissues, including cobs, leaves, and stems. Sequence analysis of the Vp1 promoter identified a potential ABA-responsive complex, consisting of an ACGT-containing ABA response element (ABRE) and a coupling element 1-like motif. Electrophoretic mobility shift assay confirmed that the ABRE and putative coupling element 1 components specifically bound proteins in embryo nuclear protein extracts. Treatment of embryos in hormone-free Murashige and Skoog medium blocked the ABRE-protein interaction, whereas exogenous ABA or mannitol treatment restored this interaction. Our data support a model for a VP1-dependent positive feedback mechanism regulating Vp1 expression during seed maturation.
Angiosperm embryo development involves three overlapping phases (West and Harada, 1993). Embryo pattern formation occurs in the early morphogenesis phase. During the maturation phase, macromolecular reserves, such as storage proteins, lipids, and carbohydrates, are synthesized and accumulated. During the final stage, tissue growth and development is arrested and embryos acquire the ability to withstand desiccation and may enter dormancy. Mature embryos remain metabolically quiescent until favorable conditions trigger germination. Proper control of maturation and germination transition is crucial for plant survival in nature and is also of great agronomic importance because of its relationship to dormancy and preharvest sprouting (for review, see Gubler et al., 2005).
Plant hormones are central regulators of the dormancy-germination transition. Abscisic acid (ABA) is well known to promote seed dormancy and repress seed germination, whereas gibberellic acid functions antagonistically to trigger seed germination (Kucera et al., 2005). During the maturation phase of seed development, ABA induces the expression of many maturation-associated genes, such as late embryogenesis abundant (Lea) genes (Marcotte et al., 1989). Mutants deficient in ABA production or sensitivity fail to enter quiescence, resulting in precocious germination or vivipary, accompanied by reduction of maturation-associated gene expression.
VIVIPAROUS1 (VP1) is a B3 domain-containing transcription factor that is central to the regulation of seed maturation in maize (Zea mays; McCarty et al., 1991). Mutations in vp1 result in kernels with reduced sensitivity to ABA, which fail to undergo normal maturation and germinate viviparously (McCarty et al., 1989). In addition, vp1 mutants fail to activate the anthocyanin regulatory gene C1, resulting in an unpigmented aleurone cell layer (McCarty et al., 1989). The Vp1 gene can complement the aba insensitive3 (abi3) mutant phenotype in Arabidopsis (Arabidopsis thaliana), which is similarly blocked in seed maturation (Giraudat et al., 1992; Suzuki et al., 2001).
Studies of ABA induction of gene expression identified the ABA-responsive element (ABRE), a cis-acting promoter element with an ACGT core sequence, which is required for ABA induction of many plant genes (for review, see Yamaguchi-Shinozaki and Shinozaki, 2005). Proteins that bind ABREs belong to a group of bZIP transcription factors, including Arabidopsis ABI5 and rice (Oryza sativa) TRAB1 (Hobo et al., 1999b; Finkelstein and Lynch, 2000; for review, see Jakoby et al., 2002). TRAB1 acts synergistically with ABA and OSVP1 to induce Osem expression in rice protoplasts (Hobo et al., 1999b). TRAB1 is phosphorylated in response to ABA, which is essential for it to induce reporter gene expression (Kagaya et al., 2002).
Although multiple copies of ABREs confer ABA inducibility to a heterologous promoter, a single-copy ABRE is not sufficient for expression. In many promoters, a second cis-acting element, called a coupling element (CE), is required to form an ABA-responsive complex (ABRC; Busk and Pages, 1998; Singh, 1998). Two types of CEs have been identified, CE1 and CE3 (Shen and Ho, 1995; Shen et al., 1996). Together with their corresponding ABREs, these elements form ABRC1 and ABRC3, respectively. ABRC1 and ABRC3 show differences in spacing requirements between the ABRE and CE (Shen et al., 2004) and in their transactivation by overexpressed VP1. ABRC3s found in maize rab28 (Busk and Pages, 1997) and in barley (Hordeum vulgare) HVA1 (Shen et al., 1996) are transactivated by VP1, whereas ABRC1 in barley HVA22 is not (Shen et al., 1996).
VP1/ABI3 and ABA regulate the expression of overlapping sets of genes, including many that require both VP1/ABI3 and ABA (Suzuki et al., 2003; Nakashima et al., 2006). VP1 regulates transcription through several mechanisms. VP1 can activate the expression of genes such as maize C1, through the direct binding to Sph/RY elements via the B3 domain (Suzuki et al., 1997). Activation of ABA-inducible genes, such as Em, is mediated by indirect interaction with ABREs via bZIP transcription factors, such as TRAB1 (Hobo et al., 1999b). Transcriptional activation functions appear to require the amino-terminal acidic domain of VP1 (McCarty et al., 1991; Bobb et al., 1995; Rojas et al., 1999). On the other hand, functional VP1 was required for the full repression of germination-related α-amylase expression in aleurone cells. Further, this repression was independent of the amino-terminal acidic activation domain, indicating that VP1 also functions as a transcriptional repressor (Hoecker et al., 1995, 1999; Suzuki et al., 1997).
Whereas substantial work has been done on the mechanisms by which VP1 and ABA regulate seed maturation, regulation of Vp1 expression is largely unknown. In Arabidopsis, the LEAFY COTYLEDON1 (LEC1), LEC2, and FUSCA3 (FUS3) genes have pleiotropic effects on embryo development and maturation and promote ABI3 expression (Kagaya et al., 2005; To et al., 2006), although it is not known whether that regulation is direct. Here, we show that in midmaturation-phase embryos, maize Vp1 expression is sensitive to ABA, salinity, and osmotic changes. β-Glucuronidase (GUS) was placed under the regulation of a Vp1 promoter fragment (Vp1∷GUS) and stably integrated into maize. The transgene displayed the expected Vp1 expression pattern and revealed unknown expression domains in vegetative tissues under stress conditions. Analysis of the Vp1 promoter region further demonstrated the nuclear protein-binding potential of an ABRC, which might mediate the regulation of Vp1 expression by ABA. Our data also point to positive feedback self regulation of Vp1 expression in maturing seeds.
RESULTS
ABA, Salt, and Osmotica Induce Vp1 Expression in Cultured Embryos
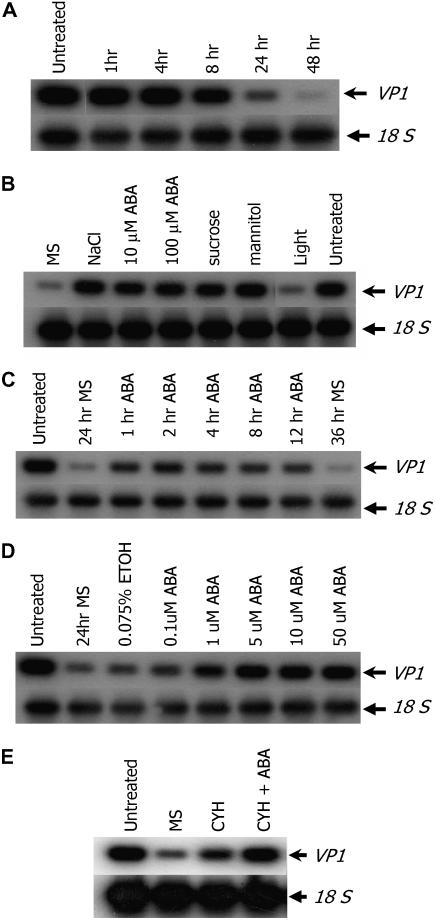
VP1 is expressed in mid- to late-phase embryos and in aleurone cells of maturing maize seeds (McCarty et al., 1989). To test whether physiological conditions regulate Vp1 expression, we performed semiquantitative reverse transcription (RT)-PCR on cultured embryos. Under our growth conditions, Vp1 expression in normal embryo development reached peak levels at around 20 d after pollination (DAP; data not shown). Therefore, 20-DAP embryos were dissected out of developing kernels and cultured in hormone-free liquid Murashige and Skoog medium. A reduction in Vp1 expression became apparent after 8 h and continued to decline to barely detectable levels at 48 h (Fig. 1A). By 24 h, expression was greatly reduced and embryos were released from dormancy and showed visible signs of germination.
Figure 1.
Vp1 transcript accumulation is induced by ABA, salt, and osmoticum in cultured zygotic embryos. Semiquantitative RT-PCR was performed to study the expression of Vp1. A, Vp1 transcript levels decrease over time during culture in hormone-free liquid medium. Untreated embryos were freshly harvested from seeds. B, Vp1 expression is maintained by ABA, salt, and osmoticum. Embryos were cultured for 24 h in the dark with or without 500 mm NaCl, 10 μm ABA, 100 μm ABA, 20% Suc, or 20% mannitol, or in Murashige and Skoog under light. C, Vp1 expression is induced by ABA within 1 h. Embryos were cultured in hormone-free medium for 24 h to decrease Vp1 transcript levels, then treated with 10 μm ABA for various times. D, Vp1 expression shows a dose response to ABA. Embryos were cultured for 24 h with 0.075% ethanol (mock treatment) or with various concentrations of ABA. E, Induction of Vp1 expression by ABA does not require de novo protein synthesis. Embryos were cultured in Murashige and Skoog medium for 24 h followed by addition of 20 μm cycloheximide (CYH). After 20 min, one sample was treated with 10 μm ABA and all samples were incubated for another hour. Untreated, Freshly harvested embryos; hr, hour. All treatments were carried out in the dark in Murashige and Skoog liquid medium containing 1% Suc, unless otherwise indicated.
Because VP1 functions in ABA-regulated processes, we checked whether Vp1 expression was regulated by ABA. Whereas Vp1 transcript levels decreased substantially after 24 h in hormone-free culture, the Vp1 expression level in embryos cultured with 10 or 100 μm of ABA was nearly as high as in freshly isolated embryos, indicating that exogenous ABA maintained Vp1 expression (Fig. 1B). As 10 μm ABA was sufficient to maintain Vp1 expression in embryos, this concentration was used in subsequent experiments. It was possible that ABA could maintain, but not induce, Vp1 expression. In a similar experiment in barley, the decline in HvVp1 expression was attributed to an indirect effect of embryo germination in hormone-free medium (Hollung et al., 1997). To test this possibility, embryos were cultured in hormone-free medium for 24 h to decrease Vp1 expression levels, after which ABA was added to the medium for various incubation times. After 1 h of treatment with ABA, Vp1 expression returned to levels nearly as high as those of freshly harvested embryos, demonstrating that ABA can induce de novo Vp1 expression (Fig. 1C). We next examined the dose response of Vp1 expression to ABA. Vp1 expression was slightly induced by 0.1 μm ABA and increased to a plateau at 10 to 50 μm (Fig. 1D). These data indicate that physiological concentrations of ABA can induce and maintain Vp1 transcript levels in maturation-phase maize embryos.
To address whether the ABA induction of Vp1 expression requires de novo protein synthesis, 20-DAP maize embryos were cultured without hormones for 24 h, then 20 μm cycloheximide was added to the medium to inhibit protein synthesis. After 20 min of cycloheximide treatment, 10 μm ABA was added and embryos were further cultured for 1 h. The cycloheximide treatment did not inhibit the induction of Vp1 expression by ABA (Fig. 1E), indicating that ABA induction of Vp1 expression in cultured embryos does not require de novo protein synthesis.
During the seed maturation phase, embryos accumulate storage reserves and develop desiccation tolerance. As such, embryos are subjected to a milieu of high concentrations of sugars, metabolites, and osmotica. We hypothesized that Vp1 expression might be regulated by these physiological factors and so examined the effects of Suc, mannitol, and salt on Vp1 expression. As shown in Figure 1B, Vp1 expression was maintained or induced in embryos cultured with 500 mm NaCl, 20% Suc, or 20% mannitol. Vp1 expression was also maintained or induced with 200 mm NaCl, 7% Suc, and/or 7% mannitol (data not shown). Thus, salt and osmotic stress can maintain or induce Vp1 expression in maturation-phase embryos. We also checked the effects of other hormones known to be involved in seed development by applying exogenous GA3, indole acetic acid, and 1-aminocyclopropane-1-carboxylic acid (ACC; a precursor for ethylene synthesis). No effects on Vp1 expression were detected with these treatments and no interaction was observed between ABA and indole acetic acid or ACC (data not shown).
Vp1 Promoter Isolation
To further study the regulation of Vp1 expression, 958 bp of the Vp1 5′ sequence was obtained by sequencing a genomic clone from the W22 inbred line (GenBank accession no. DQ886030). Like sorghum, the maize Vp1 promoter lacks a canonical TATA box (Carrari et al., 2001). To locate the start of transcription, 5′-RACE PCR was performed on mRNA from 20-DAP embryos. Vp1 transcription showed variable start sites within a 30-nucleotide region. Nine of 13 cloned RACE-PCR products identified the major transcription site, which was set as position +1, with other starts occurring at −7, −4, and +22. Thus, the 958 bp includes 891 bp of the 5′ promoter sequence and 67 bp of the transcribed 5′-untranslated region (UTR).
Rice Osvp1 and sorghum Sbvp1 promoter sequences were obtained from the National Center for Biotechnology Information and, along with maize Vp1, subjected to phylogenetic footprinting to predict conserved cis-regulatory elements (Sandelin et al., 2004). An ACGT motif resembling an ABRE was identified at position −84 to −75 in maize. Another motif resembling a CE1 was located at −24 to −16. With 50 intervening nucleotides, this spacing follows the multiples-of-10 rule, suggesting that these elements might function together as an ABRC (Shen et al., 1996). The striking sequence conservation suggests that the putative ABRC may be similarly involved in ABA regulation of Vp1 in these three species.
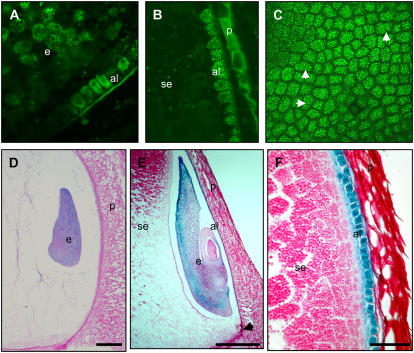
To confirm that the 958 bp of the genomic Vp1 promoter sequence contained all the regulatory elements necessary for normal transcriptional regulation, the promoter was fused to GUS or green fluorescent protein (GFP) and stably introduced into transgenic maize lines. Endogenous Vp1 is expressed by 10 DAP in maturing embryos and aleurone (McCarty et al., 1989; Miyoshi et al., 2002) and the expected expression pattern was observed for both GFP (Fig. 2, A–C) and GUS (Fig. 2, D–F) reporter genes. In developing embryos, GUS activity was detected in the transition embryo from around 7 DAP, although GUS staining was not detectable in aleurone cells at this stage (Fig. 2D). After 10 DAP, GUS stained strongly in embryos and aleurone cells, but not in starchy endosperm cells (Fig. 2, E and F). In embryos, GUS staining was strongest in the outer layer of the scutellum. Thus, the 958-bp promoter is sufficient to reproduce the normal expression pattern of Vp1.
Figure 2.
Regulation of reporter genes by the Vp1 promoter in transgenic maize. A to C, Vp1∷GFP expression in 10-DAP kernels analyzed by confocal laser-scanning microscopy. A, Embryo (e) and aleurone (al) cells expressing GFP (denoted by bright-green, grainy appearance). B, Vp1∷GFP expression in aleurone cells only (note autofluorescence of the pericarp tissue). C, GFP expression in an aleurone peel. White arrowheads indicate newly dividing cells. D to F, Vp1∷GUS expression (denoted by purple/blue precipitate) in developing maize seeds at 7 (D) and 10 (E and F) DAP. GUS expression is detected in the transition embryo from around 7 DAP (shown in D). E, GUS staining is detectable at higher levels in the scutellum at later stages of embryogenesis (indicated by arrows), as well as in the endosperm aleurone (shown here on the germinal kernel face and extending down until the basal endosperm transfer region, indicated by arrowhead). F, Cells of aleurone and subaleurone (sa) layers staining for GUS. Ten-micrometer-thick sections were stained for GUS, counterstained with periodic acid Schiff reagent (crimson color), and visualized under bright-field microscopy. Scale bars = 25 μm (D and E); 10 μm (F).
Vp1 Expression Is Induced in Vegetative Tissues by Stress
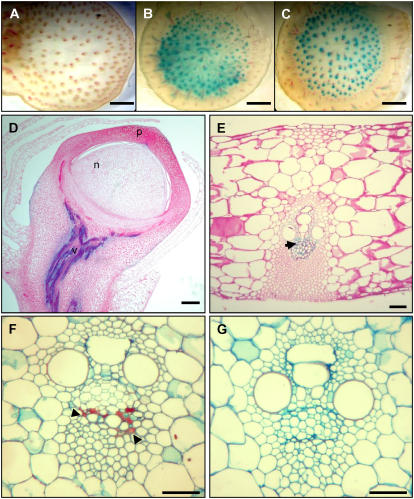
Because Vp1 expression is inducible in embryos by ABA and stress, we examined Vp1∷GUS-carrying plants to test whether transgene expression could be observed in stressed vegetative tissues. Under normal growth conditions, GUS activity was hardly detectable in transverse sections of Vp1∷GUS transgenic stems (Fig. 3A). However, GUS activity was readily detected in the vascular tissues of stem and leaf segments that had been desiccated by 6 h of air drying (Fig. 3B) or treated with 0.6 m Suc (Fig. 3C). We then examined transgene expression in different tissues and found that, upon desiccation or drought stress, GUS activity was detected in the vascular tissues of all aerial stems, including ear pedicels (Fig. 3D). Stress-induced Vp1 expression in vascular tissues was restricted to phloem companion cells (Fig. 3, D–G). Expression of Vp1 in phloem cells was further confirmed by in situ hybridization of endogenous Vp1 mRNA in transverse sections of mature stems (Fig. 3, F and G). Vp1∷GUS expression was also detected in germinating seeds, but, in established plants, no expression was observed in roots, shoot apical meristems, or floral organs (data not shown).
Figure 3.
Stress-induced Vp1∷GUS transgene and endogenous gene expression in maize vegetative tissues. A to C, GUS-stained hand-cut transverse sections of ear shank tissue from plants carrying the Vp1∷GUS reporter gene. Water control (A), 6-h desiccation (B), and 6-h incubation in 0.6 m Suc (C). GUS expression is only detected in the phloem. Sections were counterstained with phloroglucinol-HCl for detection of lignin (denoted by red precipitate). D to E, Vp1∷GUS expression after drought stress treatment in the vascular tissue (v) of an ear pedicel (D) and transverse leaf section (E). GUS staining is localized in the phloem parenchyma cells (ph), indicated by arrow. F and G, Vp1 mRNA in situ localization after drought stress treatment in phloem companion cells (denoted by red/purple staining and indicated by black arrowheads) in transverse sections of stems imaged under bright-field optics after saffranin fast-green staining. F, Antisense probe. G, Sense probe. Scale bars = 50 μm (A–C); 25 μm (D); 5 μm (E–G).
Nuclear Proteins Interact with Elements of the Vp1 Promoter Containing ABRC1
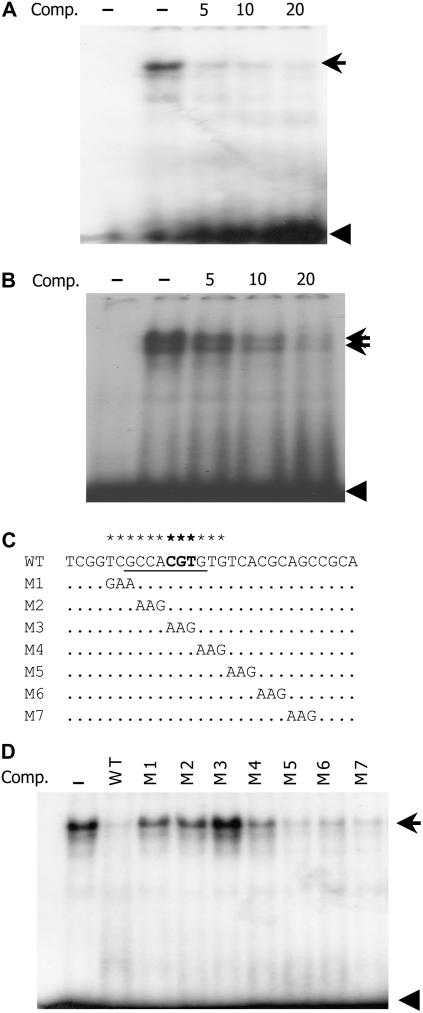
Sequence analysis showed that the Vp1 promoter contains a conserved 8-bp element with the sequence GCCACGTG, which is predicted to be an ABRE. To test whether this fragment is a nuclear protein-binding site, a 29-bp oligonucleotide containing the putative ABRE was synthesized as a probe for an electrophoretic mobility shift assay (EMSA) experiment. A major slowly migrating band was observed using nuclear protein extract from 20-DAP embryos, indicating that the oligonucleotide had bound nuclear proteins (Fig. 4A). An unlabeled fragment outcompeted the retarded band, whereas an unrelated DNA fragment (VPO4) or poly(dI-dC) could not, indicating that binding was specific (Fig. 4A; data not shown). To determine which bases are important for binding to the nuclear proteins, serial mutations were synthesized (M1–M7) and used as competitors in an EMSA experiment. The mutations in GTCGCCACGTGTG, which includes the ABRE, impaired the ability to compete with the wild-type probe. The CGT to AAG mutation in M3 completely abolished the competition capacity (Fig. 4, C and D), and radioactively labeled M3 did not produce a shifted band (data not shown). Thus, the putative 8-bp ABRE appears necessary for in vitro binding to nuclear proteins from developing embryos.
Figure 4.
Putative ABRE specifically interacts with embryo and aleurone nuclear proteins. A, The 32P-labeled ABRE probe bound nuclear proteins from 20-DAP embryos causing one major shifted band. The shifted band was outcompeted with increasing unlabeled probe. The first lane lacks nuclear protein extract. B, The ABRE probe bound nuclear proteins from 20-DAP aleurone cells, resulting in two shifted bands, both of which were outcompeted by unlabeled oligonucleotide. The first lane lacks nuclear protein extract. C and D, Mutation scanning analyses identified the ABRE sequence as critical for binding nuclear proteins. Substitutions in the GCCACGTG (especially M3) disrupted binding activity, failing to outcompete the shifted band. The unlabeled competitors were added as 10 times the labeled ABRE probe. Arrow marks the shifted band; arrowheads indicate free probe. Comp, Competitor.
Because Vp1 is expressed in aleurone cells (McCarty et al., 1989), the ABRE fragment was also tested for binding to nuclear proteins extracted from 20-DAP aleurone cells. As shown in Figure 4B, EMSA produced two distinct shifted bands of comparable intensity, indicating that this element could bind to nuclear proteins from aleurone. This result differs from that obtained in embryos, where only one major shifted band was observed. This suggests that aleurone cells might contain different, or additional, ABRE-binding factors than embryos.
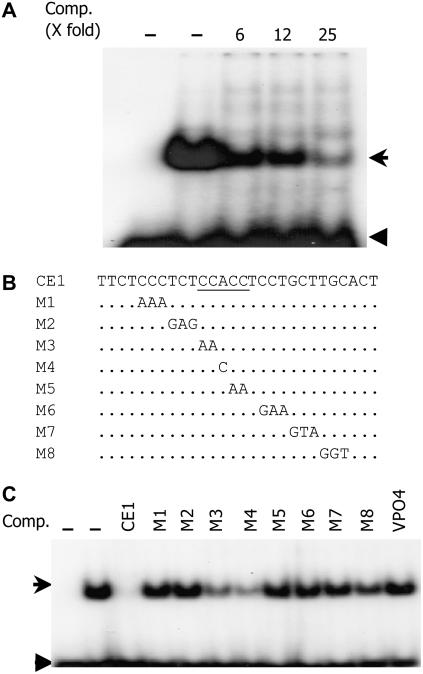
In natural promoter contexts, ABREs usually function in multiple copies or form ABRCs with CEs (Shen et al., 1993, 1996). Examination of the Vp1 promoter identified a CE1-like core sequence (CCACC) located 50 bp downstream of the ABRE. To determine whether this core sequence is a potential nuclear protein-binding site, an EMSA experiment was performed on 20-DAP embryo nuclear protein extract using a synthesized 28-bp oligonucleotide (CE1) as a probe. CE1 bound to nuclear proteins, resulting in a shifted band. The binding was outcompeted by excess unlabeled CE1, but not by the unrelated VPO4 oligonucleotide, indicating that the interaction was specific (Fig. 5, A and C).
Figure 5.
CE1 bound specifically to embryo nuclear proteins. A, CE1 bound to 20-DAP embryo nuclear proteins causing a shifted band, which was outcompeted by increasing unlabeled CE1. The first lane lacks nuclear protein extract. B and C, Mutation analysis of the CE1 binding site. B, One, two, or three base substitutions were introduced into CE1. C, Mutant CE1 oligonucleotides were used as unlabeled competitors in EMSA. All mutations were impaired in their binding activity, but there was less effect of those in the first three bases of the canonical CCACC core binding site (CE1M3 and CE1M4). Each lane contained 0.4 pmol 32P-labeled CE1 with or without 10 μg embryo nuclear proteins. The first lane lacks nuclear protein extract. Unlabeled competitors were added as 25-fold excess of radioactively labeled CE1. Arrow marks the shifted band; arrowhead indicates the free probe.
A previous study showed that maize ZmABI4 bound to CE1 from a number of ABA-related genes (Niu et al., 2002). An A to T substitution in CCACC completely abolished the binding by ZmABI4. To determine the critical binding sites of the CE1 in maize Vp1, scanning mutagenesis was conducted and substituted versions of CE1 (CE1M1–CE1M8) were used as competitors in an EMSA. Surprisingly, a similar A to T (data not shown) or A to C substitution (CE1M4) competed with the probe, indicating that the oligo was still capable of protein binding, although the reduced competition compared to wild type suggests that protein binding was impaired (Fig. 5, B and C). Similarly, CE1M3, which mutates the first two bases of the CCACC core, partially competed with the probe. Thus, it appears that the CCACC motif is not essential for CE1 binding to nuclear proteins. Rather, because other substitutions of CE1 failed to outcompete the probe, it appears that nucleotides outside this motif are more important for protein binding. Taken together, the CE1-like element of maize Vp1 does not appear to have the same binding activity as other previously described CE1s.
ABA or Osmotic Stress Induces Nuclear Protein Interaction with the ABRE
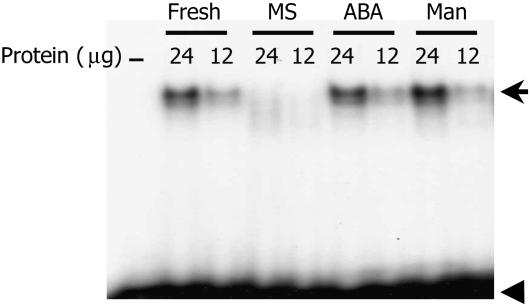
To further explore the possible basis for ABA or osmotic stress regulation of Vp1 expression, an EMSA experiment was performed using an ABRE probe on nuclear protein extracts from treated or untreated 20-DAP embryos. As shown in Figure 6, 20 h of culture in hormone-free medium greatly reduced the binding capacity of the extract, compared with extract from freshly harvested embryos. Applying 100 μm ABA or 20% mannitol in the medium restored the binding capacity comparable to extract of embryos from intact seeds. This result is consistent with the ABRE contributing to the regulation of Vp1 expression by ABA or osmotic stress. A similar EMSA experiment was performed on CE1, but no difference in binding was observed among the treatments (data not shown).
Figure 6.
Nuclear protein binding to the ABRE is induced by ABA and mannitol. Embryos were cultured for 20 h with or without 100 μm ABA or 20% mannitol. Nuclear extracts were applied to an EMSA with nuclear extract from freshly isolated embryos as a positive control. Hormone-free culture abolished the shifted band, whereas ABA or mannitol treatment maintained binding. Arrows mark the shifted band; arrowheads indicate the free probe.
Feedback Regulation of Vp1 Expression
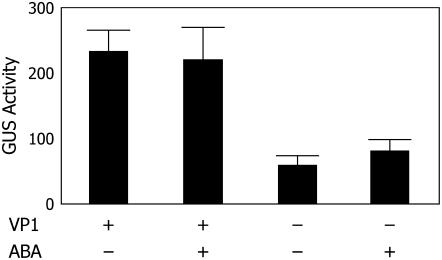
VP1 functions as a transcriptional activator required for the response of many ABA-inducible genes (Suzuki et al., 2003; Nakashima et al., 2006). Because Vp1 itself is ABA inducible, we tested whether VP1 was required for regulation of its own expression in late embryogenesis. For this, we generated plants hemizygous for the Vp1∷GUS transgene that were segregating 1:1 for normal Vp1+/vp1 and mutant vp1/vp1 kernels. A sample of aleurone tissue from each kernel was stained for GUS activity to verify the presence of the transgene and the embryos from GUS-positive kernels were subjected to fluorometric 4-methylumbilliferyl-β-d-glucuronide (MUG) assays (see “Materials and Methods”). As shown in Figure 7, VP1 protein is not required for Vp1 expression because vp1 mutant embryos showed substantial GUS activity. However, GUS activity in the normal embryos was 2- to 3-fold higher than that in the vp1 mutant embryos, indicating that Vp1 expression is positively regulated by a VP1-dependent feedback mechanism. Surprisingly, we did not observe ABA induction of GUS activity in these embryos.
Figure 7.
Feedback activation of Vp1 expression. The Vp1∷GUS transgene showed reduced expression in vp1 mutant embryos compared to wild-type embryos cultured 24 h with or without ABA. Relative GUS activity was normalized by dividing the protein concentration of each total protein extract. Error bar = 1 sd. GUS activity did not show ABA inducibility.
DISCUSSION
During late stages of seed development, plant embryos undergo maturation, acquire desiccation tolerance, and achieve quiescence. VP1, along with ABA and other factors, is required for the regulation of these processes in maize. Maize Vp1 is expressed from before 10 DAP to very late in seed development (McCarty et al., 1989; Niu et al., 2002). In developing rice embryos, Osvp1 and Osem expression is positively correlated as early as 6 DAP (Miyoshi et al., 2002). Under our growth conditions, maximal Vp1 transcript accumulation was observed around 20 DAP (data not shown). This coincides with the peak in ABA accumulation, although ABA levels subsequently decline while Vp1 transcript levels remain relatively high (McCarty et al., 1989; White et al., 2000; Niu et al., 2002). We placed 20-DAP embryos in culture to examine the physiological regulation of Vp1 expression. Vp1 transcript levels declined by 8 h of hormone-free culture, becoming barely detectable by 48 h. The cultured embryos showed visible signs of germination within 24 h. This inverse correlation between Vp1 expression and germination transition is consistent with the function of VP1 to promote seed dormancy and repress germination (McCarty, 1995). The decrease in Vp1 expression was reversed by exogenous ABA, which also arrests germination. Thus, VP1 mediates and activates ABA induction of maturation-associated gene expression and is itself positively regulated by ABA. A similar result in barley was interpreted as an indirect consequence of germination in hormone-free culture and it was concluded that barley Vp1 was not significantly modulated by ABA (Hollung et al., 1997). However, we showed that application of ABA after 24 h of culture in hormone-free medium rapidly induced Vp1 transcript levels to levels observed with continuous culture in ABA-containing medium. ABA induced Vp1 transcript accumulation at concentrations as low as 0.1 μm, which is considered to be within physiological levels (Walton and Li, 1995). Induction occurred within 1 h and did not require de novo protein synthesis because cycloheximide did not block inducibility by ABA. Cycloheximide alone caused a slight increase in Vp1 expression similar to what has been observed in the barley HVA22 gene, where ABA and cycloheximide have a synergistic effect on expression (Shen et al., 1993). Taken together, these data unambiguously demonstrate that the maize Vp1 transcript is induced by ABA in cultured embryos.
The accumulation of Vp1 transcript was also induced in embryos by NaCl, Suc, and mannitol. Whereas it remains possible that Suc might regulate Vp1 expression through either sugar or osmotic signaling, the effect of mannitol on Vp1 expression clearly indicates that an osmotic stimulus is capable of inducing Vp1 expression. Given the normal environment of developing embryos, it is not surprising that osmotic or sugar stimuli could induce a maturation-associated gene.
At this point, it appears likely that regulation of Vp1 by ABA might occur at both the transcriptional and posttranscriptional levels. Endogenous Vp1 transcript levels were clearly and consistently induced by ABA, whereas GUS expression was not induced in embryos carrying the Vp1∷GUS transgene. In transient assays, we observed only modest reporter induction by ABA and whether induction was observed was inconsistent from experiment to experiment (data not shown), suggesting that perhaps additional, as yet undefined, factors influence transcriptional regulation. Posttranscriptional and posttranslational regulation has been reported for ABI3 (Zhang et al., 2005; Bassel et al., 2006). In vegetative tissues, stress induction of the Vp1∷GUS reporter clearly indicates that regulation occurs at the transcriptional level.
Although ABA and osmoticum are able to induce or maintain Vp1 expression levels, transcript abundance is nonetheless somewhat higher in embryos freshly harvested from intact seeds. This suggests that additional extraembryonic seed factors are involved in promoting Vp1 expression in maize kernels. In tomato (Lycopersicon esculentum), signaling from extraembryonic seed tissues was also reported to regulate LeABI3 transcript levels because expression in isolated embryonic axes was lower than in intact seeds (Bassel et al., 2006).
Like sorghum, the maize Vp1 promoter lacks a canonical TATA box (Carrari et al., 2001). The Sbvp1 promoter was also reported to lack a canonical CAAT box. The maize Vp1 promoter has only one potential CAAT box located approximately 400 bp 5′ to the start of transcription (data not shown). Most CAAT boxes are located 60 to 100 bp upstream of the transcription start and it is questionable whether a CAAT box in this atypical position would be functional. Either way, it would be unusual because TATA-less promoters are usually dependent on CAAT boxes for activity (Mantovani, 1998). One common feature of TATA-less promoters is that the transcription start sites are variable (Achard et al., 2003; Chow and Knudson, 2005). Using 5′-RACE PCR, we identified a major transcription start site and several ancillary ones.
In the Arabidopsis ABI3 promoter, there are three open reading frames (ORFs) in the 5′ UTR, which negatively regulate ABI3 expression. Removing these ORFs dramatically increased promoter activity and expanded the expression domain to roots (Ng et al., 2004). We identified a region conserved among the Vp1 5′ UTRs from maize, rice, and sorghum (data not shown), but we did not find similar ORFs in the conserved region or any other region of the 5′ UTR of maize. This suggests that the mechanisms of Vp1 and ABI3 regulation might differ between maize and Arabidopsis.
Because ABREs are present in many ABA-inducible genes, it is not surprising that a predicted ABRE was identified in the Vp1 promoter. EMSA and mutagenesis experiments supported the prediction and further determined that the canonical ABRE core (CCACGTGT) was essential for binding nuclear proteins. Culture in hormone-free medium greatly reduced the binding capacity of the embryo nuclear extract to the ABRE. In contrast, ABA and osmotic stress restored ABRE protein interaction. A similar result was also reported in barley, where ABA, salt, and mannitol increased the protein-binding capacity to an ABRE-containing fragment of Lea B19.1 (Hollung et al., 1997). Considering that ABA induction of Vp1 expression does not require de novo protein synthesis, ABA or mannitol treatments most likely regulate the ABRE-binding activity through posttranslational modifications, such as phosphorylation (Lopez-Molina et al., 2001; Kagaya et al., 2002; Nieva et al., 2005).
In natural promoter contexts, ABREs are often associated with CEs to form ABRC complexes, which confer ABA responsiveness (Shen and Ho, 1995; Shen et al., 1996). Two kinds of CEs have been identified, CE1 and CE3, and are found in ABRC1 and ABRC3, respectively (Hobo et al., 1999a). The ABRC3 type can be induced by both ABA and VP1 overexpression (Guiltinan et al., 1990; Vasil et al., 1995; Busk et al., 1997). We found that the maize Vp1 promoter contains an apparent ABRC1-type complex, similar to that present in barley HVA22. Although the HVA22 promoter was not activated by transient VP1 overexpression (Shen et al., 1996), RNAi suppression of endogenous Vp1 expression abrogated ABA induction (Casaretto and Ho, 2003). Thus, VP1 appears to function in ABA regulation through both types of ABRC.
A distance with multiple of 10 bp between CE1 and the ABRE appears to be important for ABRC1 function (Shen et al., 2004). We identified a CE1-like cis-element with a CCACC core residing 50 bp downstream of the ABRE. The CE1 showed specific protein-binding activity in an EMSA experiment; however, sequence requirements for protein interactions with this element appear distinct from CE1s reported from other promoters (Niu et al., 2002), suggesting that different proteins interact with the CE1 of Vp1. Notably, protein binding still occurred when adenine in the core was mutated to thiamine or cytosine. To address the function of the ABRE and CE1 in ABA regulation of Vp1 expression, transient gene assays were carried out; however, the modest and inconsistent inducibility of the reporter precluded definitive conclusions. Experiments are in progress to identify transcription factors binding to the ABRE and CE1.
The Vp1∷GUS transgenic lines revealed a new expression domain of VP1 upon drought stress because reporter gene expression was detected in vascular cells of vegetative tissues, including stem, leaf, and cob. Reporter expression was localized to phloem companion cells and was verified by in situ hybridization of endogenous Vp1 mRNA. VP1 was believed to have no function in the plant because vp1 mutant plants are not wilty and have normal transpiration rates and ability to retain water in excised leaves (Neill et al., 1987). Additionally, vp1 mutant plants appear to be ABA sensitive, at least for stomatal responses. However, an effect of vp1 mutants on the maintenance (but not induction) of Catalase1 transcript levels under water deficit suggests that Vp1 may have a function in plant stress responses (Guan and Scandalios, 2000). This is supported by our observation of Vp1 expression in vegetative tissues. Given that vascular tissue is essential for water and solute transport, it follows that these cells would be protected during drought stress to maximize the chance of recovery. Without these functions, plants have no chance to survive. However, we did not detect a difference between normal and vp1 mutant seedlings in the rates of wilting or in the rates of survival or recovery from severe wilting (data not shown).
ABI3 and Vp1 expression have previously been reported in nonseed tissues. Vp1 is expressed in rice suspension-cultured cells (Nakagawa et al., 1996) and ABI3 is expressed in meristematic tissues of Arabidopsis and poplar (Populus spp.), under conditions of growth arrest (Rohde et al., 1999, 2002). In Arabidopsis, dark-grown plants arrest leaf initiation. Under these conditions, ABI3 expression is induced in the shoot apical meristem, where it is known to be functional because abi3 mutants fail to arrest leaf initiation (Rohde et al., 1999). In poplar, PtABI3 is expressed in foliage buds during the autumnal onset of quiescence (Rohde et al., 2002). Experimental alteration of expression levels altered the size and morphology of bud scales and leaves, indicating that PtABI3 has an important developmental function during the onset of bud quiescence. We did not detect Vp1∷GUS expression in seedling shoot apical meristems (data not shown), although it is not clear that maize meristems can become quiescent. Thus, whereas vegetative functions are known for ABI3, the vascular expression pattern and moisture stress inducibility observed for maize Vp1 are novel.
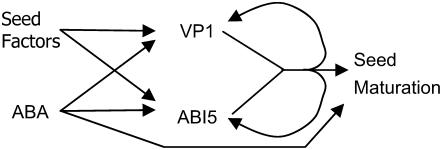
In barley aleurone, HvVP1 and HvABI5 were necessary to transactivate HvABI5 expression and together regulated ABA-inducible gene expression (Casaretto and Ho, 2003, 2005). HvABI5 itself is ABA inducible (Casaretto and Ho, 2003, 2005) and forms part of a complex feed-forward mechanism for ABA regulation of gene expression (Finkelstein et al., 2002). Similarly, our results demonstrate that maize Vp1 is ABA regulated and, because expression of a Vp1∷GUS transgene was reduced in vp1 mutant embryos, that VP1 is also feedback regulated. This feedback regulation is likely to be indirect because transient assays, where Vp1∷GUS was cobombarded with 35S∷Vp1, did not result in increased reporter expression or ABA inducibility (data not shown). In light of these findings, we propose a modified model for the function of VP1 in ABA-regulated seed maturation (Fig. 8). In this model, VP1 and ABI5 (or TRAB1) function synergistically to promote seed maturation, whereas ABA increases the expression of Vp1, Abi5, and other maturation-associated genes. In addition, VP1 and ABI5 feed back to further up-regulate their own expression. This is similar to Arabidopsis, where the expression of ABI3, ABI4, and ABI5 show an intricate system of interregulatory relationships (Söderman et al., 2000). Because VP1 is required for many ABA responses in developing seeds, it is likely that other factors regulate the initial expression of Vp1 during seed development. The involvement of additional factors is further supported by the observation that ABA alone is not sufficient to induce Vp1 transcript levels back to those found in embryos from intact seeds. Some of these other factors may include homologs of LEC1, LEC2, and FUS3, although in Arabidopsis it is unknown whether any of the regulatory interactions observed among these genes and ABI3 are direct (Kagaya et al., 2005; To et al., 2006). Information about how Vp1 is regulated, including future identification of proteins that bind the Vp1 promoter, will help to better understand the mechanisms that regulate seed maturation and germination and how the seed maturation program is coupled to seed development.
Figure 8.
Model for ABA and VP1 regulation of seed maturation. Vp1 and ABI5 expression is initiated by hypothetical unknown seed factors and then further activated by ABA. VP1, ABI5, and ABA synergistically regulate seed maturation. At the same time, VP1 and ABI5 feed back to further up-regulate their own expression, perhaps indirectly in the case of Vp1 (modified from Casaretto and Ho, 2005).
MATERIALS AND METHODS
Plant Materials
The B73/W22 hybrid plants used in Vp1 expression studies and transient expression assays were grown in a greenhouse at 28°C under 16-h light, with light supplemented by sodium halide lamps, and 8-h dark. The aleurone cells used in EMSA were collected from field-grown thick* mutants, which contain multiple aleurone layers (P. Becraft and D. McCarty, unpublished data). Pericarp layers of 20-DAP thick* mutant kernels were removed, following by peeling the aleurone cell layers, which were frozen in liquid nitrogen. The embryos used in EMSA were collected from B73/W22 hybrids grown in the field and stored frozen.
Expression Studies in Cultured Embryos
Embryos collected from B73/W22 hybrids at 20 DAP were cultured in 50-mm petri dishes with 30-mL liquid Murashige and Skoog medium containing 1% Suc under continuous gentle shaking in the dark. Total RNA was isolated by grinding tissues in liquid nitrogen followed by extraction with TRIzol reagent (Invitrogen) and purified with the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was synthesized using 3 μg total RNA. RT was performed using the SuperScript II first-strand synthesis system for RT-PCR (Invitrogen) with 50 pmol oligo(dT) and 10 pmol gene-specific primer for the maize (Zea mays) 18S ribosomal RNA gene (ZMU42796). PCR was performed using Taq DNA polymerase (Promega) with gene-specific primers: Vp1, CAAGAGCAAGGCAGTGGTTCCAG (forward) and CAAATTTAGCGTCACACAGCGGGTAG (reverse); 18S rRNA, AGCAGCCGCGGTAATTCCAGCTC (forward) and CTGGTGGTGCCCTTCCGTCAATTC (reverse). For Vp1, amplification conditions were 94°C for 2 min followed by 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min. For 18S rRNA, amplification conditions were 94°C for 2 min followed by 22 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min. PCR products were run in a 0.8% agarose gel, blotted to Duralon-UV membrane (Stratagene), hybridized with the corresponding gene-specific probes, and autoradiographed.
5′ RACE-PCR
Total RNA was isolated from 2 g of B73/W22 hybrid embryos at 20 DAP using a hot phenol method (Schmitt et al., 1990). The PolyATtract mRNA isolation system II (Promega) was used to prepare mRNA according to the manufacturer's protocol. 5′ RACE cDNA was synthesized from 200 ng mRNA using the SMART RACE cDNA amplification kit (CLONTECH). PCR amplification was carried out with 2.5 μL 5′ RACE-ready cDNA in a 50-μL reaction system. Amplification products were purified with the QIAquick PCR purification kit (Qiagen) and cloned into pBluescript (Stratagene). Plasmid DNA was prepared from 13 individual clones and sequenced at the Iowa State University DNA facility.
Bioinformatic Analysis of the Vp1 Promoter
The maize Vp1 promoter sequence was obtained by sequencing a genomic λ clone from the W22 inbred line. Osvp1 and Sbvp1 (Carrari et al., 2001) promoter sequences were obtained from the National Center for Biotechnology Information (AP003436: bases 71,033 and approximately 72,060 complementary strand; and AF317201, respectively). ConSite analysis was applied to the sequences with 74% conservation cutoff, window size 20, and 80% TF score threshold (Sandelin et al., 2004). PLACE database search was also performed on the Vp1 promoter sequence to identify potential transcription factor binding sites (http://www.dna.affrc.go.jp/PLACE; Higo et al., 1999).
Analysis of Transgenic Lines Carrying Promoter Vp1∷GUS or GFP Transcriptional Fusions
Transcriptional fusions comprised 958 bp of the Vp1 promoter fused to the maize sh1 intron, GUS, or GFP, with a Nos terminator. Stably transformed maize plants were generated according to Gutierrez-Marcos et al. (2004), and transgenic lines were backcrossed into A188 and W22 for a minimum of three generations.
Visualization of GFP reporter gene expression in thin-cut hand sections of maize seeds was achieved using a Zeiss LSM510 META confocal laser-scanning microscope with a 405-nm argon laser. GUS detection in developing seeds was performed as described in Costa et al. (2003), although sections were counterstained with periodic acid Schiff reagent (Spence, 2001), mounted under coverslips, and viewed under bright-field conditions. Stems from transgenic Vp1∷GUS plants were hand cut into thin discs and incubated in media or desiccated for 6 h prior to GUS staining. Samples were then counterstained with phloroglucinol-HCL for lignin detection and observed under a dissection microscope. To verify GUS staining patterns in vascular tissue of desiccated stems, mRNA in situ hybridization was performed on 10-μm thick wax-embedded sections using an antisense Vp1 probe, as described (Costa et al., 2003). Sections were then counterstained with Saffranin-Fast Green (Spence, 2001), mounted under coverslips, and viewed with DIC5-3 optics.
EMSA
Nuclear protein extracts were prepared from 20-DAP embryos or aleurone by isolation of nuclei using a glycerol-based method (Dorweiler et al., 2000) followed by isolation of nuclear proteins in protein isolation buffer (10 mm Tris-HCl, 1 mm dithiothreitol, 0.4 m NaCl, pH 7.5; Escobar et al., 2001). DNA probes were generated by annealing the complementary oligonucleotides (synthesized at the Iowa State University DNA facility) and by filling in the single-strand overhangs with α-[32P]dCTP (Perkin-Elmer) using the Klenow fragment of DNA polymerase I (Promega) or by PCR amplification (Sambrook et al., 1989). The labeled probes were purified with QIAquick nucleotide removal kit (Qiagen). Probe sequences are provided in Figures 5 and 6 and the VP04 negative control had the sequence CCGGCGACACGGACATCCTACGGCGCCGAAGCAGC.
The protein-DNA-binding reactions were performed in 25 μL containing 20 ng/100 bp PCR-labeled probes or 0.8 pmol oligonucleotides, 1 μg sonicated pBlueScript II SK (Stratagene), and 10 μg nuclear proteins in 1× binding buffer (10 mm Tris-HCl, pH 7.4, 50 mm NaCl, 2 mm EDTA, 2.5 mm dithiothreitol, 1.25 μg bovine serum albumin, 0.05% [v/v] NP-40, 10% glycerol). The binding reactions were incubated at room temperature for 20 min and separated on a nondenaturing 4.5% polyacrylamide gel. Electrophoresis was carried out at 70 V of constant voltage for 4 h with 0.5× 45 mm Tris-borate, 0.5 mm EDTA, pH 8.0 buffer in a 4°C cold room. Gels were vacuum dried and autoradiographed. In competition assays, nuclear protein extract was incubated with unlabeled oligonucleotides in 1× binding buffer on ice for 10 min prior to adding the radioactive probe and continuing with incubation for a further 20 min at room temperature.
GUS Activity Assay
Homozygous vp1 mutant plants were obtained by rescuing vp1 mutant kernels in the greenhouse. A Vp1∷GUS transgenic line, heterozygous for the vp1 mutant allele, was crossed to homozygous vp1 mutants to generate ears segregating 1:1 for vp1 mutant kernels. Kernels carrying the Vp1∷GUS transgene were identified by staining the crown region of each kernel for GUS activity in the aleurone. The crown was excised and incubated in GUS-staining solution (50 mm sodium phosphate [pH 7.0], 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA, 0.05% [v/v] Triton X-100, 0.35 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) for 24 h at 37°C (Jefferson et al., 1987).
Individual embryos of GUS-positive kernels were homogenized in 250 μL 1× cell culture lysis reagent (Promega) and centrifuged for 10 min at 10,000g at 4°C. Protein concentration was determined using the Bio-Rad protein assay system (Bio-Rad). Fluorometric MUG assays were carried out essentially as described (Kosugi et al., 1990). Briefly, 100-μL protein extracts were added to preincubated 600-μL 1 mm MUG in 1× cell culture lysis reagent with 20% methanol; 100-μL reaction mixtures were added into 900 μL 0.2 m Na2CO3 at various time points. The 4-methylumbelliferone was read on a Synergy HT plate reader (Bio-Tek). The relative GUS activity was normalized according to the protein concentration.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ886030.
Acknowledgments
We thank Becraft lab members and Andrea Eveland for critical reading of the manuscript.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (grant no. 2006–01163) and by the National Science Foundation (grant no. 0077676).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Philip W. Becraft (becraft@iastate.edu).
Open Access articles can be viewed online without a subscription.
References
- Achard P, Lagrange T, El-Zanaty A-F, Mache R (2003) Architecture and transcriptional activity of the initiator element of the TATA-less RPL21 gene. Plant J 35 743–752 [DOI] [PubMed] [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD (2006) ABI3 expression ceases following, but not during, germination of tomato and Arabidopsis seeds. J Exp Bot 57 1291–1297 [DOI] [PubMed] [Google Scholar]
- Bobb AJ, Eiben HG, Bustos MM (1995) PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. Plant J 8 331–343 [DOI] [PubMed] [Google Scholar]
- Busk PK, Jensen AB, Pages M (1997) Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J 11 1285–1295 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pages M (1997) Protein binding to the abscisic acid-responsive element is independent of VIVIPAROUS1 in vivo. Plant Cell 9 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37 425–435 [DOI] [PubMed] [Google Scholar]
- Carrari F, Frankel N, Lijavetzky D, Benech-Arnold R, Sanchez R, Iusem ND (2001) The TATA-less promoter of VP1, a plant gene controlling seed germination. DNA Seq 12 107–114 [DOI] [PubMed] [Google Scholar]
- Casaretto J, Ho TD (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto JA, Ho TD (2005) Transcriptional regulation by abscisic acid in barley (Hordeum vulgare L.) seeds involves autoregulation of the transcription factor HvABI5. Plant Mol Biol 57 21–34 [DOI] [PubMed] [Google Scholar]
- Chow G, Knudson W (2005) Characterization of promoter elements of the human HYAL-2 gene. J Biol Chem 280 26904–26912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LM, Gutierrez-Marcos JF, Brutnell TP, Greenland AJ, Dickinson HG (2003) The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130 5009–5017 [DOI] [PubMed] [Google Scholar]
- Dorweiler JE, Carey CC, Kubo KM, Hollick JB, Kermicle JL, Chandler VL (2000) mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12 2101–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar C, Aristizábal F, Navas A, Campo FFD, Fenoll C (2001) Isolation of active DNA-binding nuclear proteins from tomato galls induced by root-knot nematodes. Plant Mol Biol Rep 19 375a–375h [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan LM, Scandalios JG (2000) Catalase transcript accumulation in response to dehydration and osmotic stress in leaves of maize viviparous mutants. Redox Rep 5 377–383 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8 183–187 [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250 267–271 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O'Sullivan DM, Wormald M, Perez P, Dickinson HG (2004) maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T (1999. a) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19 679–689 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999. b) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR (1995) Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev 9 2459–2469 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR (1999) Signaling from the embryo conditions Vp1-mediated repression of alpha-amylase genes in the aleurone of developing maize seeds. Plant J 19 371–377 [DOI] [PubMed] [Google Scholar]
- Hollung K, Espelund M, Schou K, Jakobsen KS (1997) Developmental, stress and ABA modulation of mRNA levels for bZip transcription factors and Vp1 in barley embryos and embryo-derived suspension cultures. Plant Mol Biol 35 561–571 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46 399–406 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y (1990) An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci 70 133–140 [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15 281–307 [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R (1998) A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res 26 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte WR Jr, Russell SH, Quatrano RS (1989) Abscisic acid-responsive sequences from the em gene of wheat. Plant Cell 1 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR (1995) Genetic control and integration of maturation and germination pathways in seed development. Annu Rev Plant Physiol Plant Mol Biol 46 71–93 [Google Scholar]
- McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66 895–905 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Kagaya Y, Ogawa Y, Nagato Y, Hattori T (2002) Temporal and spatial expression pattern of the OSVP1 and OSEM genes during seed development in rice. Plant Cell Physiol 43 307–313 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Ohkura E, Ohmiya K, Hattori T (1996) The seed-specific transcription factor VP1 (OSVP1) is expressed in rice suspension-cultured cells. Plant Cell Physiol 37 355–362 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60 51–68 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Horgan R, Rees AF (1987) Seed development and vivipary in Zea mays L. Planta 171 358–364 [DOI] [PubMed] [Google Scholar]
- Ng DW, Chandrasekharan MB, Hall TC (2004) The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol Biol 54 25–38 [DOI] [PubMed] [Google Scholar]
- Nieva C, Busk P, Domínguez-Puigjaner E, Lumbreras V, Testillano P, Risueño M-C, Pagès M (2005) Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Mol Biol 58 899–914 [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ (2002) Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell 14 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W (2002) PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14 1885–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W (1999) The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ 22 261–270 [Google Scholar]
- Rojas A, Almoguera C, Jordano J (1999) Transcriptional activation of a heat shock gene promoter in sunflower embryos: synergism between ABI3 and heat shock factors. Plant J 20 601–610 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sandelin A, Wasserman WW, Lenhard B (2004) ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res 32 W249–W252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Ho TH (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho TH (1993) Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J Biol Chem 268 23652–23660 [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho THD (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QJ, Casaretto JA, Zhang P, Ho T-HD (2004) Functional definition of ABA-response complexes: the promoter units necessary and sufficient for ABA induction of gene expression in barley (Hordeum vulgare L.). Plant Mol Biol 54 111–124 [DOI] [PubMed] [Google Scholar]
- Singh KB (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol 118 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J (2001) Plant histology. In C Hawes, B Satiat-Jeunemaitre, eds, Plant Cell Biology: A Practical Approach. Oxford University Press, Oxford, pp 189–206
- Suzuki M, Kao CY, Cocciolone S, McCarty DR (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28 409–418 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li Q-B, McCarty DR (2003) Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol 132 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton DC, Li Y (1995) Abscisic acid biosynthesis and metabolism. In PJ Davies, ed, Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer, Norwell, MA, pp 140–157
- West M, Harada JJ (1993) Embryogenesis in higher plants: an overview. Plant Cell 5 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ (2000) Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua N-H (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]