Abstract
This study describes the identification of over 150 target proteins of the five 14-3-3 isoforms in 7-d-old barley (Hordeum vulgare) cv Himalaya seedlings using yeast two-hybrid screens complemented with 14-3-3 protein affinity purification and tandem mass spectrometry. Independent experiments for a subset of genes confirmed the yeast two-hybrid interactions, demonstrating a low false positive identification rate. These combined approaches resulted in the identification of more than 150 putative targets; 15% were previously reported to be 14-3-3 interactors, including, for example, Serpin, RF2A, WPK4 kinase, P-type proton-translocating adenosine triphosphatase, EF1A, glutamine synthetase, and invertases. The affinity purification resulted in 30 interactors, of which 44% function in metabolism, while the yeast two-hybrid screens identified 132 different proteins, with 35% of the proteins involved in signal transduction. A number of proteins have a well-described function in hormonal signaling, such as the auxin transport protein PIN1 and NPH3 and components of the brassinosteroid pathway, such as the receptor kinase BAK1 (OsPERK1) and BRI1-kinase domain-interacting protein 129. However, 14-3-3 interactions with these signal mediators have not been confirmed in the affinity purification. Confirmations of the 14-3-3 interaction with the three ABF-like transcription factors are shown using far western analysis. Also, a REPRESSION OF SHOOT GROWTH ortholog named RF2A was identified; these transcription factors play important roles in the abscisic acid and gibberellin pathways, respectively. We speculate that 14-3-3 proteins have a role in cross talk between these hormonal pathways. The specificity and complementary nature of both the affinity purification and the yeast two-hybrid approaches is discussed.
Phosphorylation-dependent protein-protein interactions play crucial roles in the execution of various biological functions (Mukherji, 2005). Most widespread among proteins that interact with phospho-Ser or phospho-Thr motifs are members of the 14-3-3 family (Van Heusden, 2005). 14-3-3 proteins are acidic regulatory proteins, first identified as highly abundant proteins in bovine brain extract (Moore and Perez, 1967). Later, it was found that 14-3-3 proteins are present in all eukaryotes (de Vetten et al., 1992; Hirsch et al., 1992; van Heusden et al., 1992). 14-3-3 proteins belong to a conserved protein family consisting of seven isoforms in human, two isoforms in yeast (Saccharomyces cerevisiae), 15 isoforms in Arabidopsis (Arabidopsis thaliana), and eight isoforms in rice (Oryza sativa; van Heusden et al., 1995; Van Hemert et al., 2001; Sehnke et al., 2002; Chen et al., 2006).
The crystal structure of the 14-3-3 dimer revealed that it forms a clamp shape structure with dimerization occurring at the N terminus. Co-crystallization of the 14-3-3 dimer, together with a 14-3-3 interacting peptide derived from the kinase c-Raf-1, showed that the clamp shape structure has two binding grooves that both can bind a peptide (Yaffe et al., 1997). Therefore, one 14-3-3 dimer can bind two target proteins at the same time or can bind one target protein at two specific sites (Braselmann and McCormick, 1995). It was shown that the 14-3-3 binding sites contain a conserved amino acid composition that belongs to the mode I (R/K-S-X-pS/T-X-P), mode II (R/K-X-Y/F-X-pS/T-X-P), or mode III [pS/pT(X1−2)-COOH] type (Muslin et al., 1996; Yaffe et al., 1997; Ganguly et al., 2005). Binding to the target protein depends on the phosphorylation of the Ser or Thr residues inside the binding site in most, but not all, cases.
14-3-3 proteins are known to regulate a variety of different cellular processes, such as cell division, apoptosis, signaling, and carbon and nitrogen metabolism (Van Hemert et al., 2001; Huber et al., 2002; Schoonheim et al., 2007). Most 14-3-3 targets identified to date in plants are metabolism-related enzymes, such as Gln synthetase, nitrate reductase (NR), glyceraldehyde phosphate dehydrogenase, Suc phosphate synthase, trehalose phosphate synthase, and invertase (Moorhead et al., 1996, 1999; Huber et al., 2002). In the study of Pozuelo Rubio et al. (2004), more than 200 14-3-3 interacting proteins in human cancer cells were identified using affinity chromatography. In this study, it was pointed out that there seems to be a large functional difference in the identified 14-3-3 targets in animal cells as compared to plant cells. For example, the number of plant 14-3-3 targets functioning in signal perception (receptors), transduction (kinases), and processing (transcription factors) is small as compared to animal cells. This difference is surprising in light of the conserved nature of 14-3-3 proteins. We hypothesize that the current picture of the plant 14-3-3 interactome is biased due to the methods used so far to identify 14-3-3 target proteins. Therefore, the aim of this study was to carry out a comprehensive identification of 14-3-3 targets present in barley (Hordeum vulgare) leaf tissue using two complementary methods: a yeast two-hybrid screen and an affinity purification strategy using all five known barley 14-3-3 proteins.
Yeast two-hybrid screens have been used to identify protein-protein interactions extensively (Fields and Song, 1989; McAlister-Henn et al., 1999; Ito et al., 2001). In the field of 14-3-3 research, yeast two-hybrid assays have been widely used to confirm and analyze interactions between one 14-3-3 protein and a specific target (Bornke, 2005). Large-scale yeast two-hybrid screens of cDNA libraries using 14-3-3 as bait have been carried out as well, although the number of interacting proteins reported was rather low (Zhang et al., 1997; Yan et al., 2002; Bornke, 2005; Mackie and Aitken, 2005). An important advantage of the yeast two-hybrid screen is that large amounts of cDNA clones can be checked with relative ease in a short time. A second, and maybe even more important, advantage is that with this method, low-abundance transcripts isolated from plant tissue stand a good chance to be coexpressed with the 14-3-3-GAL4 binding domain, and, therefore, its protein product can be identified as 14-3-3 targets. This is because cDNA clones are under the control of the constitutive ADH1 promoter and are therefore highly expressed in the yeast. This exact same feature of the yeast two-hybrid method can also be interpreted as a disadvantage, namely, the identification of false positives. The high concentration of proteins present in the yeast cells might result in interactions that will not occur in the plant cell. Nevertheless, the yeast two-hybrid method is a good tool to study the putative interaction between two proteins, which subsequently have to be studied in more detail in vivo by, for example, performing far-western or FRET-FLIM analysis.
The second method to study protein-protein interactions, also extensively used in the field of 14-3-3 biology, is affinity chromatography with 14-3-3 as bait, followed by identification using mass spectrometry (MS; Moorhead et al., 1999; Pozuelo Rubio et al., 2003, 2004; Alexander and Morris, 2006). Due to the dynamic nature of the 14-3-3/target interaction, it is possible to selectively compete off target proteins with a peptide such as the so-called R18 peptide mimicking a phosphorylated mode I or mode II motif (Moorhead et al., 1999; Wang et al., 1999). In this way, false positives are to a large extent eliminated. Moreover, other than in a yeast two-hybrid screen, affinity chromatography can purify protein complexes when a 14-3-3 target protein is part of that complex. This can provide more comprehensive insight into protein networks involving 14-3-3 proteins but has the disadvantage that it is not clear whether the protein binds indirectly or directly to the 14-3-3 proteins.
In this study, we show that the yeast two-hybrid screen is a good tool to identify putative 14-3-3 targets. We identified 132 proteins that interact with at least one of the five barley 14-3-3 isoforms. The affinity chromatography approach yielded 30 14-3-3 target proteins with the majority having a function in primary metabolism, possibly reflecting a bias of this method to more abundant proteins. Most of the proteins identified in the two-hybrid screen are signal mediators, providing evidence that plant 14-3-3 proteins not only play an important role in regulation of the Calvin cycle, glycolysis, and nitrogen metabolism, but also are important intermediates in signaling cascades. Combining published targets and novel targets identified in this study, an interaction map for the plant 14-3-3 proteins is emerging.
RESULTS
Novel Interaction Partners for Barley 14-3-3 Proteins Identified in a Yeast Two-Hybrid Screen
To identify proteins that interact with each of the five known barley 14-3-3 isoforms (A–E), we took advantage of the sensitive yeast two-hybrid protein-protein interaction assay to screen a barley leaf cDNA library (Robertson, 2004). Clearly, plant 14-3-3 proteins are functional and properly folded in yeast because they are able to complement the lethal yeast double knockout mutants for BMH1 and BMH2 (van Heusden et al., 1995). Because 14-3-3 proteins can form homo- and heterodimers, we first addressed the question whether positive clones might show up through dimerization of one 14-3-3 in the activation domain (14-3-3-AD) and another (or the same) 14-3-3 in the binding domain (14-3-3-BD). A yeast two-hybrid assay between the 14-3-3 isoforms showed weak homodimerization of 14-3-3B, whereas the other 14-3-3 isoforms showed no homo- or heterodimerization (data not shown). These data show that dimerization of the 14-3-3 isoforms would not overwhelm us with the identification of a majority of 14-3-3-AD fusions. Next, we tested whether auto-activation of one of the reporter genes might occur. When the 14-3-3 proteins were fused to the GAL4 binding domain (14-3-3-BD) and double yeast transformants of the 14-3-3-BD versus empty-AD were tested on the activation of the three reporter genes viz. HIS3, ADE2, and LacZ, indeed auto-activation of the HIS3 reporter gene was observed; therefore, this reporter was not used in the two-hybrid screen. In contrast, no auto-activation of the other two reporter genes was measured.
Thus, for the two-hybrid screens, a total of 1.7 × 106 double transformants were screened for activation of the reporter genes ADE and LacZ. Each 14-3-3 isoform was used in a separate screen, and, in these five screens, a total of 132 unique proteins were identified as positive clones for both reporter genes, ADE and LacZ, and thus putative new 14-3-3 target proteins (Table I). All positive clones (132) were checked for the presence of an “in frame” cDNA-AD fusion. Sizes of the clones varied from approximately 600 bp to 1,700 bp. In general, clones lack an N-terminal part of the coding region, although for some clones, for example, for the membrane bound P-type proton-translocating adenosine triphosphatase (H+-ATPase), just a small part of the protein was encoded by the identified clone, namely, the C-terminal 101 amino acids. The clones were then retransformed into yeast containing the 14-3-3 bait to confirm their interaction on selective minimal medium plates without the amino acids Leu, Trp, His, and Ala (SD minus LWHA) in combination with the presence of β-galactosidase activity (Fig. 1).
Table I.
Identified cDNA clones from the 14-3-3 yeast two-hybrid screen
The clones that showed activation of the reporter genes were selected and DNA was isolated. Subsequent sequences were BLAST searched against the TIGR barley expressed sequence tag (TIGR 9.0) database (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=barley).
| Names of Homologous Proteins | TC No.a | Spot No. |
|---|---|---|
| Carbohydrate metabolism | ||
| Rubisco small subunit (wheat) | 131295 | 3 |
| Neutral invertase (Arabidopsis) | 137088 | 43 |
| Cytosolic 6-phosphogluconate dehydrogenase (rice) | 146849 | 84 |
| Dihydrolipoamide S-acetyltransferase (Arabidopsis) | 148282 | 107 |
| Oxidative stress | ||
| Manganese superoxide dismutase (wheat) | 138696 | 5 |
| GCN5-related N-acetyltransferase (rice) | 139926 | 10 |
| Glutathione S-transferase (rice) | 151451 | 70 |
| Cytosolic glutathione reductase (wheat) | 131783 | 132 |
| Signaling | ||
| BRI1-KD interacting protein 129 (rice) | 132082 | 12 |
| Hv14-3-3B (barley) | 132090 | 15 |
| Putative bZIP transcription factor ABI5 (Arabidopsis) | 144090 | 13 |
| ABA response element binding factor (wheat) | 141259 | 20 |
| ABA-responsive element binding factor (Arabidopsis) | 134245 | 49 |
| Phytoene desaturase (barley) | 139909 | 26 |
| Mybst1 (rice) | 142444 | 21 |
| Serpin (barley) | 151425 | 22 |
| WPK4 protein kinase (wheat) | CA019527* | 28 |
| Vacuolar proton-inorganic pyrophosphatase (barley) | 131805 | 11 |
| Adr11 (auxin down-regulated gene 11; soybean) | 147470 | 27 |
| Putative stearoyl-acyl-carrier protein desaturase (rice) | 139480 | 76 |
| Rf2a (rice) | 152053 | 29 |
| Mybs3 (rice) | 140561 | 111 |
| GA-stimulated transcript 1-like protein (rice) | 148126 | 97 |
| RRJ1 (jasmonic acid responsive; rice) | 140455 | 82 |
| WERBP-1 protein (MYB-like DNA-binding domain; tobacco) | 140267 | 118 |
| Adenine phosphoribosyltransferase (barley) | 139680 | 62 |
| Orf2 (Sh1orf2) | BQ463442* | 32 |
| Helix-loop-helix protein (rice) | 148621 | 39 |
| Xyloglucan endotransglycosylase (barley) | 139640 | 46 |
| Putative NPH3 protein (rice) | 136796 | 48 |
| GT-1 (Arabidopsis) | CB883300* | 60 |
| Katanin (rice) | 140752 | 59 |
| Jasmonate-induced mRNA (barley) | 131671 | 66 |
| Phospholipase DΔ isoform 1b-like protein (rice) | 143006 | 116 |
| PIR (GTPase activating-like protein; Arabidopsis) | 140036 | 38 |
| GTP-binding protein (GTP1; rice) | 146421 | 37 |
| RING-H2 zinc protein | BQ460881* | 67 |
| Putative nonphototropic hypocotyl 3 (NPH3) | BE421015* | 65 |
| PHD finger transcription factor-like (rice) | 135529 | 51 |
| Putative Golgi-localized protein (rice) | BU993045* | 52 |
| PHD finger transcription factor-like | XM476784* | 75 |
| Glycosyl hydrolase family (rice) | 146693 | 36 |
| Mpv17 transgene-like (Arabidopsis) | 140218 | 77 |
| Oswrky47 (rice) | 152887 | 78 |
| Putative lipase class 3 family (rice) | 152325 | 24 |
| Zeta1-COP (rice) | 134145 | 81 |
| Similar to adenylate cyclase (trypanosoma) | 140534 | 71 |
| Similar to putative receptor protein kinase PERK1 (rice) | BE519502* | 72 |
| Putative AP2/EREBP domain transcription factor (Arabidopsis) | 145625 | 44 |
| Probable auxin transport protein (PIN1; Arabidopsis) | BG309386* | 93 |
| Putative guanylate kinase (Arabidopsis) | 142364 | 94 |
| Guanine nucleotide-binding protein β-subunit-like protein (rice) | 130838 | 105 |
| Nph3 (barley) | BF617760* | 127 |
| 12-Oxophytodienoate reductase 3 (rice) | 140942 | 133 |
| Putative P23 co-chaperone (rice) | 139577 | 56 |
| Protein synthesis/DNA- and RNA-binding proteins | ||
| 40S ribosomal protein S30-like (rice) | 131072 | 14 |
| Putative 60S ribosomal protein L31 (rice) | 130806 | 55 |
| Eukaryotic translation initiation Factor 6 (Arabidopsis) | 139393 | 89 |
| 9S ribosomal protein | BU981956* | 99 |
| Nonribosomal peptide synthetase modules and related proteins-like (rice) | 141489 | 103 |
| Elongation factor 1α (wheat) | 146580 | 123 |
| Rnase S-like protein (barley) | 140096 | 23 |
| Homolog to histone H3.2 protein (human) | 131253 | 61 |
| RNA-binding protein | BI960230* | 115 |
| Chloroplast proteins | ||
| Inositol phosphatase-like protein (rice) | 146317 | 7 |
| PSI reaction center subunit XI, chloroplast precursor (barley) | 146765 | 19 |
| Chlorophyll a/b-binding protein (barley) | 138667 | 33 |
| PRF light-harvesting (barley) | 139104 | 31 |
| Thylakoid lumenal 35.8-kD protein, chloroplast precursor (Arabidopsis) | 141769 | 58 |
| Chlorophyll a/b-binding protein of LHCII type III (barley) | 146726 | 68 |
| Chlorophyll a/b-binding protein (Hvlhca1; barley) | 138676 | 108 |
| Chlorophyll a/b-binding protein (Lhcb1-2; potato) | 146500 | 219 |
| Putative PSII core complex proteins Psby, chloroplast (rice) | 130812 | 86 |
| Chlorophyll a/b-binding protein (wheat) | 146521 | 25 |
| Tic62 protein | BF267751* | 83 |
| Putative Crp1 protein (rice) | 148944 | 119 |
| Signal recognition particle 54-kD subunit (barley) | 132186 | 126 |
| Ferredoxin, chloroplast precursors (wheat) | 146610 | 17 |
| Ferredoxin-thioredoxin reductase subunit A (maize) | 134446 | 63 |
| Stress-related proteins | ||
| Putative aldehyde dehydrogenase (rice) | BI953625* | 50 |
| Fatty acid hydroperoxide lyase (barley) | 141847 | 100 |
| Chitin-binding lectin 1 precursor (Chlamydomonas) | 137023 | 95 |
| Carboxymethylenebutenolidase-like protein (Arabidopsis) | 139656 | 64 |
| Cell division and apoptosis | ||
| Pelota (rice) | 134531 | 104 |
| Cytokinesis 2 protein (yeast) | 149830 | 47 |
| P14arf protein | CA018141* | 114 |
| Annexin (rice) | 134070 | 131 |
| Proteasome-related proteins | ||
| 26S proteasome regulatory particle non-ATPase subunit-8 (rice) | 147090 | 85 |
| F-box family protein-like (rice) | 145485 | 45 |
| Ubiquitin-conjugating enzyme E2-17 kD (wheat) | 139190 | 110 |
| 26S protease regulatory subunit 7 (rice) | 131810 | 2 |
| Proteolysis | ||
| Degp2 protease (Arabidopsis) | 149647 | 124 |
| ATP-dependent Clp protease (rice) | 139897 | 34 |
| ATPase | ||
| P-type H+-ATPase (wheat) | 139045 | 54 |
| ATPase-like protein (sorghum) | 150845 | 18 |
| Cell wall-associated proteins | ||
| Putative protein (rice) | 132497 | 16 |
| Extensin (Volvox carteri) | BG365257* | 79 |
| Pherophorin-Dz1 protein | BI948976* | 74 |
| Putative β-1,3-glucanase (rice) | 148136 | 106 |
| 3-Glucanase | BF625748* | 102 |
| β-1,3-Glucanase (rice) | 152166 | 69 |
| Putative extensin (rice) | 135984 | 91 |
| Vegetative cell wall protein Gp1 precursor | AV834747* | 109 |
| Extensin-like protein precursor (maize) | 141562 | 117 |
| Hydroxy-Pro-rich glycoprotein DZ-HRGP precursor (rice) | 148615 | 120 |
| Storage proteins | ||
| Putative bark storage protein (rice) | 140155 | 96 |
| Unknowns | ||
| Unknown protein | BU997476* | 6 |
| Unknown protein | CA031470* | 8 |
| Similar to GP|20160854 P0677H08.8 | BQ463267* | 9 |
| Putative protein | 139132 | 35 |
| Putative protein | 141575 | 41 |
| Putative protein | 148192 | 30 |
| Putative protein | 139730 | 128 |
| Putative protein | 134288 | 40 |
| Unknown protein | 151201 | 53 |
| Hypothetical protein | 151986 | 57 |
| Mucin-like protein (Arabidopsis) | 151069 | 73 |
| No name | 148644 | 101 |
| Hypothetical protein P0461A06.20 | 146050 | 87 |
| No name | BG300785* | 121 |
| No name | 146647 | 90 |
| Unknown protein | 134959 | 92 |
| Unknown protein | 134162 | 98 |
| No name | AU090225* | 113 |
| No name | CA017615* | 80 |
| Osjnba0086b14.2 protein (DUF341 domain) | 138807 | 112 |
| No name | BF260149* | 4 |
| No name | BU999220* | 88 |
| Integral membrane protein (unidentified) | BQ460992* | 122 |
| Similar to Arabidopsis chromosome II BAC T3F17; unknown protein | 146432 | 125 |
| Putative protein | CD663282* | 42 |
| Putative protein | 136440 | 130 |
Asterisk denotes NCBI accession number.
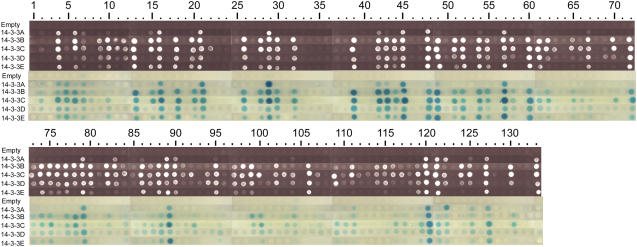
Figure 1.
Yeast two-hybrid assays between five barley 14-3-3 isoforms and the 132 identified 14-3-3 interactors. All the 132 unique 14-3-3 interactors identified in the yeast two-hybrid screens were retransformed into yeast carrying one of the barley 14-3-3 isoforms and are numbered from spot 2 to 133. Interaction is confirmed by growing the double transformants on selective media (top; SD minus LWHA) or performing β-galactosidase assay (bottom). As control, empty-BD constructs were used against the 14-3-3 interactors (top lane) and empty-AD against the 14-3-3-BD (spot 1).
Isoform specificity in 14-3-3/target interaction is an important issue in 14-3-3 biology (Bornke, 2005; Sinnige et al., 2005), and, therefore, we tried to evaluate differences in the strength of interaction between each putative target and each individual 14-3-3 isoform. To create more stringent conditions, diluted cultures were spotted on selective plates (SD minus LWHA) and grown for only 1 d. As a consequence, some weaker interacting clones did not grow at all under these conditions with any of the 14-3-3 proteins and did not turn blue in the β-galactosidase assay (Fig. 1). However, it should be noted that these clones did grow with at least one of the five 14-3-3 isoforms when undiluted cultures were spotted and turned blue when longer incubation times were used (data not shown). Therefore, all 132 clones have to be interpreted as putative 14-3-3 targets even though there is no or little growth in Figure 1. The first column in Figure 1 shows that none of the 14-3-3 proteins are auto-activators of the used reporter genes because double transformants of 14-3-3-BD and empty-AD (spot 1) show no growth and no blue color on β-galactosidase assay. Vice versa, none of the interacting proteins showed auto-activation when transformed with an empty-BD vector (Fig. 1; first row). Importantly, clear isoform-specific interactions were observed; some targets showed high affinity for all five 14-3-3 proteins (Fig. 1; spots 21, 29, 57, 79, 89, and 120), whereas others interacted with only one or two isoforms (Fig. 1; spots 22, 63, 66, 84, 92, 100, and 117).
Identification of Isolated Clones and Validation
About 10% of the identified genes represent previously identified and characterized 14-3-3 targets. The presence of these known 14-3-3 targets in our screen can be considered as positive control. One of the best characterized 14-3-3 targets was identified among these positive controls, namely, the P-type H+-ATPase (Fig. 1; spot 54). The cDNA that was identified consists of the soluble C-terminal cytosolic auto-inhibitory domain; this part is not membrane bound, thus explaining its presence in the yeast two-hybrid screen. The cDNA encodes for the last C-terminal 101 amino acids, including the 14-3-3 binding site YTV-COOH.
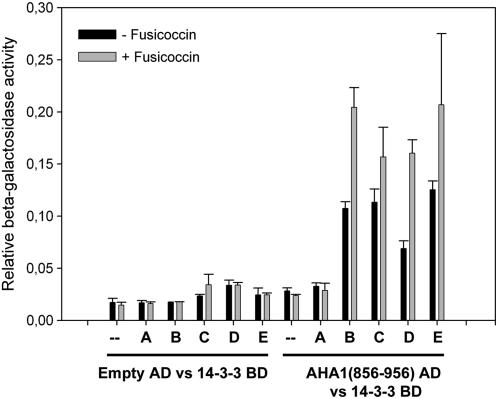
It is well known that the phytotoxin fusicoccin (FC) stabilizes the complex formed by 14-3-3 proteins and P-type H+-ATPase and increases the affinity between these two proteins (Fuglsang et al., 1999). We used the 14-3-3/ATPase interaction to demonstrate that our two-hybrid assay is capable of detecting small differences in strength of interaction between 14-3-3 and a target. So, addition of FC (10 μm) to yeast cells containing the 14-3-3 proteins as bait and the C terminus of the P-type H+-ATPase as prey should enhance the β-galactosidase activity. Figure 2 shows that FC addition indeed results in a 1.5- to 2-fold induction of the LacZ reporter gene for the interaction between the 14-3-3 proteins and the P-type H+-ATPase. This result fits with the literature and suggests that the interaction measured with the yeast two-hybrid correlates with the interactions that occur in planta. Moreover, the yeast two-hybrid principle is potentially a good method to screen for novel FC receptors. However, some caution should be taken for the membrane-bound targets identified in the yeast two-hybrid screens. Puzzlingly, for example, the PIN1 clone identified in the yeast two-hybrid screen lacks the first N-terminal 200 amino acids that contain the first five transmembrane helices, but it does contain the five transmembrane helices at the c-terminus. Another example of a membrane-bound 14-3-3 interacting clone is the pyrophosphatase (TC131805) that contains 14 transmembrane helices equally divided over the protein. These proteins will theoretically be inserted into the membrane and will not be able to activate the nuclear-located reporter genes. In our hands, these proteins do activate the reporter genes and thus are located in the nucleus. We hypothesize that these proteins are posttranslationally modified and cleaved in such a way that the GAL4 activation domain together with the cytosolic part of the protein is imported into the nucleus, where it activates the reporter genes.
Figure 2.
14-3-3 interactions with the P-type H+-ATPase is induced by FC. β-Galactosidase assays of double transformants of the 14-3-3 isoforms together with the C-terminal tail of the P-type H+-ATPase show that FC enhances β-galactosidase activity of double transformants. FC (10 μm) treatments were given for 24 h, and, subsequently, β-galactosidase assays were performed (n = 3 ± sd). β-Galactosidase activities are normalized to the OD600. FC treatment did not have an effect on yeast growth.
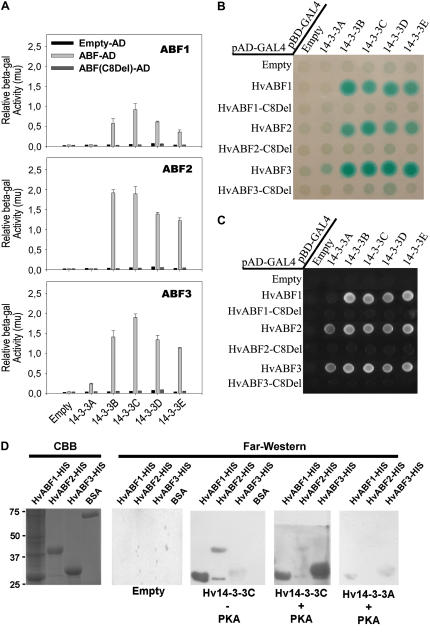
It is beyond the scope of this work to validate all 132 putative 14-3-3 targets, and, therefore, we focused on three members of the AREB/ABF/ABI5 family (HvABF1, HvABF2, and HvABF3) that we isolated in this screen (Fig. 1; spots 13, 20, and 49). Most members of the ABF/AREB/ABI5 protein family contain a canonical mode II 14-3-3 interaction motif (RRTLT350GPW-COOH) that is also conserved in the related HvABI5 protein. Mutation of Thr-350 in the HvABI5 protein showed that this residue is essential for 14-3-3 interaction (Schoonheim et al., 2007). Here, we show that deletion of the conserved C-terminal putative 14-3-3 binding motif in the ABF transcription factors (ABF-C8Del) disrupts the interaction with the 14-3-3 proteins in a yeast two-hybrid assay (Fig. 3, A and B). Moreover, we confirmed the interaction between the ABF proteins and 14-3-3C by performing far-western analysis using recombinant ABF proteins on blot and recombinant biotinylated Hv14-3-3A and Hv14-3-3C in the overlay buffer (Fig. 3D). Equal amounts of bovine serum albumin were used to show that the 14-3-3/ABF interactions on blot are not due to nonspecific protein-protein interactions. Moreover, phosphorylation of the recombinant ABFs by protein kinase A (PKA) shows that the interaction between Hv14-3-3C and HvABF3 is greatly enhanced (Fig. 3D). Puzzlingly, the interaction between ABF1 and 14-3-3 is not affected by the PKA treatment, whereas the interaction between 14-3-3C and HvABF2 is reduced by the PKA treatment. The interaction between the ABFs and Hv14-3-3A shows that the yeast two-hybrid data correlate with the far-western analysis, namely, 14-3-3C interacts strongly with the ABFs in contrast to 14-3-3A, which only weakly interacts with the ABFs. Finally, 10 of the 132 identified proteins were also identified in the affinity chromatography experiment, as shown below.
Figure 3.
Three ABF transcription factors interact with 14-3-3 proteins. A, Quantitative β-galactosidase activity of yeast cells carrying the 14-3-3 proteins as bait and the ABF transcription factors as prey. Eight amino acid deletions at the C terminus (C8Del) were tested for their interaction with the 14-3-3 proteins. B, Semiquantitative β-galactosidase activity assay of double-transformed yeast cells carrying 14-3-3 proteins as bait and the ABF transcription factors (wild type and C8Del) as prey. C, Qualitative assays of the 14-3-3 interactions with the ABF transcription factors (wild type and C8Del). D, Interaction between recombinant protein of the transcription factors HvABF1, HvABF2, and HvABF3 and recombinant 14-3-3C is shown by performing far-western analysis. Glutathione S-transferase-R18 was taken along as positive control. [See online article for color version of this figure.]
14-3-3 Proteins Interact with a Variety of Proteins Involved in Signaling
As discussed in the introduction, the list of known 14-3-3 targets in plants contains far fewer proteins with a function in signal transduction than in animals. Table I and Figure 6A show that the main group of proteins identified in this yeast two-hybrid screen belongs to the class of signal mediators (35%). Interestingly, many of these signal mediators belong to pathways of hormonal signal transduction. A few examples are: (1) BRI1-kinase domain (KD)-interacting protein 129 (spot 12), a protein that can be phosphorylated by BRI1-KD brassinosteroid receptor and likely has a function in brassinosteroid signal transduction (Hirabayashi et al., 2004); (2) the bZIP transcription factor family of the ABFs (spots 13, 20, and 49), which function in the abscisic acid (ABA) pathway (Schoonheim et al., 2007); (3) the bZIP transcription factor RF2A, the barley ortholog of the tobacco REPRESSION OF SHOOT GROWTH that plays a role in the GA biosynthesis (Igarashi et al., 2001; RF2A shows strong interactions with all five barley 14-3-3 isoforms; spot 29); and (4) NPH3, a protein with a function in lateral auxin transport during phototropism (Haga et al., 2005) showing strong interaction with four out of five 14-3-3s as well (spot 48, Fig. 1).
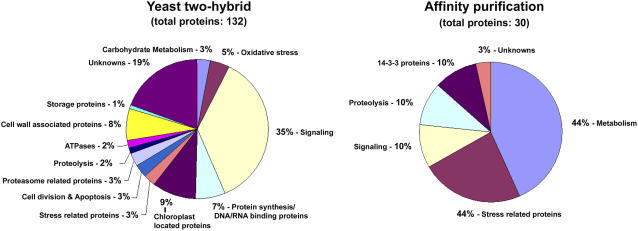
Figure 6.
Classification of the novel putative 14-3-3 interactors. The 132 identified proteins from the yeast two-hybrid screen were divided and classified into 13 classes. The 30 proteins identified from the affinity purification were classified into six classes. Signaling-associated proteins were found to be the major class of proteins identified in the yeast two-hybrid screens, whereas metabolism-related proteins were predominantly found in the 14-3-3 affinity purification. [See online article for color version of this figure.]
Hormone-responsive transcripts were found as well, including the jasmonic acid-responsive RRJ1 (TC140455) and jasmonate-induced mRNA (TC131671); for GA, the GA-stimulated transcript1 (TC148126); and for auxin, Adr11 (TC147470).
14-3-3 Affinity Purification and Tandem MS Identifies Novel 14-3-3 Targets
In addition to the yeast two-hybrid screens, we performed a large-scale affinity chromatography experiment using recombinant proteins of all five barley 14-3-3 isoforms as bait. The five barley 14-3-3 proteins (A–E) were cloned into the Invitrogen pRSET vector, resulting in N-terminal-tagged 14-3-3-hexahistidine fusions. Recombinant proteins of all five 14-3-3 isoforms were produced in BL21-DE3 cells and purified using Ni2+-NTA affinity chromatography (Fig. 4A). These hexahistidine-tagged 14-3-3 isoforms were used as bait.
Figure 4.
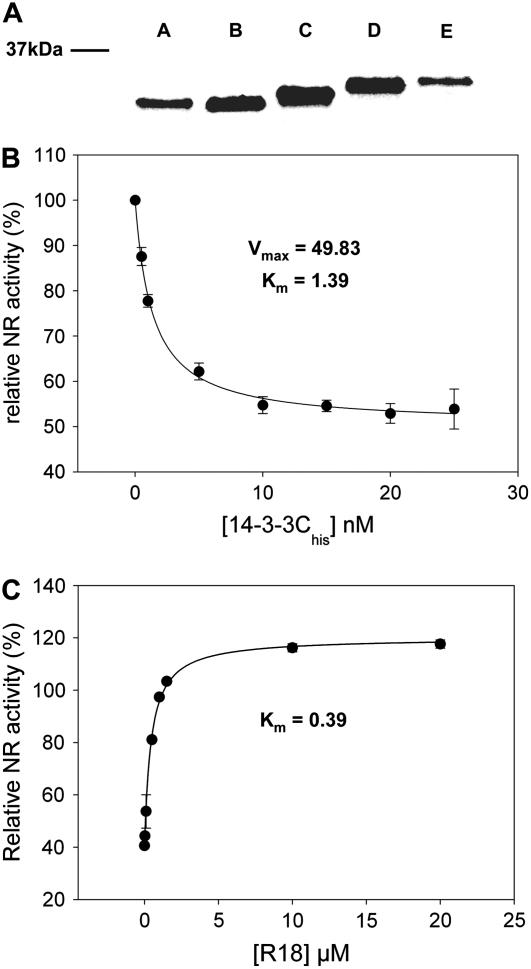
Quality control of recombinant 14-3-3 protein and R18 competing peptide. A, Recombinant His-tagged protein was prepared of all five barley 14-3-3 isoforms. Two micrograms of each isoform was run on SDS-PAGE and stained with Bio-Rad BioSafe colloidal Coomassie. B, Functionality of the recombinant Hv14-3-3C protein was tested using the inhibition of 14-3-3 proteins on NR. NR was semipurified from barley leaf extract. The affinity of Hv14-3-3C for NR was tested using different concentrations of recombinant Hv14-3-3C, ranging from 1 to 25 nm (molecular mass 14-3-3C = 29.9 kD). C, Functionality of the competing peptide R18 was tested using 10 nm of Hv14-3-3C inhibition on the NR enzyme. Concentration range of the R18 peptide was between 10 nm and 20 μm.
Our interaction analysis was based on two selective steps: specific binding of target proteins into the 14-3-3 groove and selective elution of proteins that interact with 14-3-3 through this groove with a peptide that has a high affinity for the binding groove, called R18 (Wang et al., 1999). R18 is a peptide that specifically binds to 14-3-3 proteins without isoform selectivity. Scatchard analysis of R18 binding to empty 14-3-3 showed Kd values in the range of 0.1 μm (Wang et al., 1999). Because we wanted to use the R18 peptide to compete off bound proteins, we first carried out a competition assay using the well-characterized 14-3-3 inhibition of the enzyme NR as reporter system. First, the affinity of NR for 14-3-3C proteins was determined (Fig. 4B). From this graph, we determined that 10 nm of 14-3-3C was sufficient to inhibit NR for more than 85%. This concentration of 14-3-3 was chosen to study the affinity of the R18 peptide for 14-3-3C. As shown in Figure 4C, R18 peptide abolishes the inhibition of NR by 14-3-3C proteins with a Ki of 0.39 μm. This is close to the Kd, as determined by Scatchard analysis (Wang et al., 1999), and shows that we can use R18 as a high-affinity competing peptide for specific elution of 14-3-3 target proteins.
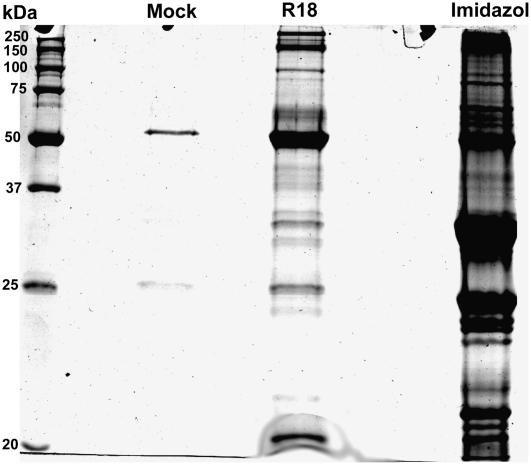
Protein extract from leaves of 7-d-old barley cv Himalaya plants (identical to the source of the yeast two-hybrid cDNA library) was prepared. A total of 500 mg of cytosolic proteins was mixed with 10 mg of recombinant His-tagged 14-3-3 in a total volume of 25 mL. After incubation at 4°C, the mixture was run through a nickel column. After rinsing extensively, a control elution was performed solely with the injection of buffer. Next, bound target proteins were eluted from the column using 0.5 mm of the R18 peptide. This relatively high R18 concentration (1,250 times Km) was chosen because the time of exposure is relatively short (flow rate 0.5 mL min−1). Higher concentrations of R18 (1 mm) did not elute more bands (data not shown). Eluted proteins were separated on SDS-PAGE gel, and the R18 elution resulted in many bands ranging from high molecular mass (250 kD) to low molecular mass (15 kD) on a colloidal Coomassie-stained gel (Fig. 5).
Figure 5.
Elutions of 14-3-3 interactors. 14-3-3 interacting proteins were specifically eluted using 0.5 mm R18 peptide. The mock elution, R18 elution, and the imidazol strip were concentrated, and 10% of the samples were separated on a 12% SDS-PAGE gel. Proteins were stained with Bio-Rad Biosafe colloidal Coomassie.
Mock and R18 eluate were run out on a one-dimensional SDS-PAGE gel, and the R18 lane was sliced into six pieces, proteins were in-gel digested with trypsin, and peptides were identified using nano liquid chromatography (LC)-electrospray ionization-MS/MS. The MS/MS data were searched against National Center for Biotechnology Information (NCBI) green plants and a recent version of the annotated rice genome (the Institute for Genomic Research [TIGR] OsGI version 4) and maize (Zea mays) unigene assembly (TIGR ZmGI version 16) using Mascot.
Identified proteins are listed in Table II, ordered according to the gel slices ranging from high to low molecular mass (gel slices 1–6). The molecular mass of proteins identified from the respective gel slices correlates well with the predicted molecular mass range (based on the protein markers) of each gel slice that they originated from. Intriguingly, 45% of the 30 eluted targets are metabolism-related proteins, and many of these are part of either glycolysis or the Calvin cycle (Table II).
Table II.
Protein identification of the R18 eluate analyzed by LC-MS/MS
Mass spectral data were searched against three plant databases. Accession numbers are presented for different databases where the superscript letters denote: A, NCBI-Plants; B, OSGI; and C, ZMGI. Predicted molecular masses correlate with the gel slices from which the proteins were extracted.
| No. | Protein Name | Accession No. | Molecular Mass | Mowse Score (Peptides Matched MS/MS) | Gel ID | Function | Reported | 14-3-3 Motif |
|---|---|---|---|---|---|---|---|---|
| kD | ||||||||
| 1 | Acetyl-CoA carboxylase | gi:2827150A | 254.956 | 110 (2) | 1 | Multifunction | No | KEGTSAP |
| 2 | Gly dehydrogenase | 9629.m04981B | 113.767 | 320 (7) | 1 | Nitrogen metabolism | No | KPSDTFP |
| 3 | Elongation factor 2, putative | 11972.m08296B | 94.015 | 57 (2) | 1 | Translation | No | RLAKSDP |
| 4 | Cytosolic heat shock protein 90 | gi:32765549A | 80.413 | 129 (2) | 1 | Chaperone | Yes (human) | KHFSV/KNDKSV |
| 5 | β-Fructofuranosidase | gi:79326306A | 65.032 | 1,138 (17) | 1 | Glycolysis | Yes | RRSNSW |
| 6 | Neutral invertase-like | gi:18395144A | 63.446 | 581 (8) | 1 | Glycolysis | Yes | KRSASWT |
| 7 | Ulp1 protease family, C-terminal catalytic domain-containing protein | 11981.m05812B | 108.167 | 46 (2) | 2 | Proteolysis | No | RDHFSV/KRSTV |
| 8 | TCP-1/cpn60 chaperonin family | 9634.m00140B | 64.081 | 349 (5) | 2 | Chaperone | No | RRALSYP |
| 9 | Catalase-1 | gi:3929924A | 56.171 | 52 (2) | 2 | Antioxidant | No | RRGSSP |
| 10 | Rubisco large subunit | gi:31087879A | 53.141 | 1,827 (32) | 2 | Calvin cycle | No | – |
| 11 | Elongation factor 1α | TC299030C | 49.137 | 76 (2) | 2 | Translation | No | KMDATTP/KMIPTKP |
| 12 | Transferase family | 9632.m05549B | 47.971 | 60 (2) | 2 | Unknown | No | – |
| 13 | Gln synthetase, catalytic domain | 9632.m05496B | 47.407 | 73 (2) | 2 | Nitrogen metabolism | Yes | KHSSV/KLWSSV/RRPASNM |
| 14 | Phosphoribulokinase | gi:21839A | 45.032 | 388 (6) | 2 | Calvin cycle | No | KLTCSYP |
| 15 | Sedoheptulose-1, 7-bisphosphatase | gi:14265A | 42.057 | 202 (4) | 3 | Calvin cycle | No | KVISV |
| 16 | Fructose-1,6-bisphosphate aldolase precursor | gi:8272480A | 41.919 | 93 (2) | 3 | Glycolysis | No | RTVVSIP/RIPPSVP |
| 17 | GAP dehydrogenase, cytosolic | gi:120668A | 33.233 | 65 (2) | 3 | Glycolysis | Yes | KVIISAP |
| 18 | Short-chain alcohol dehydrogenase | gi:7431022A | 31.644 | 118 (3) | 3 | Signaling | No | – |
| 19 | 14-3-3-like protein | gi:37903393A | 28.880 | 312 (5) | 3 | Signaling | Yes | – |
| 20 | 14-3-3 protein homolog | gi:22607A | 29.262 | 836 (17) | 3 | Signaling | Yes | |
| 21 | Peroxidase, putative | 9635.m04996B | 42.112 | 111 (2) | 4 | Antioxidant | No | RRVLSHP/KIESRP/RVLGSYP |
| 22 | Carbonic anhydrase | gi:558499A | 35.072 | 448 (9) | 4 | Calvin cycle | No | KLIRTTP |
| 23 | Putative acid phosphatase | gi:52346236A | 29.603 | 630 (8) | 4 | Signaling | No | RDWKTVP/RTFSRP |
| 24 | Brain-specific protein | gi:303859A | 29.127 | 114 (2) | 4 | Signaling | Yes | – |
| 25 | Bas1 protein | gi:861010A | 29.090 | 315 (6) | 4 | Antioxidant | No | KSSLSSP/RTLSSP |
| 26 | Thioredoxin peroxidase | gi:3328221A | 28.131 | 325 (6) | 4 | Antioxidant | No | – |
| 27 | ATP-dependent Clp protease proteolytic subunit (endopeptidase Clp) | TC299023C | 27.951 | 78 (2) | 4 | Proteolysis | No | – |
| 28 | Ascorbate peroxidase | gi:15808779A | 27.637 | 252 (5) | 4 | Antioxidant | Yes | KVLLTDP |
| 29 | Rubisco small subunit | gi:4090291A | 19.512 | 383 (7) | 5 | Calvin cycle | No | – |
| 30 | Ser carboxypeptidase S10 family protein, similar to Glc acyltransferase | TC293082C | 51.683 | 51 (2) | 6 | Proteolysis | No | RWLSGE |
The two approaches that were used in this study identified a large group of putative 14-3-3 interactors that have to be studied in more detail in future studies. From the group of proteins identified using the 14-3-3 affinity purifications, 30% of the interacting proteins were confirmed in the yeast two-hybrid screen.
DISCUSSION
Classification of Identified 14-3-3 Targets
The yeast two-hybrid screens and the affinity purification show a large difference in the representation of different protein classes (Fig. 6). More than one-third (50) of the interacting proteins identified in the yeast two-hybrid screens fall in the class of signaling proteins; 17 of these are transcription factors. In contrast, only 10% of the proteins identified in the affinity purification could be assigned a function in signaling, and none of these was a transcription factor. The 14-3-3 affinity purification method identified mostly metabolic enzymes (42%). A similar observation was made by Alexander and Morris (2006), who purified 14-3-3 interacting proteins from developing barley grains using a 14-3-3 affinity chromatography strategy. Metabolic proteins are abundant proteins and are therefore probably preferentially identified when using 14-3-3 affinity chromatography. In contrast, signaling intermediates are low-abundance proteins and are therefore less likely to be detected in the affinity purification.
Overlap between Yeast Two-Hybrid and Affinity Purification
Although many differences can be found between the yeast two-hybrid screen and the affinity purification, also a large portion of the identified proteins overlap. Thirty percent of the identified proteins from the affinity purification were also identified in the yeast two-hybrid screens, like elongation factor 1A, neutral invertase, acyl-transferase, ATP-dependent Clp protease, Rubisco small subunit, and 14-3-3 proteins themselves. In addition, several proteins identified only by affinity purification were reported before as 14-3-3 interacting proteins: ascorbate peroxidase, Gln synthetase, and glyceraldehyde phosphate dehydrogenase (Zhang et al., 1997; Moorhead et al., 1999; Cotelle et al., 2000). Likewise, many proteins previously reported in the literature as 14-3-3 interacting proteins were identified in the yeast two-hybrid screen: RF2A, Serpin, WPK4, and the P-type H+-ATPase (Ikeda et al., 2000; Igarashi et al., 2001; Alexander and Morris, 2006). This shows that these two different methods are complementary.
Functional Network of Identified 14-3-3 Interactors
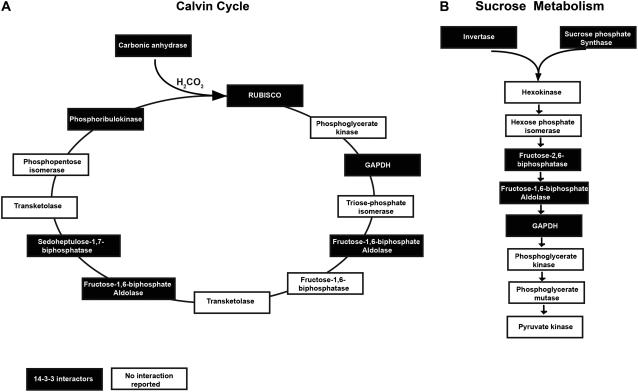
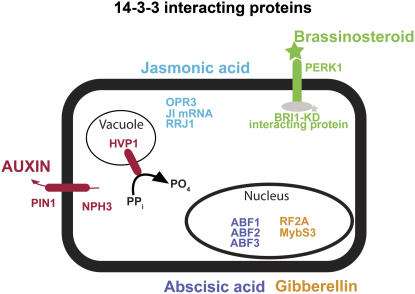
From the barley 14-3-3 interacting proteins that we have identified here, three interesting cellular processes can be highlighted. First, the Calvin cycle, according to the textbooks, consists of 10 main enzymes, six of which have been identified as published 14-3-3 targets (Fig. 7A). In addition to these six Calvin cycle enzymes, another novel 14-3-3 target was identified that does not belong to the Calvin cycle but is necessary for the Calvin cycle, viz. carbonic anhydrase. This enzyme plays an important role in the conversion of CO2 into carbonic acid (Werdan and Heldt, 1972), which is then converted into glyceraldehyde-3-P (GAP) by the Calvin cycle. The second process emerging from the 14-3-3 target analysis that is densely populated with 14-3-3 targets is situated in sugar metabolism, namely, glycolysis. As shown in Figure 7B, the two enzymes that can break down Suc into Glc and Fru are 14-3-3 targets: invertase and Suc synthase. Furthermore, previously identified proteins together with proteins identified in this study show that more 14-3-3 targets are present downstream of Suc hydrolysis, like Fru-2,6-bisphosphatase, Fru-1,6-bisphosphate aldolase, and cytosolic GAP dehydrogenase (Pozuelo Rubio et al., 2003). In line with a major function for 14-3-3 in animal cells (Darling et al., 2005), it becomes clear from the analysis of our yeast two-hybrid screen that plant 14-3-3 proteins are also important regulators of hormonal signaling intermediates (Tables I and II; Fig. 8). It is noteworthy that there are interesting relationships between these 14-3-3 protein targets. For example, PERK1 (BAK1) interacts with the BRI1 brassinosteroid receptor in a brassinosteroid- and phosphorylation-dependent manner (Wang and Chory, 2006). Interestingly, the BRI1-KD-interacting protein 129 has been shown to be phosphorylated by the BRI1 receptor (Hirabayashi et al., 2004). Also, the PIN1 protein (polar transport of auxin; Wisniewska et al., 2006) and NPH3 proteins (lateral transport of auxin; Haga et al., 2005) play a key role in auxin transport. Moreover, three members of the ABF transcription factor family, transcriptional regulators of ABA-responsive gene expression (Choi et al., 2000), interact with the 14-3-3 protein family. In addition, a large group of putative 14-3-3 interactors was identified that belongs to the cell wall-associated proteins (Table I). This indicates that 14-3-3 proteins are most likely transported out of the cell, where they regulate the function of cell wall-associated proteins as has been shown before (Voigt and Frank, 2003).
Figure 7.
Calvin cycle and glycolysis are densely populated with 14-3-3 interactors. Schemes of the Calvin cycle (A) and glycolysis (B), where 14-3-3 interactors are in black boxes and white text, show that both pathways are densely populated with 14-3-3 interactors.
Figure 8.
14-3-3 interactors that play a role in the hormonal signal transduction. A schematic representation of the cell showing the novel identified 14-3-3 interactors that play a role in plant hormone signal transduction. The five hormones together with the signal mediators that were identified in this study as putative 14-3-3 targets are depicted in different colors. [See online article for color version of this figure.]
In conclusion, our screens have revealed many established and novel putative 14-3-3 interacting proteins. The two methods employed in this study are complementary in the sense that different classes of proteins prevail in one or the other screen. In this study, a wide array of targets with a function in signal transduction has been identified in plants. Just like in animal cells, 14-3-3 proteins may form a platform for cross talk in the signal transduction pathways of the different hormones. Moreover, a picture is emerging that, like in animal cells, a saturation of 14-3-3 targets in the specific cellular processes, whether cyclic or linear in nature, is observed.
The cataloging, functional characterization, and networking of 14-3-3 targets will in the end allow the formation of a functional and dynamic map of the 14-3-3 interactome that will bring us closer to understanding cellular functioning.
MATERIALS AND METHODS
Yeast Two-Hybrid Screen
The Stratagene GAL4 two-hybrid phagemid vector pBD-GAL4 was used to prepare 14-3-3-GAL4 binding domain fusions using the BamHI and EcoRI restriction sites. The open reading frame (ORF) from the Hv14-3-3 genes was amplified using PFU DNA polymerase, and restriction sites were introduced into the primers. The constructs were checked for correct DNA sequence by performing sequence reactions. A CLONTECH MATCHMAKER two-hybrid cDNA library prepared from mRNA isolated from 5- to 7-d-old leaf tissue from barley (Hordeum vulgare) cv Himalaya (Robertson, 2004) was a kind gift of Masumi Robertson (CSIRO Plant Industry, Canberra, Australia). The improved yeast (Saccharomyces cerevisiae) strain PJ69-4A with genotype MATa, trp-901, leu2-112, ura3-200, his3-200, gal4D, gal80D, GAL-ADE2, LYS∷GAL1-HIS3, met∷GAL7-LacZ, which carries an extra reporter selecting for adenine auxotrophy (ADE2), was used for the yeast two-hybrid screens (James et al., 1996). Yeast transformations were performed using the lithium acetate protocol according to Gietz et al. (1992). Briefly, PJ69-4A yeast cells carrying the bait construct were grown up in SD minus LW broth for 16 h at 30°C. The cells were pelleted and resuspended in 1 L of YAPD (1% [w/v] yeast extract, 2% [w/v] peptone, 2% [w/v] d-Glc, and 0.01% [w/v] Ade) broth and grown for an additional 4 h. Cells were pelleted, washed with 1 volume of water, pelleted again, and resuspended in 100 mL of 0.2 m lithium acetate/Tris-EDTA. Again, cells were pelleted and resuspended in 10 mL end volume of 0.2 m lithium acetate. Aliquots of 2 mL yeast cells were divided in five 15-mL tubes and the following was added: 10 μg of cDNA, 20 μg of linearized salmon sperm DNA (Invitrogen), and 12 mL of 40% polyethylene glycol-4000/ 0.2 m lithium acetate. Tubes were mixed and incubated at 30°C for 45 min; subsequently, the tubes were transferred to 42°C for 25 min. Aliquots of 100 μL were spotted on SD minus LWHA plates and incubated at 30°C for 7 d. In total, 1.7 million colony-forming units were checked on interaction with the five barley 14-3-3 proteins. Double transformants were grown on selective plates (SD minus LWHA) for 7 d. Positive clones were streaked on plates lacking LW, one plate was used to test for LacZ assay and the other plate for storage. Clones that showed activation of both reporter genes (ADE2 and LacZ) were selected, and DNA was isolated according to Stratagene's protocol. To prepare large quantities of DNA for sequencing reactions, the DNA isolated from yeast was electroporated into Escherichia coli, and DNA mini-preps were performed. To identify the positive clones, sequence reactions were performed using the pACT2 sequencing primer (5′-TGATGAAGATACCCCACCAAACC-3′). The resulting sequences of the cDNA clones were BLAST searched against the TIGR barley database.
Preparation of Recombinant His-Tagged Proteins
The 14-3-3 genes were cloned from cDNA using PCR and T/A cloning into the Invitrogen pGEM-T easy vector. Primers were designed on the 14-3-3 genes introducing BamHI sites at both sites, and amplified 14-3-3 genes were subsequently cloned in frame into the Invitrogen pRSET-C vector. The ABF-His fusions were prepared by digestion of the pACT2-cDNA constructs isolated from the yeast two-hybrid screen using BamHI and BglII. Digested insert was ligated into BamHI-digested pRSET-C vector, resulting in an N-terminal-tagged in-frame fusion protein. Recombinant proteins were prepared by electroporation of the appropriate DNA construct into BL21 E. coli cells. Positive BL21 cells were grown up in YT medium for 16 h at 37°C, then isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mm. The culture was grown for another 8 h, and, subsequently, E. coli cells were pelleted and recombinant protein was isolated using the Amersham His-trap column (1 mL) according to the manufacturer's protocol. The quality of the recombinant proteins was checked by performing SDS-PAGE and western blotting using the Novagen T7-tag antibody. Far-western analysis was performed according to Moorhead et al. (1999).
Protein Extraction
Barley plants were grown in the greenhouse for 7 d. Leaves of 7-d-old seedlings were harvested (200 g) and ground in a blender using 500 mL of homogenization buffer (50 mm HEPES-KOH, pH 7.5, 50 mm NaF, 5 mm NaPPi, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 tablet Roche protease inhibitor cocktail per 50 mL, 1% polyvinylpolypyrrolidone). The homogenate was centrifuged for 20 min at 12,000g, and the supernatant was filtered through four layers of Miracloth. ATP was added to a final concentration of 0.5 mm, and, subsequently, proteins were fractionated by adding solid ammonium sulfate to a final concentration of 70% (w/v). Precipitated proteins were pelleted at 12,000g for 20 min and the pellet was resuspended in 10 mL buffer C (50 mm HEPES-KOH, pH 7.5, 1 μm cantharidin). To remove remaining salts, the sample was dialyzed in a dialysis tube (NMWL = 3,500 D) against 1 L of buffer C during 3 h, which was refreshed every hour (total of 3 L). After dialysis, the protein extract was cleared by centrifugation at 33,000g for 20 min, and protein concentration was determined using the Bradford method (Takahashi et al., 1999).
14-3-3 Affinity Purification
14-3-3 affinity purification was performed using a modified protocol of Moorhead et al. (1999). The protein sample, collected as described above, was diluted with buffer C to reach a protein concentration of 20 mg/mL. To 25 mL of this sample, ATP, leupeptin, and phenylmethylsulfonyl fluoride were added to reach a final concentration of 2 mm, 4 μg mL−1, and 0.5 mm, respectively, and this was incubated at 30°C for 10 min. Finally, 10 mg of His-tagged recombinant 14-3-3 proteins (2 mg of each isoform) and NaCl (final concentration 150 mm) was added, and the tube was mixed end-over-end for 1 h at 4°C. Thereafter, the sample was pumped through an Amersham His-trap column to bind the 14-3-3His proteins (and putative targets), using a flow rate of 0.5 mL min−1. The column was washed extensively (flow rate 3 mL min−1) with buffer C (50 mm HEPES-KOH, pH 7.5, 500 mm NaCl, 50 mm imidazol) until the protein concentration of the flow-through was lower than 5 μg protein mL−1. A mock elution was performed using 1 mL of buffer C without the competing peptide (using the same tubing and injection procedure as used for the specific elution and strip). Subsequently, 1 mL of buffer C containing 0.5 mm R18 peptide was injected into the column with a flow rate of 0.5 mL min−1 to specifically elute interacting proteins. The synthetic R18 peptide (PHCVPRDLSWLDLEANMCLP) was purchased from Genscript. Finally, the column was stripped by injecting buffer C plus 500 mm imidazol. Both the mock and the R18 elution were collected in fractions of 0.5 mL and concentrated using Millipore Amicon Ultra-15 tubes to a final volume of 200 μL.
MS
To identify the 14-3-3 interacting proteins, 50 μL of each of the mock, R18 elution, and the imidazol strip were run out on one-dimensional SDS-PAGE gel (12% acrylamide). Because no protein was detected in the mock elution using Coomassie Brilliant Blue R250 (note that Bio-Rad, Biosafe colloidal Coomassie was used for Fig. 5), only the gel lane from the R18 eluate was analyzed. This lane was cut in six gel slices, which were then washed, digested with trypsin, and peptides were extracted, as described by Friso et al. (2004). Peptides from the in-gel digests were resuspended in 5% formic acid and analyzed using nano LC-electrospray ionization-MS/MS using a quadrupole time-of-flight mass spectrometer (Waters) using gradients as described by Friso et al. (2004). Spectral data were extracted using Mascot distiller and searched with Mascot (Matrix Science) against NCBI nonredundant green plants: rice (Oryza sativa; OsGI version 4 from TIGR at http://www.tigr.org/), maize (Zea mays; ZmGI version 6; TIGR), and Arabidopsis (Arabidopsis thaliana; TAIR version 6 at http://www.arabidopsis.org/). Only proteins with two or more matching peptides matched each with a minimal ion score of 22 and a total protein MOWSE score higher than 50 were considered in this first identification step. The accession numbers of the four databases were then compared using directional BLAST alignments to remove redundancies and to find the best annotations.
GenBank accession numbers of the genes used in this study are as follows (in parentheses): 14-3-3A (X62388), 14-3-3B (X93170), 14-3-3C (Y14200), 14-3-3D (DQ295785), 14-3-3E (DQ295786), HvABF1 (DQ786408), HvABF2 (DQ786409), and HvABF3 (DQ786410).
Acknowledgments
We thank Masumi Robertson for the kind gift of the barley yeast two-hybrid cDNA library. Thanks are also due to Erik Souer, Walter Verweij, and Ronald Koes for their advice on the yeast two-hybrid work.
This work was supported by the INTAS Aral Sea Programme (project no. 00–1021 to A.H.d.B. and P.J.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Albertus H. de Boer (bert.de.boer@falw.vu.nl).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alexander RD, Morris PC (2006) A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics 6 1886–1896 [DOI] [PubMed] [Google Scholar]
- Bornke F (2005) The variable C-terminus of 14-3-3 proteins mediates isoform-specific interaction with sucrose-phosphate synthase in the yeast two-hybrid system. J Plant Physiol 162 161–168 [DOI] [PubMed] [Google Scholar]
- Braselmann S, McCormick F (1995) Bcr and Raf form a complex in vivo via 14-3-3 proteins. EMBO J 14 4839–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Li Q, Sun L, He Z (2006) The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13 53–63 [DOI] [PubMed] [Google Scholar]
- Choi H-i, Hong J-h, Ha J-o, Kang J-y, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275 1723–1730 [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SEM, Provan F, Milne FC, Morrice N, MacKintosh C (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J 19 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling DL, Yingling J, Wynshaw-Boris A (2005) Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol 68 281–315 [DOI] [PubMed] [Google Scholar]
- de Vetten NC, Lu G, Ferl RJ (1992) A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell 4 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O-k (1989) A novel genetic system to detect protein-protein interactions. Nature 340 245–246 [DOI] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier J-B, Rudella A, Sun Q, van Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem 274 36774–36780 [DOI] [PubMed] [Google Scholar]
- Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC (2005) Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci USA 102 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Takano M, Neumann R, Iino M (2005) The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 17 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Matsushita Y, Sato M, Oh-I R, Kasahara M, Abe H, Nyunoya H (2004) Two proton pump interactors identified from a direct phosphorylation screening of a rice cDNA library by using recombinant BRI1 receptor kinase. Plant Biotechnol 21 35–45 [Google Scholar]
- Hirsch S, Aitken A, Bertsch U, Soll J (1992) A plant homologue to mammalian brain 14-3-3 protein and protein kinase C inhibitor. FEBS Lett 296 222–224 [DOI] [PubMed] [Google Scholar]
- Huber SC, MacKintosh C, Kaiser WM (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol Biol 50 1053–1063 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Ishida S, Fukazawa J, Takahashi Y (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Koizumi N, Kusano T, Sano H (2000) Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J Biol Chem 275 31695–31700 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie S, Aitken A (2005) Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J 272 4202–4210 [DOI] [PubMed] [Google Scholar]
- McAlister-Henn L, Gibson N, Panisko E (1999) Applications of the yeast two-hybrid system. Methods 19 330–337 [DOI] [PubMed] [Google Scholar]
- Moore BW, Perez VJ (1967) Specific acidic proteins of the nervous system. In F Carlson, ed, Physiological and Biochemical Aspects of Nervous Integration. Prentice Hall, Woods Hole, MA, pp 343–359
- Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, Deiting U, Stitt M, Scarabel M, Aitken A, et al (1999) Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J 18 1–12 [DOI] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C (1996) Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr Biol 6 1104–1113 [DOI] [PubMed] [Google Scholar]
- Mukherji M (2005) Phosphoproteomics in analyzing signaling pathways. Expert Rev Proteomics 2 117–128 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84 889–897 [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, MacKintosh C (2004) 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation, and trafficking. Biochem J 379 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Peggie M, Wong BHC, Morrice N, MacKintosh C (2003) 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J 22 3514–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M (2004) Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol 136 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, Quatrano RS, De Boer AH (2007) 14-3-3 adapter proteins are intermediates in ABA signal transduction during barley seed germination. Plant J (in press) [DOI] [PubMed]
- Sehnke PC, DeLille JM, Ferl RJ (2002) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14 S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnige MP, Roobeek I, Bunney TD, Visser AJWG, Mol JNM, de Boer AH (2005) Single amino acid variation in barley 14-3-3 proteins leads to functional isoform specificity in the regulation of nitrate reductase. Plant J 44 1001–1009 [DOI] [PubMed] [Google Scholar]
- Takahashi E, Abe J-i, Gallis B, Aebersold R, Spring DJ, Krebs EG, Berk BC (1999) p90RSK is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem 274 20206–20214 [DOI] [PubMed] [Google Scholar]
- Van Hemert MJ, Steensma HY, van Heusden GP (2001) 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23 936–946 [DOI] [PubMed] [Google Scholar]
- van Heusden GP, Griffiths DJ, Ford JC, Chin-A-Woeng TF, Schrader PA, Carr AM, Steensma HY (1995) The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur J Biochem 229 45–53 [PubMed] [Google Scholar]
- Van Heusden GPH (2005) 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life 57 623–629 [DOI] [PubMed] [Google Scholar]
- van Heusden GPH, Wenzel TJ, Lagendijk EL, de Steensma HY, van den Berg JA (1992) Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett 302 145–150 [DOI] [PubMed] [Google Scholar]
- Voigt J, Frank R (2003) 14-3-3 proteins are constituents of the insoluble glycoprotein framework of the Chlamydomonas cell wall. Plant Cell 15 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BC, Yang HZ, Liu YC, Jelinek T, Zhang LX, Ruoslahti E, Fu H (1999) Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38 12499–12504 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313 1118–1122 [DOI] [PubMed] [Google Scholar]
- Werdan K, Heldt HW (1972) Accumulation of bicarbonate in intact chloroplasts following a pH gradient. Biochim Biophys Acta 283 430–441 [DOI] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC (1997) The structural basis for 14-3-3: phosphopeptide binding specificity. Cell 91 961–971 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Zhang H (2002) An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29 193–202 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang J, Nickel U, Allen RD, Goodman HM (1997) Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol Biol 34 967–971 [DOI] [PubMed] [Google Scholar]