Abstract
Regulated protein degradation contributes to plant development by mediating signaling events in many hormone, light, and developmental pathways. Ubiquitin ligases recognize and ubiquitinate target proteins for subsequent degradation by the 26S proteasome. The multisubunit SCF is the best-studied class of ubiquitin ligases in Arabidopsis (Arabidopsis thaliana). However, the extent of SCF participation in signaling networks is unclear. SCFs are composed of four subunits: CULLIN 1 (CUL1), ASK, RBX1, and an F-box protein. Null mutations in CUL1 are embryo lethal, limiting insight into the role of CUL1 and SCFs in later stages of development. Here, we describe a viable and fertile weak allele of CUL1, called cul1-6. cul1-6 plants have defects in seedling and adult morphology. In addition to reduced auxin sensitivity, cul1-6 seedlings are hyposensitive to ethylene, red, and blue light conditions. An analysis of protein interactions with the cul1-6 gene product suggests that both RUB (related to ubiquitin) modification and interaction with the SCF regulatory protein CAND1 (cullin associated and neddylation dissociated) are disrupted. These findings suggest that the morphological defects observed in cul1-6 plants are caused by defective SCF complex formation. Characterization of weak cul1 mutants provides insight into the role of SCFs throughout plant growth and development.
The ubiquitin/26S proteasome pathway affects many aspects of plant development by precisely regulating the degradation of key proteins in response to environmental or biological cues. Ubiquitination of proteins involves the action of three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligating enzyme (E3). Several classes of E3 ligases allow for the recognition and degradation of a diverse array of substrates (Moon et al., 2004). A major class of E3s in plants, SCF complexes, have been implicated in a variety of growth and developmental processes, including cell cycle, hormone response, circadian rhythm, and photomorphogenesis (Gray et al., 1999; Woo et al., 2001; del Pozo et al., 2002; Guo and Ecker, 2003; Harmon and Kay, 2003; McGinnis et al., 2003; Wang et al., 2003; Moon et al., 2004). The SCF is composed of four protein subunits: CULLIN 1 (CUL 1), Arabidopsis (Arabidopsis thaliana) SKP1 (ASK), RING-box 1 (RBX1), and an F-box protein. The scaffold-like CUL1 binds both ASK and RBX1. The substrate specificity factor for the SCF is the F-box protein, which is bound to the complex through its association with ASK.
Each of the subunits of the SCF complex is encoded by gene families. For example, there are 21 predicted ASK genes and over 700 F-box genes in the Arabidopsis genome (Gagne et al., 2002; Risseeuw et al., 2003). Although 11 CUL-like genes have been identified in Arabidopsis, only CUL1 and CUL2 have been shown to participate in an SCF complex (Risseeuw et al., 2003; J. Moon and M. Estelle, unpublished data). The combinatorial possibilities of the SCF subunits, such as ASK and F-box proteins, contribute to the ability of SCF to target a wide variety of substrates.
The activity of the SCF complex is regulated by the attachment of RUB (related to ubiquitin) to a conserved Lys residue within the C terminus of the CUL1 subunit. Although the precise function of RUB modification is not clear, the cycling of RUB attachment and cleavage from CUL1 is necessary for SCF activity (Wu et al., 2005). CAND1/TIP120A has been shown to interact preferentially with unmodified CUL1 (Zheng et al., 2002a; Oshikawa et al., 2003; Chuang et al., 2004; Feng et al., 2004) and may modulate SCF activity by affecting complex formation. CAND1 binds both the C- and N-terminal domain of CUL1 and shares a similar binding site with ASK. Indeed, recent evidence suggests that ASK and CAND1 binding is mutually exclusive (Liu et al., 2002; Goldenberg et al., 2004). CAND1 may negatively regulate SCF activity by preventing formation of the CUL1-ASK-RBX1 complex. RUB modification of CUL1 appears to displace CAND1 and allow subsequent ASK binding. CAND1 is a nonessential gene in Arabidopsis, but mutations in CAND1 display severe defects in several signaling pathways, suggesting that it is necessary for proper SCF activity (Chuang et al., 2004). However, it is still unclear how CAND1-CUL1 interaction affects SCF function.
Mutations in F-box and ASK genes confer diverse defects, suggesting that CUL1 is required throughout development. However, specifying the role of CUL1 in later stages of development has been difficult due to the gene's essential functions. The embryo-lethal phenotype of cul1 null alleles underlines the importance of SCF-mediated protein degradation during embryogenesis (Shen et al., 2002; Hellmann et al., 2003). The axr6-1 and axr6-2 alleles of CUL1 have dominant-negative characteristics (Hobbie et al., 2000; Hellmann et al., 2003). Whereas axr6-1 and axr6-2 homozygous plants are seedling lethal, heterozygous plants display reduced apical dominance and curled leaves indicative of auxin defects. The complex genetic behavior of these mutants and the lethality of the homozygotes early in development prevent analysis of CUL1 function in adult plants. Finally, the study of CUL1 cosuppression lines, which display meristem defects and fail to initiate organs, provides some insight into the function of CUL1 (Hellmann et al., 2003). However, using these cosuppression lines for genetic study is difficult due to the instability of the transgenic lines.
A significant advance in CUL1 characterization was recently reported (Quint et al., 2005). This group described the identification of a new allele of CUL1 isolated in a tir1-1 enhancer screen. This allele, called axr6-3, is temperature sensitive. Mutant plants are sterile at 22°C, but fertile at 18°C. Although axr6-3 plants have numerous defects, some pathways known to have an SCF component are unaffected by the axr6-3 mutation. One possibility is that the SCF complexes involved in these responses are assembling with CUL2, which has been shown to interact with SCF components (Risseeuw et al., 2003; J. Moon and M. Estelle, unpublished data). Another explanation is that the CUL1 protein encoded by the axr6-3 gene retains sufficient function for these pathways. Therefore, additional viable alleles of cul1 would be valuable tools in understanding the full extent of CUL1 function in plant growth and development.
We have isolated a new allele of CUL1 (cul1-6) from a genetic screen for mutants resistant to sirtinol. Sirtinol is a synthetic compound that induces expression of auxin response genes. Like axr6-3, the cul1-6 allele is a viable allele. Although the cul1-6 mutation affects a residue that is a few amino acids away from that affected by the axr6-1 mutation, it has a remarkably different phenotype than axr6-1 plants. Characterization of cul1-6 plants indicates that CUL1 is required for diverse signaling pathways throughout development.
RESULTS
The cul1-6 Mutant Is Resistant to Sirtinol
A genetic screen was undertaken to identify mutants resistant to the synthetic compound sirtinol, which has been shown to activate auxin response pathways. This screen appears to be very sensitive because almost all of the known auxin-resistant mutants were identified in addition to several novel mutants (Zhao et al., 2003; Cheng et al., 2004; Dai et al., 2005; Perry et al., 2005; Li et al., 2006). One of the recessive sirtinol-resistant mutants was mapped to the top of chromosome IV. Sequencing of candidate genes in this region identified a C-to-T mutation in the CUL1 gene. The C-to-T missense mutation results in an amino acid change from Leu to Phe at position 115.
cul1-6 Plants Have Diverse Morphological Defects
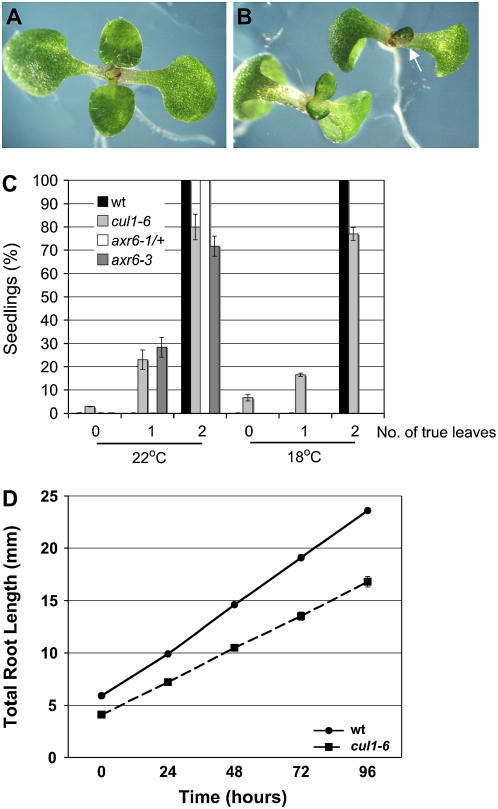
Early defects in cul1-6 seedlings include altered timing of leaf emergence (Fig. 1, A–C) and slower root growth (Fig. 1D) compared to wild-type seedlings. After 11 d in 24-h light at 22°C, 80% of cul1-6 seedlings and approximately 72% of axr6-3 seedlings had produced two true leaves, whereas 100% of the wild-type and segregating population of axr6-1 seedlings produced two true leaves (Fig. 1C). At 18°C, the timing of leaf initiation was similarly delayed in cul1-6 seedlings, indicating this phenotype is not temperature sensitive. The remaining cul1-6 seedlings either had no leaves (approximately 7%) or only one leaf (approximately 17%; Fig. 1C). Subsequent rosette leaves emerge normally (data not shown). Delayed leaf emergence suggests that cul1-6 seedlings grow more slowly than wild-type seedlings or that cul1-6 seedlings have a defect in the shoot apical meristem. Cross sections of 11-d-old cul1-6 seedlings appeared normal (data not shown). To further evaluate cul1-6 seedling growth rate, the root length of 4-d-old seedlings was measured every 24 h for 4 d. Data show that cul1-6 roots grow more slowly than wild-type roots (Fig. 1D).
Figure 1.
cul1-6 seedlings have abnormal leaf emergence and growth rate. A, Fourteen-day-old wild-type (wt). B, cul1-6 seedlings with emerging true leaves. Arrow indicates a cul1-6 seedling with one true leaf emerging instead of two true leaves. C, Table showing percentage of 11-d-old wt, axr6-1/+ (segregating population), axr6-3, and cul1-6 seedlings with zero, one, or two true leaves grown at 22°C or 14-d-old wt and cul1-6 seedlings grown at 22°C. n = 100 seedlings, in triplicate; error bars ± sd. D, Total root length of wt and cul1-6 seedlings was measured every 24 h beginning 4 d after germination. n ≥ 48 seedlings; error bars ± se.
Under our growing conditions, 30-d-old cul1-6 plants were dwarfed with curled leaves (Fig. 2, A and B). Later in development, cul1-6 plants produced more lateral shoots and secondary inflorescences, indicative of reduced apical dominance (Fig. 1B), which is consistent with the role of CUL1 in auxin response (Hellmann et al., 2003).
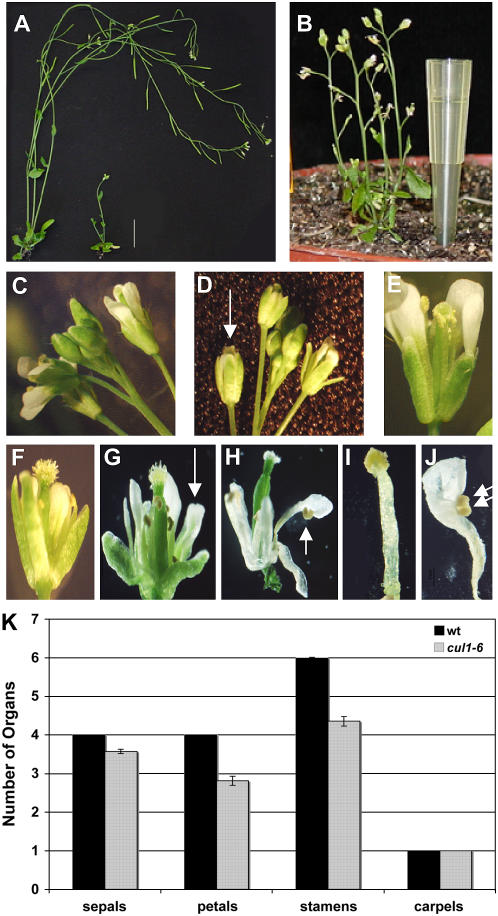
Figure 2.
cul1-6 adult plants have reduced stature and floral defects. A, Thirty-day-old wild-type (wt; left) and cul1-6 (right) plants. White bar = 2 cm. B, cul1-6 plant at 55 d. Floral phenotypes: C and E, wt flowers; D, F, G, and H, cul1-6 flowers. Defects commonly seen in cul1-6 flowers include gynoecium protruding from immature flower (D), fewer petals (F), sepal-petal fusions (G), petal-anther fusions (H). I, wt stamen. J, cul1-6 anther-petal fusion showing two anthers. Arrows in D, G, H, and I point to examples of defects described. K, Table showing frequency of cul1-6 defects in floral organ number. n = 110 flowers; error bars ± se.
Floral morphology was also altered in cul1-6 plants (Fig. 2, D, F–H, and J) compared to wild-type plants (Fig. 2, C, E, and I). Some of the common floral defects included sepals that do not completely enclose the carpels (Fig. 2D), shorter stamens (Fig. 2, F–H), fewer petals (Fig. 2F), petal-like sepals (Fig. 2G), and anthers fused to petals (Fig. 2, H and J). Further, the number of floral organs was reduced in cul1-6 flowers compared to wild-type flowers (Fig. 2K). These floral phenotypes suggest that floral organ identity genes are misregulated in cul1-6 plants. Many aspects of floral meristem and organ identity are regulated by SCFUFO of which CUL1 is a subunit (Kuroda et al., 2002; Wang et al., 2003). Thus, the defects in floral development observed in cul1-6 flowers are consistent with known SCF function.
Auxin Signaling Is Disrupted in cul1-6 Plants
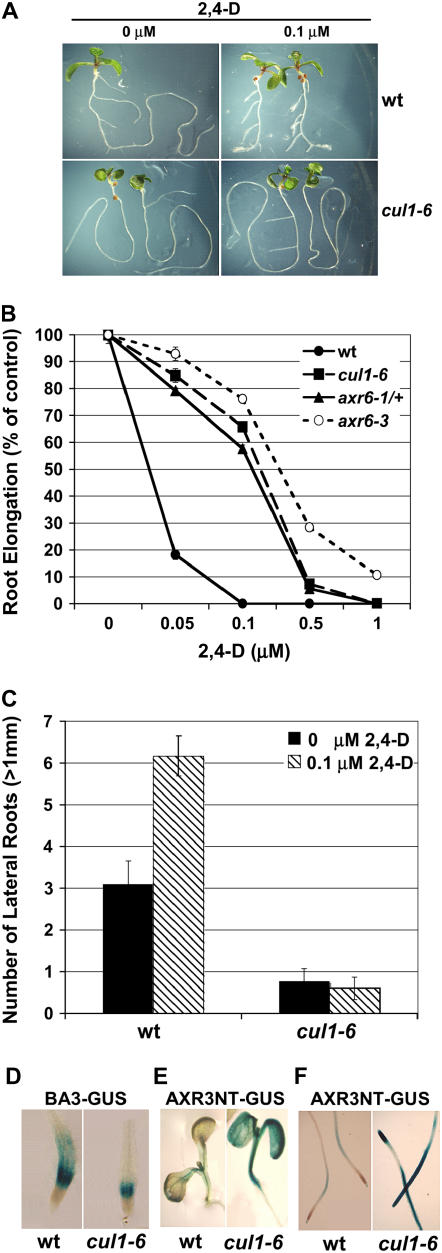
The cul1-6 mutant was identified in a sirtinol resistance screen. Sirtinol is a synthetic compound that induces auxin response genes (Zhao et al., 2003; Dai et al., 2005). Previous reports have shown that SCFTIR1 mediates auxin signaling by promoting the degradation of Aux/IAA proteins, negative regulators of auxin response factors (Gray et al., 1999, 2001). To confirm that auxin response is altered in cul1-6 seedlings, root growth assays were performed on increasing concentrations of the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D). Data show that cul1-6 seedling roots are less sensitive to auxin than wild-type (Fig. 3A) and heterozygous axr6-1 seedling roots (Fig. 3B). Because cul1-6 root growth is resistant to auxin, it seemed likely that other auxin responses might also be affected, such as lateral root initiation (Xie et al., 2000). High concentrations of exogenous auxin induce lateral root production in wild-type seedlings. To determine whether lateral root development is defective in cul1-6 seedlings, mutant seedlings were grown with and without 0.1 μm 2,4-D and the number of lateral roots longer than 1 mm was counted. Data show that cul1-6 seedlings formed fewer lateral roots than wild-type seedlings in the absence of exogenous auxin. In the presence of exogenous auxin, cul1-6 seedlings did not form additional lateral roots (Fig. 3, A and C).
Figure 3.
cul1-6 seedlings display reduced auxin response. A, Four-day-old wild-type (wt) and cul1-6 seedlings were transferred to medium with or without 0.1 μm 2,4-D and grown for an additional 4 d. B, Root growth of wt, cul1-6, axr6-1/+, and axr6-3 seedlings on increasing concentrations of 2,4-D was measured and expressed as percentage of root length on control plates. n = 20; error bars ± se. C, Number of lateral roots produced by 10-d-old wt and cul1-6 seedlings grown on plates with and without 0.1 μm 2,4-D for 5 d. n = 10; error bars ± se. D, BA3-GUS expression induced by 20 μm auxin treatment in wt or cul1-6 seedling roots. E and F, HS-AXR3NT-GUS expression in cotyledons (E) or roots (F) of wt seedlings or cul1-6 seedlings.
To confirm that auxin-responsive gene expression is compromised in the cul1-6 background, the auxin-inducible BA3-GUS transgene was crossed into cul1-6 plants. Our expectation was that less GUS staining would be present in the transgenic BA3-GUS cul1-6 seedlings because Aux/IAA proteins would be more stable, thus reducing expression of the auxin reporter. Indeed, after treatment with exogenous auxin, less GUS staining was detected in transgenic cul1-6 seedling roots compared to BA3-GUS staining in the wild-type background (Fig. 3D). This suggests that the cul1-6 mutation results in reduced SCF activity and stabilization of Aux/IAA proteins.
Many of the Aux/IAA proteins have a very short half-life and are rapidly degraded in response to auxin, in part through SCFTIR1 (Worley et al., 2000; Zenser et al., 2001, 2003). To determine whether targets of SCFTIR1 were more stable in the cul1-6 background, the cul1-6 allele was crossed into plants containing the HS-AXR3NT-GUS reporter gene. The N-terminal portion of AXR3 (domains I and II) is sufficient for nuclear localization and for targeting the protein for degradation by SCFTIR1 (Gray et al., 2001; Ramos et al., 2001). The cul1-6 plants expressing HS-AXR3NT-GUS were exposed to heat shock to induce GUS expression. Seedlings were incubated at room temperature for 20 min to allow for AXR3-GUS degradation and subsequently stained for GUS activity. More GUS staining was detected in the cul1-6 background compared to the wild-type control, indicating that AXR3NT-GUS was more stable in the cul1-6 background (Fig. 3, E and F). Therefore, it is likely that increased stability of Aux/IAA proteins in the cul1-6 background contributes to the auxin-resistant phenotype.
Cytokinin, Jasmonic Acid, and Ethylene-Signaling Pathways Are Altered in the cul1-6 Background
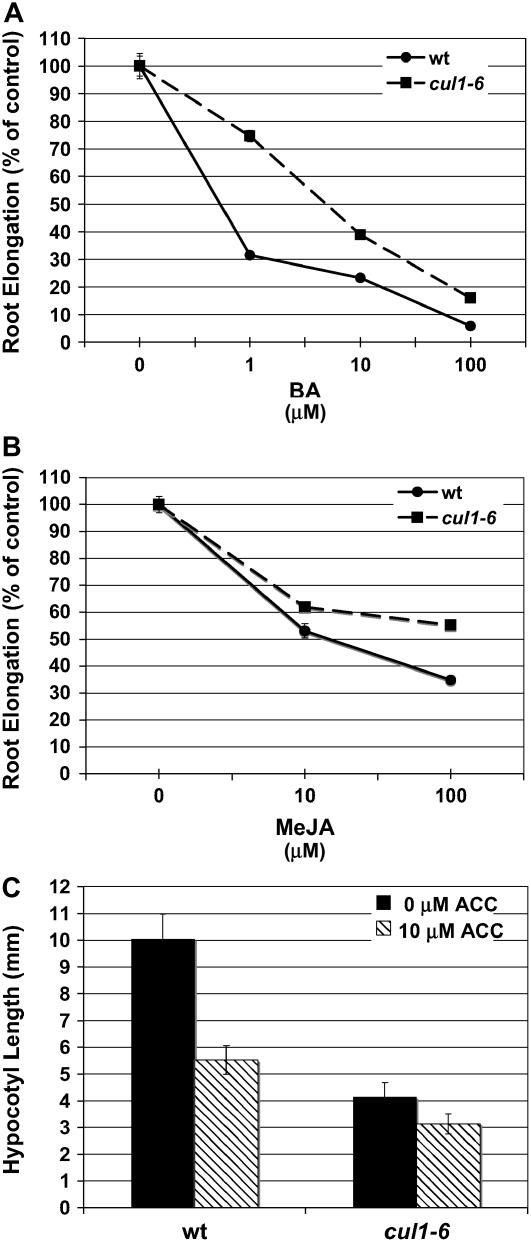
Because SCF-mediated degradation has been shown to play a role in the signaling pathways of many hormones, we investigated how cul1-6 plants respond to jasmonic acid (JA) or 6-benzyladenine (BA), a cytokinin. Root elongation assays using BA or JA showed that cul1-6 seedling roots were less sensitive to both of these hormones than wild-type seedlings (Fig. 4, A and B). Whereas JA signaling is known to be SCF mediated (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002), involvement of SCF complexes in the cytokinin response has not been shown.
Figure 4.
Reduced cytokinin, JA, and ethylene response in cul1-6 seedlings. Wild-type (wt) and cul1-6 root growth assays on increasing concentrations of BA (cytokinin; A) and methyl JA (Me-JA; B). Four-day-old seedlings were grown on exogenous hormone for an additional 5 d. n = 15; error bars ± sd. C, Hypocotyl length of wt and cul1-6 seedlings grown in the dark for 6 d with or without ACC. n = 20; error bars ± se.
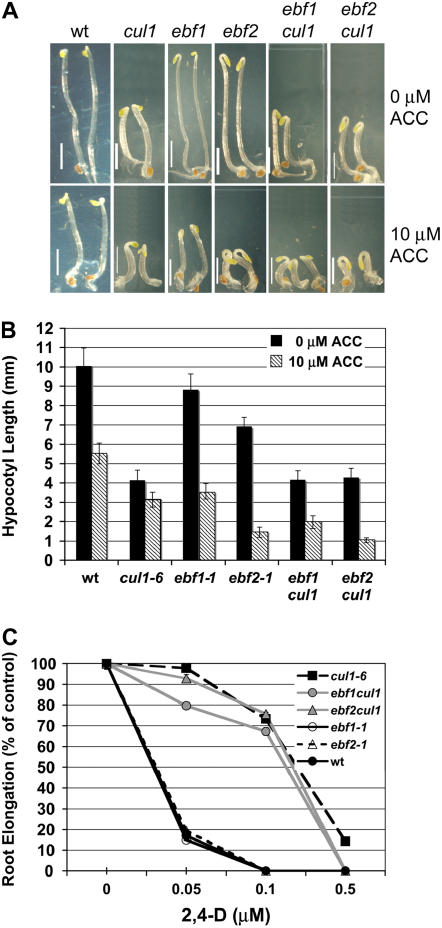
The F-box proteins EBF1 and EBF2 mediate ethylene response by promoting the degradation of the transcription factor EIN3 in the absence of ethylene (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). To investigate ethylene response in cul1-6 seedlings, we grew wild-type and mutant seedlings in the dark on medium with or without the ethylene precursor 1-aminocyclopropane 1-carboxylic acid (ACC). The wild-type triple-response characteristics of dark-grown Arabidopsis seedlings germinated on ethylene include an exaggerated apical hook, short roots, and a short hypocotyl. Our data show that dark-grown cul1-6 seedlings have shorter hypocotyls than dark-grown wild-type seedlings (Fig. 3C). In the presence of ACC, the cul1-6 dark-grown phenotype was only slightly enhanced compared to the change in wild-type seedlings (Fig. 4C). Notably, cul1-6 seedlings exposed to ACC failed to form an exaggerated apical hook (Fig. 5A). Because apical hook formation requires differential elongation on one side of the tissue, which is an auxin and gibberellic acid-mediated process (Lehman et al., 1996; Raz and Ecker, 1999; Li et al., 2004; Vriezen et al., 2004), we reasoned that the cul1-6 phenotype on ethylene may be an indirect result of reduced auxin or GA response.
Figure 5.
Triple response in cul1-6 and double-mutant seedlings is compromised. Wild-type (wt), cul1-6, ebf1, ebf2, ebf1 cul1, and ebf2 cul1 seedlings were germinated and grown in the dark for 6 d. A, Representative phenotypes of seedlings grown on minimal medium or on medium supplemented with 10 μm ACC. White bar = 2 mm. B, Hypocotyl length of seedlings grown in the dark for 6 d with or without 10 μm ACC. Error bars ± se. C, Four-day-old wt, cul1-6, ebf1, ebf2, ebf1 cul1, and ebf2 cul1 seedlings were transferred to auxin for an additional 4 d. Root measurements are expressed as percentage of root growth without auxin. Error bars ± se. [See online article for color version of this figure.]
To determine whether cul1-6 could suppress the exaggerated hook formation of hypersensitive ethylene mutants ebf1 and ebf2, we analyzed the ethylene sensitivity of double mutants ebf1 cul1-6 and ebf2 cul1-6. The response of ebf1 cul1-6 and ebf2 cul1-6 hypocotyls to ACC was enhanced over that of the single mutants (Fig. 5, A and B). However, ebf1 cul1-6 and ebf2 cul1-6 double mutants did not form an exaggerated apical hook, indicating that auxin or GA signaling may be required to mediate this ethylene response. To determine whether auxin response was altered in the single ebf1 and ebf2 mutants as well as in the double mutants, we performed a root elongation assay using increasing concentrations of auxin. ebf1 and ebf2 seedlings displayed wild-type sensitivity to auxin (Fig. 5C). This suggests that EBF1 and EBF2 do not participate in auxin-mediated root elongation. The root elongation data also show that ebf1 cul1-6 seedlings were slightly more sensitive to auxin than cul1-6 seedlings (Fig. 5C). This may be a result of changes in auxin or GA sensitivity in a background that constitutively responds to ethylene.
CUL1 Is Necessary for Gravitropic Response in Dark-Grown Hypocotyls
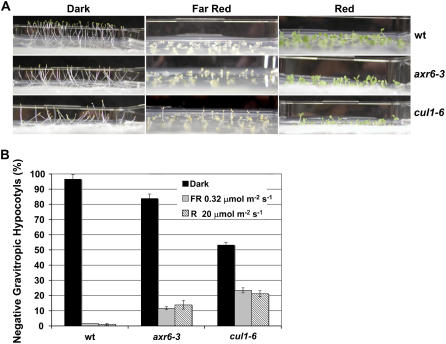
In plants, light and gravity cues contribute to the direction of growth. Dark-grown Arabidopsis hypocotyls have a strong negative-gravitropic response, which promotes seedling growth toward the soil surface. To determine whether this aspect of hypocotyl gravitropism was defective in cul1-6 seedlings, wild-type and cul1-6 seedlings were grown on a horizontal plate in the dark for 6 d. Data show that cul1-6 hypocotyls were more randomly oriented with respect to gravity than wild-type hypocotyls. About 50% of cul1-6 hypocotyls were completely prone on agar (Fig. 6A). This is consistent with a role for CUL1 in hypocotyl gravitropism, which is dependent on auxin signaling (Fig. 3; Stowe-Evans et al., 1998). However, because some light response mutants also show altered hypocotyl negative gravitropism (pif1, pif3, and hfr1; Fairchild et al., 2000; Oh et al., 2004; Shen et al., 2005), it may also indicate reduced response to light signaling. In wild-type seedlings, exposure to red (R) or far-red (FR) light suppresses negative gravitropism. To determine whether phytochrome (phy)-mediated suppression of negative gravitropism was affected in cul1 mutants, we grew wild-type, axr6-3, and cul1-6 seedlings for 6 d in 20 μmol m−2 s−1 R or 0.32 μmol m−2 s−1 FR light. With exposure to R and FR light, the gravity response is attenuated in cul1 seedlings, although not to the extent seen in wild-type seedlings (Fig. 6B). Therefore, both the auxin- and light-signaling pathways are affected by the cul1 mutation, indicating a functional role for cul1 in these pathways.
Figure 6.
Dark-grown cul1-6 seedlings show reduced gravitropic response. Wild-type (wt), cul1-6, and axr6-3 seedlings were grown in the dark, 20 μm m−2 s−1 R, or 0.32 μm m−2 s−1 FR light for 6 d. A, Representative phenotypes of wt, cul11-6, and axr6-3 seedlings grown in the dark, R, or FR light. B, Chart showing percentage of negatively gravitropic hypocotyls in wt, cul1-6, axr6-3 seedlings. n = 180; error bars ± se. [See online article for color version of this figure.]
CUL1 Is Important for Phy-Mediated Suppression of Hypocotyl Length
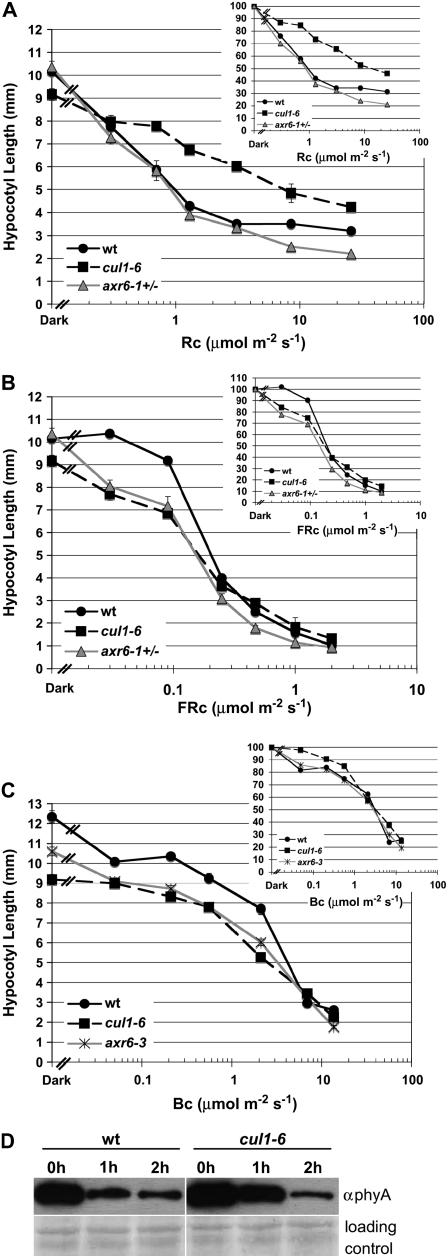
Previous work has shown that SCFEID1 and SCFAFR are involved in phyA-mediated light signaling (Buche et al., 2000; Dieterle et al., 2001; Harmon and Kay, 2003). The F-box mutant eid1 is hypersensitive to FR light, whereas afr mutants are hyposensitive to FR light (Harmon and Kay, 2003). Because F-box proteins are involved in light signaling, we reasoned that mutations in other components of SCF complexes, such as CUL1, would also show defects in light signaling. To examine the role of CUL1 in light signaling, we measured the hypocotyl lengths of wild-type, cul1-6, and segregating axr6-1 seedlings grown for 3 d under increasing fluence rates of R, FR, or blue (B) light. In R light, cul1-6 hypocotyls were longer than wild-type hypocotyls, indicating that cul1-6 seedlings are hyposensitive to phy-mediated inhibition of hypocotyl elongation in R light (Fig. 7A and inset). In FR light, hypocotyl lengths of cul1-6 seedlings were shorter than wild-type seedlings, particularly at low fluence rates (Fig. 7B and inset). This suggests that cul1-6 seedlings are hypersensitive to FR light, possibly due to reduced SCFEID1 function in degrading a positive regulator of FR light response. In addition, cul1-6 seedlings showed moderate hyposensitivity to B light compared to wild-type seedlings (Fig. 7C, inset). In contrast, axr6-3 seedlings show a wild-type response to B light (Fig. 7C and inset). The cul1-6 phenotypic data suggests that SCF may also be involved in B light signaling, which is a novel finding.
Figure 7.
cul1-6 seedlings show altered response to R and FR light. Wild-type, cul1-6, and axr6-1 seedlings were grown for 24 h in darkness, then transferred to various fluence rates of R, FR, or B light for 3 d. The fluence rate response curve was generated by measuring the hypocotyl length of 4-d-old seedlings in continuous R light (A), FR light (B), or B light (C). n = 30; error bars ± se. A to C, Inset shows hypocotyl length as a percentage of dark-grown hypocotyl length. D, Western blot probed with antibody to phyA showing the relative amounts of phyA in wt and cul1-6 seedlings after 0-, 1-, and 2-h exposure to 20 μm m−2 s−1 R light. Loading control (bottom) shows unidentified proteins on same blot stained with Ponceau S.
The hypersensitivity to FR light and hyposensitivity to R light observed in cul1-6 seedlings may be due to changes in the degradation kinetics of the phyA protein. Alternatively, regulation of downstream signaling factors may be defective in cul1-6 mutants. To investigate whether phyA protein levels are altered in the cul1-6 background, we extracted total protein from dark-grown seedlings exposed to increasing amounts of R light because there have been reports that phyA is degraded in R light by ubiquitin-mediated degradation (Shanklin et al., 1987; Clough et al., 1999). A western blot probed with antibody to phyA shows that more phyA is present in the dark and 1-h cul1-6 samples compared to phyA protein levels in the wild-type samples (Fig. 7D). Therefore, the hypersensitive phenotype of cul1-6 under FR light might be due to increased stability of phyA, as observed in the axr6-3 mutant background (Quint et al., 2005). This result also suggests that CUL1 and SCF complexes play a significant role in phyA degradation in R light.
SCF Complex Formation Is Compromised in the cul1-6 Background
The cul1-6 allele results from a point mutation located in a region of the protein that binds ASK1 and CAND1, substituting a conserved Leu for a Phe at position 115 (Fig. 8). The substitution is four residues away from the amino acid change in the axr6-1 gene product. However, unlike the cul1-6 allele, axr6-1 is a seedling-lethal, semidominant mutation. Further, we noted that many of the phenotypes described for cul1-6 were not reported in the other characterized CUL1-recessive allele, axr6-3 (Figs. 1, A–C and 4–7, A and C). We sought to determine the nature of the phenotypic discrepancy between these alleles by examining the SCF complex formation and RUB modification of CUL1 in the cul1-6 background and comparing these findings to previously published results.
Figure 8.
Location of the cul1-6 mutation. Partial amino acid sequence of CUL1 is shown with numbers indicating amino acid position. The mutation in cul1-6 changes a Leu (L) to a Phe (F) at position 115. This site is four amino acids away from the mutation site of axr6-1, which results in a change from a Phe (F) to a Val (V; axr6-1) or to an Ile (I; axr6-2). Black boxes indicate amino acids important for ASK1 binding as derived from the crystal structure (Zheng et al., 2002b); light-gray boxes indicate amino acids important for CAND1 binding (Goldenberg et al., 2004); and dark-gray boxes indicate amino acids important for RBX1 binding (Zheng et al., 2002a).
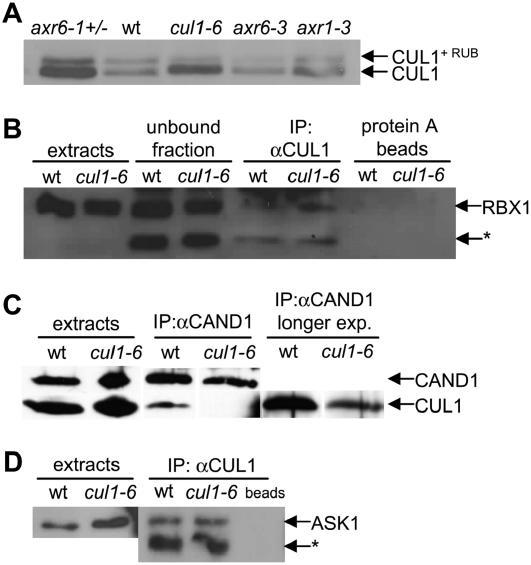
Defects in the RUB modification pathway not only reduce the level of CUL1 modified by RUB1, but also result in accumulation of unmodified CUL1 protein (Hellmann et al., 2003). To compare the level of CUL1 modification in the cul1-6 background, we probed a western blot of total protein with antibody to CUL1. The results show that, whereas slightly less CUL1 was modified in the cul1-6 background, more unmodified CUL1 was present compared to the wild-type samples (Fig. 9A). Accumulation of unmodified CUL1 was also seen in the RUB E1 enzyme mutant axr1-3 and, more dramatically, in axr6-1/+ samples. Increased levels of CUL1 protein in RUB pathway mutants have been reported previously (Hellmann et al., 2003). Because the level of CUL1 RNA in these mutants is not increased, protein accumulation is probably due to changes in posttranslational processing (Hellmann et al., 2003).
Figure 9.
RUB modification and protein interaction of CUL1. A, Western blot of total protein (20 μg/lane) probed with antibody to CUL1. Bands show both relative amounts of RUB-modified CUL1 and unmodified CUL1, as indicated by arrows, in axr6-1/+, wild-type (wt), cul1-6, axr6-3, and axr1-3 seedlings. B, CUL1 was immunoprecipitated from 3 mg wt and cul1-6 protein extract using antibody to CUL1. The western blot was probed with antibody to RBX1. Extracts, Total protein sample; unbound fraction, protein extract that did not bind to protein A beads; protein A beads, protein extract that bound the protein A beads without primary antibody; *, unidentified cross-reacting band. C, CAND1 was immunoprecipitated from wt and cul1-6 seedlings using antibody to CAND1. Western blot was probed with antibody to CUL1 or CAND1, as indicated. Extracts, Total protein sample. Far right image shows longer exposure of the blot pictured in the middle. D, CUL1 was immunoprecipitated from wt and cul1-6 seedlings using antibody to CUL1. A western blot was probed with antibody to ASK1. Extracts, Total protein sample; *, unidentified cross-reacting band.
The RBX1 protein facilitates RUB modification of CUL1 (Gray et al., 2002). Because cul1-6 plants show reduced CUL1 modification, we sought to determine whether the CUL1-6 protein is able to bind RBX1. CUL1 was immunoprecipitated and the western blot probed with antibody to RBX1. The blot showed RBX1 is bound to CUL1 in both wild-type and cul1-6 samples (Fig. 9B). These results suggest that RBX1 binding may not be altered in the cul1-6 background.
CAND1 is believed to regulate SCF formation by binding unmodified CUL1. RUB modification by RBX1 displaces CAND1 from CUL1, which permits ASK1 to interact with CUL1 and complete SCF assembly. To determine whether CAND1/CUL1 binding was altered in the cul1-6 background, CAND1 was immunoprecipitated and the western blot probed with antibody to CUL1. CUL1-6 shows dramatically reduced interaction with CAND1 compared to wild type (Fig. 9C). The reduced interaction between CAND1 and CUL1 may have consequences for efficient CUL1 modification and assembly into active SCF complexes.
To determine whether CUL1-6 is able to participate in an SCF complex, we asked whether CUL1-6 could bind ASK1 in vivo. CUL1 was immunoprecipitated from cul1-6 and wild-type seedlings and immunoblotted with antibody to ASK1. The data show that a similar amount of ASK1 coimmunoprecipitated with CUL1-6 as compared to CUL1 in the wild-type background (Fig. 9D).
DISCUSSION
SCF ubiquitin protein ligases participate in many developmental and regulatory pathways. Genetic analysis of mutants affected in SCF subunits is a useful method to determine the extent to which ubiquitin-mediated proteolysis is involved in various aspects of plant growth and development. Because null mutations of CUL1 are lethal in embryogenesis (Shen et al., 2002; Hellmann et al., 2003), weak cul1 alleles provide an important means to study CUL1 function throughout the life cycle of the plant. Recently, a viable, temperature-sensitive allele of CUL1, called axr6-3, was reported. This mutant displayed many of the defects expected based on previous analyses of F-box and ask mutants. In addition, Quint and colleagues reported a previously undocumented role for CUL1 in Suc and pH response (Quint et al., 2005). FR light signaling is also disrupted in the axr6-3 background, probably a result of the increased stability of phyA (Quint et al., 2005). However, some developmental pathways that are suspected to involve an SCF based on ask1 and F-box protein mutant analysis were not altered by the axr6-3 mutation. By characterizing another viable allele of CUL1, cul1-6, we show that CUL1 is required for ethylene and light responses.
CUL1 Regulates Organ Development in Shoots and Flowers
Several aspects of the cul1-6 phenotype, including the reduced size of the seedling, the delay in leaf initiation, and the decreased rate of root growth, all suggest that the mutation has a general effect on plant growth rate (Figs. 1 and 2, A and B). This effect could involve the role of SCFs in cell cycle progression (del Pozo et al., 2002; Hartig and Beck, 2006) and/or hormone-regulated cell elongation (Sun and Gubler, 2004; Vriezen et al., 2004).
Floral organs in cul1-6 plants are often fused (Fig. 2, C–J) or present in abnormal numbers (Fig. 2K). This phenotype is consistent with defects in the regulation of organ identity genes. The F-box protein UFO has been implicated in the regulation of the organ identity gene AP3 (Lee et al., 1997; Ni et al., 2004). In ufo mutants, basal flowers display petal-stamen fusion, although most flowers lack petals (Durfee et al., 2003; Hepworth et al., 2006). Mutations in ask1, a core component of the SCF, also display sepal-petal fusions and short stamen filaments (Zhao et al., 1999, 2001). The petal-stamen fusions seen in cul1-6 flowers may be due to misexpression of AP3, although additional studies are required to confirm this possibility. Additionally, auxin is known to affect the number and identity of floral organs (Cheng et al., 2006). Some of the cul1-6 floral phenotypes described may be caused by compromised auxin response.
The cul1-6 Mutation Leads to Altered Hormone Responses
Numerous reports have described the role of SCFTIR1 in auxin response (for review, see Nemhauser and Chory, 2005). As expected, the weak cul1-6 allele also displayed auxin defects, such as reduced apical dominance and curled leaves (Fig. 2, A and B), fewer lateral roots (Fig. 3, A and C), and reduced sensitivity to exogenous auxin (Fig. 3, A–C) compared to wild-type seedlings. In addition, the cul1-6 mutation reduced expression of the BA3-GUS reporter (Fig. 3D) and stabilized AXR3NT-GUS, a known target of SCFTIR1 (Fig. 3, E and F). These findings confirm that SCF activity is attenuated in the cul1-6 background. Similarly, our finding that cul1-6 seedlings are resistant to JA is consistent with previous results showing JA resistance in axr6-3 seedlings (Fig. 4B; Quint et al., 2005).
In contrast, the effect of cul1-6 on ethylene response is different from that of axr6-3 (Quint et al., 2005). Hypocotyl length in cul1-6 seedlings was not significantly decreased by ACC (Fig. 4C). Further, unlike in wild-type seedlings, ACC did not promote an exaggerated apical hook in cul1-6 seedlings (Fig. 5A) or in ebf1 cul1 or ebf2 cul1 double-mutant seedlings (Fig. 5B). Interpretation of these results is complicated by the fact that SCF-mediated proteolysis regulates a number of hormone-signaling pathways, many of which are highly integrated. For example, SCFEBF1/EBF2 mediates ethylene response by targeting the transcription factor EIN3 for degradation in the absence of ethylene (Guo and Ecker, 2003). Therefore, reduced SCFEBF1/EBF2 activity in cul1-6 plants would likely result in hypersensitivity to ethylene. However, failure to form an exaggerated apical hook on ACC implies that cul1-6 seedlings are hyposensitive to ethylene. Perhaps this contradiction can be explained by examining the interaction between ethylene signaling and other hormones, such as GA and auxin, both involved in hypocotyl cell elongation and both mediated by SCFs (Gray et al., 1998, 2002; Dill et al., 2004; Fu et al., 2004; Sun and Gubler, 2004). Recent evidence suggests that ethylene-mediated suppression of gibberellic acid synthesis leads to differential cell elongation, resulting in apical hook formation (Vriezen et al., 2004), which implies that compromised GA signaling could lead to the failure to form an apical hook in cul1-6 seedlings grown on ACC. Additional studies are required to determine the nature of the apical hook defect in cul1-6 seedlings.
Defects in the cytokinin response of cul1-6 seedlings imply that cytokinin signaling is also regulated by the ubiquitin/26S proteasome (Fig. 4A). However, it is possible that cul1-6 seedlings are resistant to cytokinin as an indirect result of auxin resistance. Signaling events in response to auxin or cytokinin are highly integrated (Aloni et al., 2006; Tanaka et al., 2006; Werner et al., 2006). Furthermore, some auxin response mutants, such as the auxin influx carrier aux1, are also resistant to BA (Timpte et al., 1995). Therefore, additional study is required to determine the basis of the cytokinin-resistant phenotype in cul1-6 roots.
The cul1-6 Mutation Results in Altered Gravitropic and Light Responses
Ubiquitin/26S-mediated degradation of positive regulators of growth in the dark and negative regulators in the light plays a major role in photomorphogenic development (Shen et al., 2005; Yang et al., 2005). COP1, a RING-type E3 ligase, has been shown to ubiquitinate LAF1, HY5, and HFR1 and induce their degradation in the dark (Osterlund et al., 2000; Seo et al., 2003; Duek et al., 2004; Yang et al., 2005). COP1 also induces degradation of phyA in R light (Seo et al., 2004). Although SCF complexes have been proposed to function in FR light signaling (Buche et al., 2000; Dieterle et al., 2001; Harmon and Kay, 2003; Quint et al., 2005), involvement of SCFs in R and B light signaling has not been reported. Our results show that cul1-6 seedlings have altered gravitropism in the dark, R, and FR light (Fig. 6, A and B). The aberrant gravitropic response of dark-grown cul1 seedlings is likely a consequence of auxin response defects (Fig. 3; Stowe-Evans et al., 1998). Interestingly, R and FR light attenuate the gravity response in both wild-type and cul1 seedlings, but the extent to which light reduces the negative gravitropic response in cul1 is less than in wild-type seedlings (Fig. 6B). Although these data suggest that light-signaling events are affected by the cul1 mutation, it is possible the light-signaling defect reported is ultimately a consequence of the failure of cul1 seedlings to respond normally to auxin. For example, light-mediated attenuation of gravity response may result from a redistribution of auxin in the hypocotyl. Defects in auxin response, such as those observed in cul1 seedlings, may contribute to phenotypes observed in light-mediated suppression of negative gravitropism studies.
Hypocotyl length assays show that cul1-6 seedlings are markedly hyposensitive to R light (Fig. 7A) and marginally hyposensitive to B light (Fig. 7C, inset), indicating that R and B light suppression of hypocotyl length is reduced in the cul1-6 background. Notably, axr6-3 seedlings display wild-type sensitivity to B light (Fig. 7C), suggesting that the cul1-6 mutation confers a B light response phenotype. Hypersensitivity of cul1-6 seedlings to FR light (Fig. 7B) may be a result of increased stability of phyA in this background (Fig. 7D), which is consistent with the previous findings reported for axr6-3 seedlings (Quint et al., 2005). Taken together, these data suggest that, in addition to COP1, multiple SCF complexes are involved in regulating the stability of photoreceptors and light-signaling factors to fine tune photomorphogenic development.
The cul1-6 Mutation and Consequences for SCF Activity
The cul1-6 allele results in a single amino acid change from L to F at position 115 (Fig. 8). Previous work characterized two other mutations in CUL1: a substitution at position 111, which results in a semidominant gain-of-function allele (axr6-1; Hellmann et al., 2003) and a substitution at position 159 (E to L), resulting in a weak, recessive allele (axr6-3; Quint et al., 2005). Current models in yeast (Saccharomyces cerevisiae) and animal systems suggest that SCF formation favors RUB-modified CUL1 over unmodified CUL1 (Osaka et al., 2000; Read et al., 2000; Kawakami et al., 2001). Figure 9A shows that both axr6-1 and cul1-6 have an excess of unmodified CUL1. In contrast, Quint and colleagues have shown that, at 20°C, CUL1 modification is unaffected by the axr6-3 mutation (Quint et al., 2005). Accumulation of unmodified CUL1 in axr6-1 and cul1-6 backgrounds suggests that either less CUL1 is modified in these lines or that unmodified CUL1 is more stable or both. Higher levels of unmodified CUL1 may result from failure of CUL1-6 to participate in an active SCF complex and be subsequently degraded.
RBX1 binds CUL1 at the C terminus and facilitates RUB modification. We conclude from our data that CUL1 binds the RUB E3 RBX1 in the cul1-6 background (Fig. 9B). CAND1 preferentially binds unmodified CUL1, but is displaced upon RUB modification of CUL1, which allows ASK1 to access the modified CUL1 to form an active SCF complex. Our data show that dramatically less CUL1 is coimmunoprecipitated with CAND1 in the cul1-6 background (Fig. 9C). Interestingly, the CUL1-CAND1 interaction is relatively unaffected in the axr6-3 background (Quint et al., 2005), which may suggest a basis for the phenotypic differences between cul1-6 and axr6-3 plants. It is possible that the disruption in CAND1 binding may result in increased amounts of unmodified CUL1, although the reason for this is not understood because CUL1 modification is unaffected by mutations in cand1 (Chuang et al., 2004).
CONCLUSION
We conclude that discrepancies in the phenotype between axr6-3 and cul1-6, two weak CUL1 alleles, may be due to differences in SCF complex formation. Perhaps pathways that are affected by the cul1-6 mutation in CUL1, such as ethylene and R light signaling, are more sensitive to CAND1 regulation of CUL1 modification than other processes. The phenotypic discrepancies between the null allele axr6-1 and the weak allele cul1-6 are interesting because these mutations are within four amino acids of each other. Unlike the axr6-1 mutation, the cul1-6 mutation leads to disruptions in the interaction between CUL1 and CAND1 (W. Zhang and W.M. Gray, unpublished data). At this time, we are uncertain what role CAND1 plays in the cycling of RUB modification of CUL1.
Because CUL1 plays a major role in many plant processes, studies on the characterization of cul1 mutant alleles have advanced our understanding of plant growth and development. The cul1-6 allele, together with the axr6 alleles, provides useful tools for determining nuances in the regulation of SCF activity and, more broadly, the role of SCF in plant growth and development.
MATERIALS AND METHODS
Plant Cultivation
All plants used for this study were in the Arabidopsis (Arabidopsis thaliana) Columbia-0 background. Plants were cultured in Metro-Mix 200 soil (Sun Gro Horticulture) under 24-h light at 24°C ± 0.5°C. Seeds for experiments other than light fluence rate experiments were surface sterilized, stratified, plated on ATS medium [5 mm KNO3, 2.5 mm KH2PO4 (pH 5.6), 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm CuSO4, 1 μm ZnSO4, 0.2 μm NaMoO4, 10 μm NaCl, and 0.01 μm CoCl2] supplemented with 1% Suc and 0.8% agar, and grown at 22°C under continuous light, unless otherwise stated. Seeds for light fluence rate experiments were surface sterilized and sown on Murashige and Skoog growth medium containing 0.9% agar as described by Huq and Quail (2002). After stratification at 4°C for 4 d, seeds were exposed to 1 h of white light at room temperature to induce germination before placing them in the dark at 21°C. R, FR, and B light treatments were performed in growth chambers equipped with light-emitting diodes (model E30LED; Perceival Scientific). Light fluence rates were measured using a spectroradiometer (model EPP2000; StellarNet).
Sirtinol Screen
Genetic screening for mutants resistant to sirtinol was undertaken as described in Blackwell and Zhao (2003).
Floral Organ Phenotypes
To quantify floral organ phenotypes in cul1-6 plants, floral organs in the first 10 flowers of 11 wild-type or cul1-6 individual plants were counted.
Double Mutants and Ethylene Response
The ebf1 and ebf2 mutants were crossed in cul1-6 lines. The F2 generation was genotyped using PCR primers: EBF1 F, 5′-CGGCTTTTCGCTTGAGAAATCAAGCGTT-3′ and EBF1 R, 5′- GAGACTTGATAAACGAACTTGGACGGACT-3′; MD17, 5′-GCGTGGACCGCTTGCTGCAACT-3′; EBF2 F, 5′-GTCTGGAATCTTCAGATTTAGTG-3′ and EBF2 R, 5′-TCCGTGATCTGAGACCAAAG-3′; LB1, 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′.
For triple response experiments with double mutants, seeds were plated on ATS medium with or without 10 μm ACC and exposed to light for 24 h to induce germination. Seedlings were grown in the dark at 22°C for 6 d. Hypocotyls were measured and imaged using a Nikon SMZ 1500 dissecting microscope with a Nikon digital DXM 1200 camera. Images were taken immediately upon exposing hypocotyls to light. Hypocotyl measurements were made using ImageJ freeware (version 1.32; http://rsb.info.nih.gov/ij/index.html).
Root Growth Assays
Seeds were sterilized and stratified as above, plated onto ATS plates supplemented with 1% Suc, and grown vertically in continuous light at 22°C for 5 d. Seedlings with approximately the same size root length were transferred to plates containing ATS or ATS supplemented with hormone. The length of the root at the time of transfer was indicated by a mark on the plate. After 4 d, measurements were taken of the new root growth. Hormone stock solutions were prepared in ethanol (2,4-D or JA) or dimethyl sulfoxide (BA and ACC). Subsequent dilutions were made in sterile, distilled water. For lateral root measurements, 5-d-old seedlings were transferred to medium with or without 0.1 μm 2,4-D for an additional 5 d. Lateral roots longer than 1 mm were counted with the aid of a dissecting microscope.
For growth rate assay, seedlings were germinated on ATS and grown for 4 d. Four-day-old seedlings were then transferred to a fresh ATS plate and the root length measured. A mark was made on the plate to indicate root length at time zero. Measurements were taken every 24 h for 4 d. The total root length was calculated at each interval and graphed over time.
GUS Histochemical Staining
BA3-GUS and HS-AXR3NT-GUS transgenic lines have been described in Gray et al. (2001). BA3-GUS and HS-AXR3NT-GUS were crossed into cul1-6 lines. F2s were screened for auxin resistance and stained for GUS expression and propagated by selfing. F3 lines were screened again for GUS expression.
Six-day-old cul1-6 seedlings containing one or more copies of HS-AXR3-GUS were exposed to heat shock at 37°C for 2 h in ATS solution and then transferred to 25°C for 20 min. Seedlings were washed in 100 mm Na2HPO4, pH 7, for 20 min at 25°C. After incubation in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid overnight at 37°C, tissues were destained in ethanol and mounted in 50% glycerol for imaging.
Five-day-old cul1-6 seedlings with one or more copies of BA3-GUS were incubated in ATS liquid medium containing 20 μm 2,4-D for 2 h at 25°C. Seedlings were washed in 100 mm Na2HPO4, pH 7, for 20 min at 25°C, then vacuum infiltrated with β-glucuronic acid solution for 10 min before 16-h incubation at 37°C in the dark. Tissues were destained in ethanol and mounted in 50% glycerol/phosphate buffer solution for imaging.
Negative Gravitropism Experiments
Seedlings were grown on Murashige and Skoog plates without Suc in 20 μmol m−2 s−1 R or 0.32 μmol m−2 s−1 FR or dark for 4 d. The number of negative gravitropic hypocotyls was counted and expressed as a percentage of the total number of seedlings. Hypocotyls were counted as agravitropic if they were lying completely flat on the agar surface as described (Oh et al., 2004; Shen et al., 2005).
phyA Western Blot
Four-day-old wild-type and cul1-6 seedlings were exposed to 20 μmol m−2 s−1 R light for up to 4 h. After grinding tissue in liquid nitrogen, total protein was extracted in buffer (750 μL g−1 tissue; 100 mm MOPS, pH 7.6, 50% ethylene glycol, 5 mm Na4 EDTA, 14.3 mm β-mercaptoethanol) supplemented with protease inhibitors (Complete Mini; Roche Diagnostics), 10 mm iodoacetamide, and 1 mm phenylmethylsulfonyl fluoride. Samples were centrifuged at 4°C, 13,000g for 20 min. A 5-μL aliquot of the supernatant was removed for Bradford protein quantification. The remaining supernatant was mixed with 6× Laemmli buffer, boiled for 3 min, and 20 μg of each sample were loaded onto an 8% PAGE gel. A western blot was probed with polyclonal antibody to phyA in Tris-buffered saline + 0.1% Tween followed by horseradish peroxidase-conjugated goat anti-rabbit secondary antibody and visualized by chemiluminescence (SuperSignal West Pico chemiluminescent substrate; Pierce Biotechnology).
Native Protein Extraction and Immunoprecipitation
Proteins were extracted from plant tissue in a glass homogenizer using 1 mL native extraction buffer (100 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20, protease inhibitor cocktail; Complete Mini; Roche Diagnostics) and 1 mm phenylmethylsulfonyl fluoride per 250 mg tissue. After 10 min on ice, samples were centrifuged at 16,000g for 10 min. Up to 30 μg protein were used for western-blot analysis. Up to 3 mg protein were used for immunoprecipitation (IP). IP was performed as described in Hellmann et al. (2003). Protein extraction and IP protocol for the CUL1-CAND1 co-IP were performed as described in Quint et al. (2005).
Crude Protein Extractions for RUB Modification Analysis
Crude protein extracts were performed by grinding midsized rosette leaves (or other tissue as indicated in text) in 100 μL extraction buffer (125 mm Tris HCl, pH 8.8, 1% SDS, 10% glycerol, and 50 mm Na2S2O3). Samples were spun for 10 min and the supernatant was mixed with 4× loading buffer (200 mm Tris, pH 6.8, 8% SDS, 0.4% bromphenol blue, 40% glycerol), boiled for 5 min, and loaded onto a 10% PAGE gel. Western blots were probed with antibody to CUL1 as described in Hellmann et al. (2003).
Acknowledgments
We thank Joseph Ecker of the Salk Institute for the ebf1 and ebf2 mutant seeds.
This work was supported by the National Science Foundation (grant nos. MCB–0519970 [to M.E.] and IBN–0418653 [to E.H.]), the National Institutes of Health (grant nos. GM43644 [to M.E.], GM067203 [to W.M.G.], and GM68631 [to Y.Z.]), and the University of Texas (setup fund to E.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mark Estelle (maestell@indiana.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot (Lond) 97 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell HE, Zhao Y (2003) Chemical genetic approaches to plant biology. Plant Physiol 133 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buche C, Poppe C, Schafer E, Kretsch T (2000) eid1: a new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12 547–558 [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2004) AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol 135 1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM (2004) Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, Gatz C, Vierstra RD (1999) Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J 17 155–167 [DOI] [PubMed] [Google Scholar]
- Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y (2005) Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci USA 102 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF (AtSKP2) pathway in response to light. Plant Cell 14 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32 457–466 [DOI] [PubMed] [Google Scholar]
- Dieterle M, Zhou YC, Schafer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev 15 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14 2296–2301 [DOI] [PubMed] [Google Scholar]
- Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC (2003) The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci USA 100 8571–8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW (2004) Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119 517–528 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14 2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677 [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kay SA (2003) The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol 13 2091–2096 [DOI] [PubMed] [Google Scholar]
- Hartig K, Beck E (2006) Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol (Stuttg) 8 389–396 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M (2003) Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J 22 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Klenz JE, Haughn GW (2006) UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223 769–778 [DOI] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127 23–32 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al (2001) NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J 20 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43 1073–1085 [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7 95–104 [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR (1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7 193–204 [DOI] [PubMed] [Google Scholar]
- Li J, Dai X, Zhao Y (2006) A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol 140 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10 1511–1518 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Chory J (2005) A new FronTIR in targeted protein degradation and plant development. Cell 121 970–972 [DOI] [PubMed] [Google Scholar]
- Ni W, Xie D, Hobbie L, Feng B, Zhao D, Akkara J, Ma H (2004) Regulation of flower development in Arabidopsis by SCF complexes. Plant Physiol 134 1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka F, Saeki M, Katayama S, Aida N, Toh EA, Kominami K, Toda T, Suzuki T, Chiba T, Tanaka K, et al (2000) Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J 19 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI (2003) Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun 303 1209–1216 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466 [DOI] [PubMed] [Google Scholar]
- Perry J, Dai X, Zhao Y (2005) A mutation in the anticodon of a single tRNAala is sufficient to confer auxin resistance in Arabidopsis. Plant Physiol 139 1284–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689 [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J 43 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indole acetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Ecker JR (1999) Regulation of differential growth in the apical hook of Arabidopsis. Development 126 3661–3668 [DOI] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al (2000) Nedd8 modification of cul-1 activates SCF(β(TrCP))-dependent ubiquitination of IκBα. Mol Cell Biol 20 2326–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34 753–767 [DOI] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 424 995–999 [DOI] [PubMed] [Google Scholar]
- Shanklin J, Jabben M, Vierstra RD (1987) Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci USA 84 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell 13 1916–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans EL, Harper RM, Motchoulski AV, Liscum E (1998) NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol 118 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55 197–223 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45 1028–1036 [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M (1995) The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J 8 561–569 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, Van Der Straeten D (2004) Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J 37 505–516 [DOI] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 15 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Kollmer I, Bartrina I, Holst K, Schmulling T (2006) New insights into the biology of cytokinin degradation. Plant Biol (Stuttg) 8 371–381 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21 553–562 [DOI] [PubMed] [Google Scholar]
- Wu JT, Lin HC, Hu YC, Chien CT (2005) Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol 7 1014–1020 [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Dreher KA, Edwards SR, Callis J (2003) Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J 35 285–294 [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Yang M, Solava J, Ma H (1999) The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet 25 209–223 [DOI] [PubMed] [Google Scholar]
- Zhao D, Yu Q, Chen M, Ma H (2001) The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128 2735–2746 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H (2002. a) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 10 1519–1526 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al (2002. b) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416 703–709 [DOI] [PubMed] [Google Scholar]