Abstract
The Arabidopsis (Arabidopsis thaliana) gene MEKK1 encodes a mitogen-activated protein kinase kinase kinase that has been implicated in the activation of the map kinases MPK3 and MPK6 in response to the flagellin elicitor peptide flg22. In this study, analysis of plants carrying T-DNA knockout alleles indicated that MEKK1 is required for flg22-induced activation of MPK4 but not MPK3 or MPK6. Experiments performed using a kinase-impaired version of MEKK1 (K361M) showed that the kinase activity of MEKK1 may not be required for flg22-induced MPK4 activation or for other macroscopic FLS2-mediated responses. MEKK1 may play a structural role in signaling, independent of its protein kinase activity. mekk1 knockout mutants display a severe dwarf phenotype, constitutive callose deposition, and constitutive expression of pathogen response genes. This dwarf phenotype was largely rescued by introduction into mekk1 knockout plants of either the MEKK1 (K361M) construct or a nahG transgene that degrades salicylic acid. When treated with pathogenic bacteria, the K361M plants were slightly more susceptible to an avirulent strain of Pseudomonas syringae and showed a delayed hypersensitive response, suggesting a role for MEKK1 kinase activity in this aspect of plant disease resistance. Our results indicate that MEKK1 acts upstream of MPK4 as a negative regulator of pathogen response pathways, a function that may not require MEKK1's full kinase activity.
Mitogen-activated protein (MAP) kinase cascades are conserved signaling modules that are present in all eukaryotes. Activation of a MAP kinase kinase kinase (MAP3K) by an upstream signal leads to phosphorylation and activation of a MAP kinase kinase (MAP2K), which then phosphorylates and activates a MAP kinase (MAPK). The genome of Arabidopsis (Arabidopsis thaliana) encodes over 60 apparent MAP3Ks, 10 MAP2Ks, and 20 MAPKs (MAPK-Group, 2002). A current challenge in this research area is to determine which signaling pathways in the plant make use of which MAPK components. Recent reviews have summarized our current understanding of the roles of plant MAPK pathways in cell division, development, and stress responses (Tena et al., 2001; Jonak et al., 2002; Nakagami et al., 2005).
MEKK1 was one of the first MAP3Ks to be characterized in Arabidopsis; it is transcriptionally up-regulated in response to touch, cold, and salt stress (Mizoguchi et al., 1996; Covic et al., 1999). Studies using the yeast (Saccharomyces cerevisiae) two-hybrid method have indicated that MEKK1 can interact with the MAP2Ks MKK1 and MKK2, which in turn can interact with MPK4 (Ichimura et al., 1998; Mizoguchi et al., 1998). In addition, MPK4 was shown to interact directly with the N-terminal regulatory domain of MEKK1, suggesting that MEKK1 may posses a scaffolding function that helps assemble a complete MAPK cascade by binding to both a MAP2K and a MAPK (Ichimura et al., 1998). Experiments with a number of plant species have indicated the involvement of MAPK cascades in plant responses to pathogens (Ligterink et al., 1997; Asai et al., 2002; del Pozo et al., 2004; Lee et al., 2004; Menke et al., 2005).
Analysis of the function of MEKK1 in Arabidopsis signaling pathways indicated that it acts downstream of the flagellin receptor FLS2 (Asai et al., 2002). These experiments, performed using a protoplast transient expression system, led to the development of a model in which a MAPK cascade composed of MEKK1-MKK4/MKK5-MPK3/MPK6 is responsible for transmitting the signal initiated when a plant recognizes flagellin. Flg22 is a 22-amino acid peptide sequence derived from the N-terminal portion of Pseudomonas aeruginosa flagellin that stimulates the FLS2 receptor and is sufficient to elicit innate immune responses in plants that include callose deposition, an oxidative burst, induction of pathogenesis-related (PR) gene expression, and seedling growth inhibition (Felix et al., 1999; Chinchilla et al., 2006; Sun et al., 2006).
Although the protoplast system used to place MEKK1 upstream of MPK3/6 is an extremely valuable tool for studying signal transduction, it is important to use alternative methods to test the resulting models. In this study, we used whole-plant reverse genetic methods to investigate the role played by MEKK1 in Arabidopsis signaling pathways that respond to flagellin and pathogenic bacteria. For the latter experiments, we analyzed the response of Arabidopsis plants to Pseudomonas syringae pv tomato (Pst) DC3000 strains, which is an extensively characterized system for plant-pathogen research (Quirino and Bent, 2003; Chisholm et al., 2006).
RESULTS
Mutation of the MEKK1 Gene Causes a Dwarf Plant Phenotype
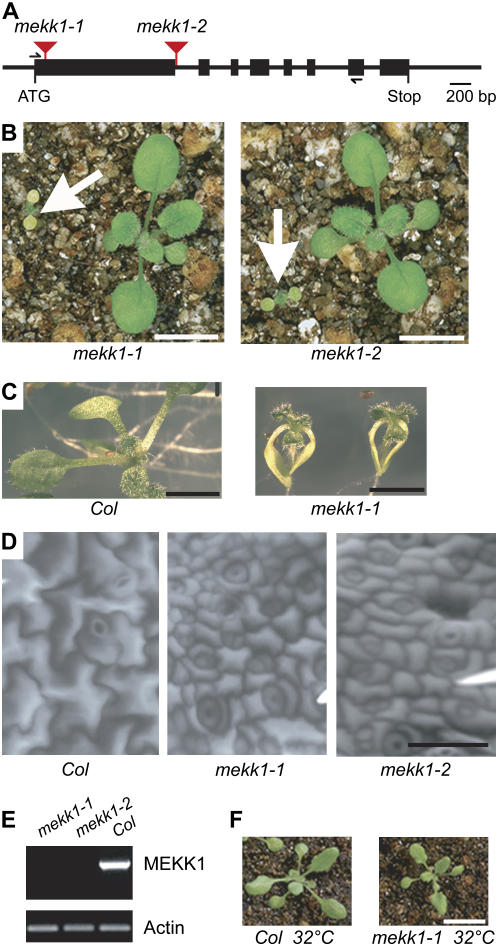
Mutant alleles of MEKK1 were obtained from two independent collections of T-DNA-transformed Arabidopsis lines. mekk1-1 is from the Salk Institute (Alonso et al., 2003), while mekk1-2 is from the University of Wisconsin's pDs-Lox collection (http://www.hort.wisc.edu/krysan/2010/). Both mutant alleles are in the Columbia (Col-0) ecotype. The T-DNA in mekk1-1 is located 80 bp downstream of the start codon in exon 1. The insertion in mekk1-2 is located in the intron 8 bp downstream of exon 1 (Fig. 1A).
Figure 1.
Mutation of mekk1 causes a dwarf phenotype. A, Location of T-DNA insertions. Thick lines are exons. Thin lines are introns and untranslated regions. B, Twelve-day-old homozygous mekk1-1 and mekk1-2 plants grown at 24°C are indicated by arrows. Larger plants are wild-type Col-0. Bar = 1 cm. C, Twelve-day-old plants grown at 24°C on agar plates. Bar = 0.5 cm. D, Environmental scanning electron microscope images of primary leaf epidermis of 10-d-old plants grown at 24°C. Bar = 50 μm. E, RT-PCR of mekk1-1 and mekk1-2 plants. PCR primer locations indicated by arrowheads in A. Actin gene Act2 used as a control. F, Plants grown on soil for 3 weeks at 32°C. Bar = 1 cm.
Reverse transcription (RT)-PCR analysis showed that both mutant lines lack full-length MEKK1 mRNA (Fig. 1E). When grown at temperatures typical for Arabidopsis propagation (22°C–24°C), mekk1 plants initially resemble the wild type. Seedling germination is normal, and the cotyledons appear normal for the first few days. Shortly thereafter, however, mekk1 cotyledons undergo premature senescence. The first true leaves of mekk1 plants emerge slowly, are curled, and have a dark-green color (Fig. 1, B and C). mekk1 plants do no reach maturity or produce seed under these growth conditions. Observation of the epidermis of primary leaves from mekk1 and wild-type plants grown at 24°C using environmental scanning electron microscopy showed that the pavement cells of mekk1 leaves are much smaller than wild type (Fig. 1D).
We observed that elevated temperature dramatically improved the growth of mekk1 plants, which appear very similar to wild type when grown in soil at 32°C for 3 weeks (Fig. 1F). Although vegetative growth is robust at this temperature, neither wild-type nor mekk1 plants are fertile at 32°C. Homozygous mekk1 plants were propagated by lowering the temperature to 28°C during the reproductive stage.
The mekk1 Dwarf Phenotype Is Caused by Constitutive Expression of Salicylic Acid-Dependent Stress-Response Genes
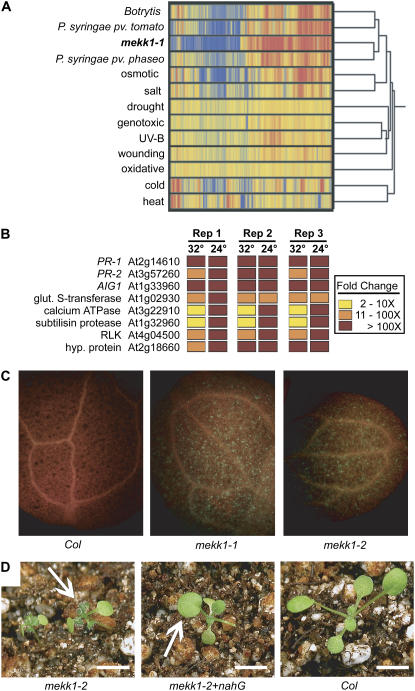
Affymetrix ATH1 whole genome microarrays were used to characterize the transcriptional profile of mekk1-1 plants. Wild-type and mutant plants were grown at 32°C for 3 weeks and then shifted to 24°C for 24 h. The initial growth at 32°C allowed us to generate sufficient tissue for RNA extraction, while the shift to 24°C was done to elicit the strongest mekk1 phenotype. A total of 1,802 genes showed a difference in expression of greater than 5-fold when comparing mekk1-1 to wild type (see Supplemental Data S1–S4). Figure 2A presents a condition tree generated by clustering the gene expression differences observed in our mekk1 experiment with 12 stress-treatment datasets obtained from public databases. The gene expression profile of mekk1-1 plants matches most closely with the three biotic stress treatments. Additional condition trees were also constructed comparing mekk1 with various hormone-treated samples, but none of these treatments showed appreciable similarity to mekk1.
Figure 2.
The mekk1 dwarf phenotype is caused by constitutive expression of stress response genes. A, Condition tree comparing the gene expression profile of mekk1-1 plants with various stress-treated Arabidopsis samples from a public database. Dark red indicates highly up-regulated genes, dark blue indicates highly down-regulated genes, and yellow indicates genes with little change in expression. B, RT-PCR of eight genes showing a high level of up-regulation in mekk1-1 plants in the microarray experiment. Three independent biological replicates were tested (Rep 1–Rep 3). 32°, Plants grown for 3 weeks at 32°C. 24°, Plants grown at 32°C for 3 weeks followed by 24 h at 24°C. Fold change indicates the relative expression level of each gene in mekk1-1 compared to wild-type Col-0 grown in the same conditions. C, Eleven-day-old seedlings stained for callose deposition and visualized using UV epifluorescence microscopy. No callose-inducing treatment was given to the seedlings. D, Two-week-old plants grown at 27°C. mekk1-2 + nahG is a homozygous mekk1-2 plant stably transformed with the nahG transgene. White arrows indicate representative primary leaves for comparison. One leaf is missing from the mekk1-2 + nahG plant, because it was harvested for genotyping. Bars = 0.5 cm.
To validate the microarray results, we selected eight highly up-regulated genes and used real-time RT-PCR to quantify their transcript levels in three independent biological samples. These genes included the PR genes PR-1, PR-2, and AIG1. RNA was collected from plants grown under two different regimens: 3 weeks at 32°C with no shift to a lower temperature and 3 weeks at 32°C followed by 24 h at 24°C. All eight genes were found to be up-regulated greater than 10-fold in all replicates that had been shifted to 24°C, validating the results obtained with the microarray (Fig. 2B). In addition, we observed that these eight genes were also up-regulated in the plants that had not been shifted to 24°C, but the level of up-regulation was lower (Fig. 2B), indicating that the phenotypic rescue afforded by growth at 32°C is not complete.
Consistent with the expression profile indicating constitutive expression of biotic stress markers, the mekk1 mutants exhibited constitutive callose deposition. When mekk1 and wild-type Col-0 plants were grown at 24°C for 11 d and not given any callose-inducing treatment, mekk1 mutants showed substantial levels of callose deposition in cotyledon tissue (Fig. 2C). As expected, no callose was detectable in the wild-type samples.
Salicylic acid (SA) is a central mediator of plant responses to biotic stress (Durrant and Dong, 2004). It has been previously reported that mutation of the Arabidopsis MAPK gene MPK4 causes a dwarf plant phenotype that can be largely rescued by the expression of the bacterial nahG gene (Petersen et al., 2000). We therefore introduced a nahG construct into mekk1 mutant plants to determine if elevated SA levels were responsible for the dwarf phenotype of mekk1 mutants. As shown in Figure 2D, the mekk1 dwarf phenotype was largely rescued by this transgene. Taken together, our results suggest that the dwarf plant phenotype of mekk1 plants is caused by constitutive expression of SA-dependent stress response genes.
Kinase-Inactive MEKK1 (K361M) Rescues the mekk1 Dwarf Phenotype
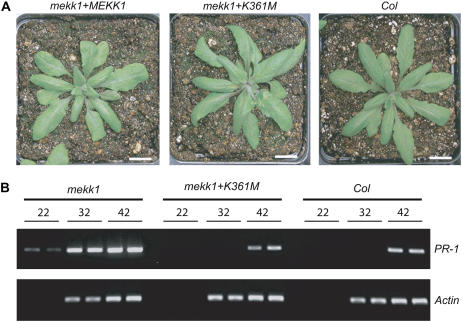
A 5.4-kb fragment of genomic DNA that included the full MEKK1 coding region, promoter, and 3′ regulatory sequences was PCR amplified, cloned into a binary vector, and introduced into mekk1-1 plants. PCR genotyping of transformed progeny identified mekk1-1 homozygotes carrying an ectopic copy of the wild-type MEKK1 gene. Eight independent lines were isolated that all demonstrated the ability of the wild-type MEKK1 gene to rescue the mekk1-1 dwarf phenotype (Fig. 3A).
Figure 3.
Kinase-inactive MEKK1 (K361M) rescues the mekk1 dwarf phenotype. A, Three-week-old plants grown on soil at 24°C. Bars = 1 cm. B, Agarose gel of RT-PCR products generated by 22, 32, or 42 PCR cycles using gene-specific primer pairs for PR-1 or an actin control gene. mekk1 + K361M, mekk1-1 mutant stably transformed with MEKK1 (K361M). mekk1 + MEKK1, mekk1-1 mutant stably transformed with the wild-type MEKK1 gene. [See online article for color version of this figure.]
We also transformed mekk1-1 plants with a modified version of the MEKK1 gene in which a single amino acid substitution was introduced into the ATP-binding pocket of the protein. The precise K361M mutation used for this experiment has been previously shown to eliminate the kinase activity of Arabidopsis MEKK1 in an in vitro assay (Asai et al., 2002). It should be noted, however, that the MEKK1 (K361M) protein may posses residual kinase activity that was not detected in those experiments. The K361M construct was introduced into mekk1-1 plants, and eight independent transgenic lines were isolated. Surprisingly, the ectopic copy of MEKK1 (K361M) rescued the mekk1-1 dwarf phenotype in each of these lines. A representative line carrying a single-locus insertion of the rescue construct was chosen for detailed phenotypic analysis (Fig. 3A). PCR and DNA sequencing were used to confirm that these lines carried the T-DNA-disrupted and K361M alleles, but no wild-type allele, of MEKK1. These K361M plants were fully fertile and grew with a morphology closely resembling wild-type plants. RT-PCR analysis showed that these plants express wild-type levels of the pathogen response gene PR-1, which is strongly up-regulated in mekk1 mutant plants (Fig. 3B). These results suggest that the kinase activity of MEKK1 may be dispensable for many of its in planta functions. It is also possible, however, that a residual level of kinase activity may be present in the MEKK1 (K361M) protein, and it is this activity that allows the mutant protein to perform its normal functions.
MEKK1 and flg22-Induced MAPK Activation
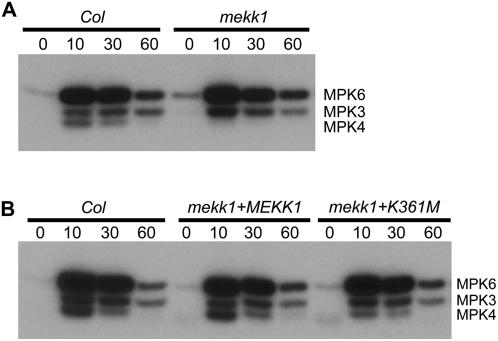
It has previously been reported that MEKK1 is responsible for the activation of MPK3 and MPK6 in response to treatment with the flagellin elicitor peptide flg22 (Asai et al., 2002). We therefore measured MAPK activity following flg22 treatment by using an in-gel kinase assay with myelin-basic protein as the embedded kinase substrate. Protein was extracted from 14-d-old seedlings at various time points following flg22 treatment. Wild-type Col-0 plants were found to activate MPK3, MPK4, and MPK6 in response to flg22 treatment (Fig. 4A). In mpk3, mpk4, and mpk6 single-knockout mutant seedlings, flg22 treatment could not activate the missing kinase, allowing us to assign the identities to the three bands detected in the in-gel kinase assays (Y. Liu and S. Zhang, unpublished data). The strongest activation occurred 10 min after flg22 treatment. By contrast, it was observed that mekk1 plants do not activate MPK4 when treated with flg22. mekk1 plants did, however, activate MPK3 and MPK6 at a similar level to that observed in wild type (Fig. 4A). mekk1 mutant plants transformed with either the wild-type or K361M version of MEKK1 displayed the wild-type profile of MAPK activation (Fig. 4B).
Figure 4.
MAPK activation in response to flg22 peptide. Crude protein extracts were prepared from 14-d-old seedlings and subjected to an in-gel kinase assay using myelin basic protein as the embedded substrate. Kinase activity corresponding to MPK3, MPK4, and MPK6 is indicated. Seedlings were harvested at the indicated number of minutes following flg22 treatment. 0, No flg22 treatment. A, mekk1 mutant versus wild type. B, mekk1 + MEKK1, mekk1-1 mutant stably transformed with the wild-type MEKK1 gene. mekk1 + K361M, mekk1-1 mutant stably transformed with MEKK1 (K361M).
MEKK1 and Macroscopic flg22 Responses
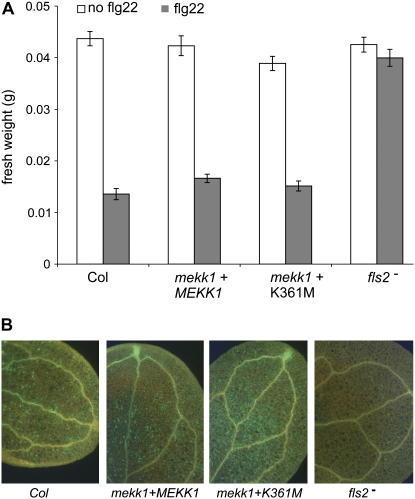
We demonstrated above that the K361M version of MEKK1 was able to support the activation of MPK4 in response to flg22 treatment. To investigate the potential role of MEKK1 kinase activity in other aspects of the flg22 response, we tested K361M plants for flg22-induced seedling growth inhibition and callose deposition. In the standard seedling growth assay for overall flg22 response, K361M plants responded similarly to wild-type controls (Fig. 5A). As a separate test of plant FLS2-mediated responses to flagellin, callose deposition was monitored in leaf cells of flg22-treated samples (Fig. 5B). K361M seedlings produced a callose response upon flg22 treatment that was indistinguishable from the response of wild-type controls. As expected, homozygous fls2 mutant plants did not develop callose when treated with flg22 (Fig. 5B). In addition, K361M plants did not constitutively produce callose in the absence of flg22 treatment (data not shown), further confirming that the K361M construct is able to rescue the mekk1 mutant phenotype. Taken together, these data indicate that MEKK1 kinase activity may not be required to activate FLS2-mediated defense responses in Arabidopsis.
Figure 5.
K361M seedlings are responsive to treatment with flg22 peptide. A, Five-day-old seedlings subsequently treated with and without 10 μm flg22 peptide for 10 d. Error bars indicate se. B, Ten-day-old seedlings treated with 10 μm flg22 peptide for 24 h and stained for callose deposition. Independent replicated experiments gave similar results for both A and B. mekk1 + MEKK1, mekk1-1 mutant stably transformed with the wild-type MEKK1 gene. mekk1 + K361M, mekk1-1 mutant stably transformed with MEKK1 (K361M). fls2, fls2-101/fls2-101 T-DNA mutant.
K361M Plants Display Modified Responses to P. syringae
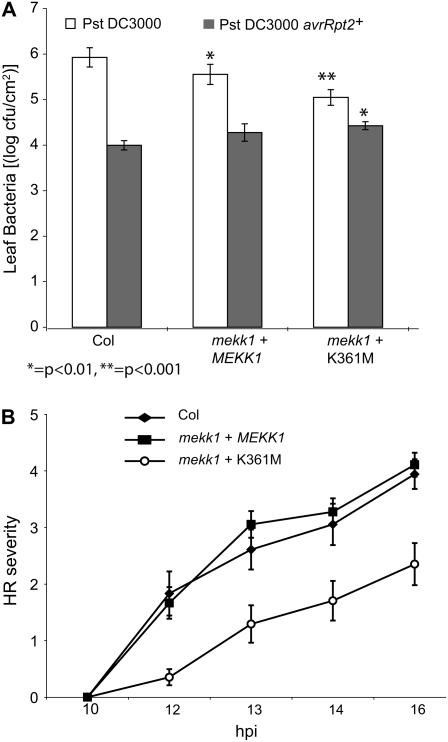
It has been reported that plants transiently expressing a constitutively active form of Arabidopsis MEKK1 exhibit enhanced resistance to both fungal and bacterial pathogens and that silencing of a tomato (Lycopersicon esculentum) MEKK1-related kinase alters responses to P. syringae pathogens (Asai et al., 2002; del Pozo et al., 2004). We therefore tested the responses of our K361M plants to Pst bacteria. K361M plants were found to be more resistant than wild-type Col-0 to the virulent strain Pst DC3000 (Fig. 6A; P < 0.001). We also observed a more subtle enhancement of resistance in mekk1-1 plants that had been rescued with the wild-type MEKK1 construct (P < 0.01).
Figure 6.
K361M plants have altered responses to Pst DC3000. A, Bacterial leaf populations 3 d after inoculation with Pst DC3000 ± avrRpt2. Four biological replicates were performed and a representative graph is shown (mean ± se of the mean). ANOVA using a generalized linear model was applied to the data from all four replicates, and Tukey's simultaneous test was used to determine the likelihood that bacterial populations were similar in a given plant genotype compared to the wild-type control treated with the same bacterial strain (*, P < 0.05; **, P < 0.01). mekk1 + MEKK1, mekk1-1 mutant stably transformed with the wild-type MEKK1 gene. mekk1 + K361M, mekk1-1 mutant stably transformed with MEKK1 (K361M). B, Leaves were tested for the HR by syringe inoculation with P. syringae pv Glya Race 4 pV288 (avrRpt+). The progression of the HR was monitored over a period of 24 h and was recorded using a scale from 0 = no tissue collapse to 5 = extensive tissue collapse. Data are mean ± se of the mean. Independent replicated experiments gave similar results. mekk1 + MEKK1, mekk1-1 mutant stably transformed with the wild-type MEKK1 gene. mekk1 + K361M, mekk1-1 mutant stably transformed with MEKK1 (K361M).
Interestingly, the K361M plants were found to be slightly more susceptible to an avirulent strain of Pst DC3000 expressing avrRpt2 when compared to the wild-type controls (Fig. 6A; P < 0.01). In separate experiments, we monitored the development of hypersensitive response (HR) programmed cell death, a hallmark of R-gene-mediated plant disease resistance against avirulent pathogen strains (Quirino and Bent, 2003; Chisholm et al., 2006). Full HR cell collapse developed in all plant lines by 33 h after inoculation, but the HR developed more slowly in the K361M plants when compared to the wild-type controls (Fig. 6B).
DISCUSSION
Using T-DNA knockout plants, we observed that absence of the MEKK1 protein prevents plants from activating MPK4 in response to flg22 treatment. The activation of MPK3 and MPK6 is not affected in mekk1 plants. These results suggest that the model of FLS2/flagellin signaling developed from the protoplast-based transient expression experiments may need to be modified (Asai et al., 2002). In this previous model, MEKK1 activates MPK3 and MPK6 in response to flg22 treatment. Whether MEKK1 activates MPK3 and MPK6 in a completely wild-type plant remains to be determined, but an alternative and primary pathway apparently exists that activates MPK3 and MPK6 to near wild-type levels in the absence of MEKK1. The protoplast experiments of Asai et al. (2002) used a truncated form of MEKK1 in which the 325-amino acid regulatory domain was not present. This regulatory domain has been previously shown to directly interact with MPK4, but not MPK3 or MPK6, in yeast two-hybrid experiments (Ichimura et al., 1998). It is therefore possible that the regulatory domain of MEKK1 dictates with which MAPK partners it is able to associate in the cell. Because MAP3Ks are not expected to directly phosphorylate MAPKs, a MAP2K would also be expected to be involved in the resulting three-kinase complex. Overexpression of the truncated form of MEKK1 in the protoplast system may allow it to promiscuously activate MPK3 and MPK6, a role that may not normally be played by full-length MEKK1 in intact plants.
Our observation that the kinase-impaired MEKK1 (K361M) is able to rescue the mekk1 mutant phenotype supports a model in which the structural properties of MEKK1 are required for proper MPK4 activation, but MEKK1's kinase activity may not be. It is possible that the kinase-impaired version of MEKK1 serves as a scaffold that allows the correct MAP2K to complex with MPK4. The kinase activity that is diminished in MEKK1 (K361M) could then be provided in trans by one of the related MAP3Ks encoded by the Arabidopsis genome. This model suggests that there is functional redundancy present in Arabidopsis for the kinase activity of MEKK1 but that there is not redundancy for the structural features of MEKK1. Comparison of the amino acid sequences of MEKK1 and its closest homolog indicates that there is 88% identity in the kinase domain but only 46% identity in the regulatory domain. Divergence of regulatory domain sequences could be a mechanism by which signaling specificity is achieved by various members of the Arabidopsis MAP3K gene family.
Disruption of the MEKK1 gene in Arabidopsis caused a dwarf phenotype, constitutive callose deposition, and constitutive expression of pathogenesis-associated genes. The mekk1 phenotype was rescued by a nahG transgene that degrades SA. It has frequently been observed that constitutive expression of biotic stress response genes in Arabidopsis leads to dwarf plant phenotypes (Greenberg, 1997). Arabidopsis mpk4 mutants have a dwarf phenotype and constitutive PR gene expression similar to that of mekk1 plants, and mpk4 mutants can also be rescued by nahG expression (Petersen et al., 2000; Brodersen et al., 2006). We observed that mekk1 mutant plants are not able to activate MPK4 in response to flg22 treatment. Because of this observation and the similar phenotypes displayed by mekk1 and mpk4 mutant plants, a simple model can be proposed where the dwarf phenotype of mekk1 plants is due to the inability of these plants to activate MPK4.
We observed that the growth of mekk1 plants was greatly improved by elevated temperature. A similar situation has been previously described for the Arabidopsis humidity- and temperature-sensitive mutant BONZAI1 (Hua et al., 2001; Jambunathan et al., 2001; Jambunathan and McNellis, 2003; Yang and Hua, 2004; Yang et al., 2006). BON1 is an infection-activated gene and encodes a protein from the copine family that has been shown to function as a negative regulator of plant defense responses. Although the mechanism by which elevated temperature is able to rescue these mutants has not been determined, the similarities between bon1 and mekk1 raises the possibility that high-temperature rescue might share some common mechanistic theme in these two instances involving stress-associated genes.
Activation of FLS2 by flg22 treatment stimulates the innate immune response in Arabidopsis, leading to changes such as PR gene expression, callose deposition, and seedling growth inhibition (Felix et al., 1999; Chinchilla et al., 2006). Because mekk1 and mpk4 mutants constitutively express these phenotypes in the absence of FLS2 activation, MEKK1 and MPK4 can formally be defined as negative regulators of the FLS2 pathway. We have shown that flg22 treatment causes activation of MPK4 in wild-type plants, and it may seem contradictory that a negative regulator of FLS2 responses is activated by flg22. However, MPK3 and MPK6 are also activated by flg22 treatment. It is possible that MPK3 and/or MPK6 positively regulate events downstream of FLS2 activation, while MPK4 serves as a negative regulator. Activating both positive and negative regulators of the downstream pathways may allow the FLS2 receptor to exert more precise control over the level of response generated by a given stimulus.
Although we did not find evidence for a role of MEKK1 kinase activity in the flg22 response pathway, we did observe significant differences in the response of K361M plants to pathogenic bacteria. The mekk1 mutants carrying the K361M transgene exhibited reduced HR and increased growth of avirulent P. syringae, indicating a positive role for MEKK1 kinase activity in these aspects of plant disease resistance. Avirulent P. syringae grew more rapidly in the K361M plants than in wild type, despite the fact that virulent P. syringae grew less well in the K361M plants than in wild type.
The fact that virulent P. syringae grew less well in the K361M plants could imply that a residual fraction of the SA-mediated responses caused by mekk1 mutation remain despite the normal appearance and absence of elevated PR gene expression in these plants. Alternatively, MEKK1 kinase activity may contribute both to HR cell death and to the susceptibility-associated cell death caused by virulent P. syringae. Cell death phenotypes are separable from SA-mediated responses, for example in Arabidopsis dnd1 (cngc2) and dnd2 (hlm1/cngc4) mutants that continue to exhibit suppression of HR cell death despite removal of SA-mediated responses by nahG expression or eds16 mutation (Yu et al., 1998; Jurkowski et al., 2004). Precedent for a MAP3K mediating the cell death associated with both immunity and disease susceptibility has been established (del Pozo et al., 2004). Virus-induced transient gene silencing of MAP3Kα in Nicotiana benthamiana and tomato plants suppressed the HR cell death response to avirulent P. syringae strains. Disease symptoms and bacterial growth were reduced in these plants when responding to virulent P. syringae (del Pozo et al., 2004). Our Arabidopsis results are consistent with a positive role for MEKK1 in activating not only the HR but also the more gradual host cell death associated with growth of virulent pathogens such as P. syringae.
Rescue of mekk1 mutants with a kinase-impaired MEKK1 (K361M) protein suggested a possible structural role for MEKK1 in pathogenesis-associated signaling, independent of its protein kinase activity. Protoplast and gene overexpression approaches provide information about nonnative systems, as do experiments done with whole plants that lack relevant proteins due to knockout mutations. Expressing a catalytically impaired form of a protein may become an increasingly important strategy for obtaining the most valid information about a protein's normal cellular function, especially for research on MAPK cascades where there is precedent for scaffold proteins, signaling complexes, and cross talk from paralogous proteins.
MATERIALS AND METHODS
Plant Material and Microscopy
Plants were grown in soil under constant light at 24°C or 32°C. PCR-based genotyping was performed using the following three primers: Mekk1-F, 5′-CTTTAGAATCCGTGGACGAGGCATACACC-3′; Mekk1-R, 5′-CAAGTACAATGAAAGTGAAATGAATCCAC-3′; and p745, 5′-AACGTCCGCAATGTGTTATTAAGTTGTC-3′. p745 recognizes the T-DNA left border. Primary leaves from 10-d-old plants were observed under a Quanta 200 environmental scanning electron microscope.
Genetic Rescue with Wild-Type and K361M Alleles
The MEKK1 genomic locus was PCR amplified and cloned from wild-type Col-0 DNA using the primers 5′-AGGTTACTAAAGTGAAGTCTAAGTG-3′ and 5′-GTAGAATAACGGATCAAAGCGAATCTAAC-3′. The K361M kinase-inactive allele was generated by site-directed mutagenesis with a Stratagene Quick Change Mutagenesis kit. The constructs were introduced into Arabidopsis (Arabidopsis thaliana) plants via Agrobacterium transformation (Clough and Bent, 1998).
cDNA Synthesis and Quantitative RT-PCR
RNA was extracted from whole-plant tissue using the RNeasy Plant Mini kit (Qiagen). First-strand cDNA was synthesized from DNAse-treated total RNA using the SuperScript II First Strand cDNA Synthesis system (Invitrogen). Reactions contained first-strand cDNA template and a pair of gene-specific primers. Primer sequences are as follows: ACT2-F, 5′-AAGGATCTGTACGGTAACATTGTGCTCAG-3′, ACT2-R, 5′-ACACTGTACTTCCTTTCAGGTGGTGCAAC-3′; H2A-F, 5′-CGATTTTTGAAAGCCGGTAAGTACGCCGA-3′, H2A-R, 5′-GCAACTTGCTTAGCTCCTCATCATTCCTC-3′; MEKK1-F, 5′-TCAACTGGACGAAGAGGAGGAGATAAGAA-3′, MEKK1-R, 5′-GATCTGACCAGTACACATTTCCAGCACAG-3′; At1g02930-F, 5′-GAAACAACCTTCTCTCAACTGGCAAGGAC-3′, At1g02930-R, 5′-AGTGATGTCAGCAACCCAAGCACTCACAT-3′; At3g57260-F, 5′-AAACCGCGTTCTCGATGTTCTGCATTGCT-3′, At3g57260-R, 5′-TCGGACGTTGTGGCTCTTTACAAACAACA-3′; At2g18660-F, 5′-TGCACAAATCTTAGCTCCAATCGCTGAAG-3′, At2g18660-R, 5′-TAACCCGAAAAGCGTCACGAGAGAGATTA-3′; At1g33960-F, 5′-GTCGGTATCTGCTGAATTTATCGGTAAAG-3′, At1g33960-R, 5′-TCAAACAGAATCATTCGTTGGCCACACAA-3′; At3g22910-F, 5′-AGTCATATCTCGCGTGACACTAACGAGCA-3′, At3g22910-R, 5′-TCACAAGCAGAGAGCTTTCTCACCATAGC-3′; At1g32960-F, 5′-TACATCTCCCAATGACACCTTAAATGTCG-3′, At1g32960-R, 5′-ACCCTTCTGCAAAAATCTGCTCTCCAAAT-3′; At2g14610-F, 5′-TTAGCCTTCTCGCTAACCCACATGTTCAC-3′, At2g14610-R 5′-CTCGAAAGCTCAAGATAGCCCACAAGATT-3′; At4g04500-F, 5′-AAGAAAAGGTTCAGGACAAGGAGGTATGG-3′, At4g04500-R, 5′-GATAAAGAAGCCCTCTTGCAACTCCTTCT-3′.
Microarray Analysis
mekk1-1 and wild-type Col-0 plants were grown on soil in constant light at 32°C for 3 weeks and then shifted to 24°C for 24 h. Total RNA extraction, cDNA synthesis, and probe preparation were performed according to Affymetrix guidelines. ATH1 genome arrays were used. Genespring v 7.1 (Agilent Technologies) was used to generate a condition tree using the standard correlation similarity measure. Public Affymetrix datasets were obtained from http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp.
In-Gel Kinase Assay
Protein was extracted from seedlings and stored at −80°C as described (Liu and Zhang, 2004). The concentration of protein extracts was determined using the Bio-Rad protein assay kit with bovine serum albumin as the standard. Myelin basic protein was used as the substrate for the in gel-kinase assay (Zhang and Klessig, 1997; Yang et al., 2001).
Seedling Growth Inhibition
Flg22-induced seedling growth inhibition assays (Gomez-Gomez et al., 1999) were performed as described (Pfund et al., 2004). Approximately 12 Arabidopsis seedlings per treatment were grown on 0.5× Murashige and Skoog agar media supplemented with 2% Suc and 1× Gamborg's vitamins for 5 d and then transferred to 24-well plates (one seedling per well) containing 400 μL of liquid 0.5× Murashige and Skoog salts, 2% (w/v) Suc, and 1× Gamborg's vitamins media with and without 10 μm flg22 peptide. Seedling fresh weight was recorded 2 weeks later.
Callose Deposition
Approximately 12 Arabidopsis seedlings per treatment were grown on 0.5× Murashige and Skoog, 2% (w/v) Suc, 1× Gamborg's vitamins media for 10 d and then transferred to 24-well plates (one seedling per well) containing 400 μL of liquid 0.5× Murashige and Skoog salts, 2% (w/v) Suc, and 1× Gamborg's vitamins media with and without 10 μm flg22 peptide. After 24 h, seedlings were fixed overnight in 1% (v/v) glutaraldehyde, 5 mm citric acid, 90 mm Na2HPO4, pH 7.4, and then cleared and dehydrated with 100% ethanol. Callose was visualized as described (Gomez-Gomez et al., 1999). Briefly, cleared seedlings were transferred sequentially to 50% ethanol and 67 mm K2HPO4, pH 12. The seedlings were then stained for 1 h at 25°C in 0.01% (w/v) aniline blue in 67 mm K2HPO4, pH 12, mounted in 70% glycerol/30% stain, and examined using UV epifluorescence microscopy.
Bacterial Growth Assays
Bacterial strains were grown at 28°C on NYGA plates. A fresh culture of Pst DC3000 (±avrRpt2) was scraped off the growth plate, resuspended in 10 mm MgCl2 with 0.005% (v/v) Silwet L-77 (OSi Specialties), and vacuum infiltrated into leaves of 4- to 6-week-old Arabidopsis plants grown under 9 h light conditions. Three days postinfection, leaf discs from four leaves were combined and ground in 10 mm MgCl2 with quadruplicate replication (16 leaves total). The samples were vortexed, serially diluted, and plated onto NYGA solid medium. Viable colonies were counted after 2 d at 28°C. Analysis of variance and Tukey's simultaneous test were performed on the combined data from separate experiments using Minitab (release 14).
HR
Four- to 6-week-old Arabidopsis plants grown under 9-h light conditions were tested for the HR by syringe inoculation of approximately 10 leaves with P. syringae pv Glya Race 4 carrying pV288 (avrRpt2+) or pVSP61 (vector with no avr gene) at 5 × 107 colony forming units (compare with μ/mL). The progression of the HR was monitored over a period of 24 h as described (Klement et al., 1964) and was recorded using a scale from 0 = no visible loss of turgidity to 5 = extensive tissue collapse.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data S1. Col wild-type microarray data.
Supplemental Data S2. mekk1-1 mutant microarray data.
Supplemental Data S3. Genes up-regulated in mekk1-1 compared to Col wild type.
Supplemental Data S4. Genes down-regulated in mekk1-1 compared to Col wild type.
Supplementary Material
Acknowledgments
The authors thank Suraphon Chaiwongsar for assistance with microscopy.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2003–02593 to P.J.K.) and the U.S. Department of Energy, Energy Biosciences (grant no. DE–FG02–02ER15342 to A.F.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Patrick Krysan (fpat@biotech.wisc.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Bjorn Nielsen H, Zhu S, Newman MA, Shokat KM, Rietz S, Parker J, Mundy J (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J 47 532–546 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Covic L, Silva NF, Lew RR (1999) Functional characterization of ARAKIN (ATMEKK1): a possible mediator in an osmotic stress response pathway in higher plants. Biochim Biophys Acta 1451 242–254 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Pedley KF, Martin GB (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18 265–276 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18 277–284 [DOI] [PubMed] [Google Scholar]
- Greenberg JT (1997) Programmed cell death in plant-pathogen interactions. Annu Rev Plant Physiol Plant Mol Biol 48 525–545 [DOI] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K (1998) Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem Biophys Res Commun 253 532–543 [DOI] [PubMed] [Google Scholar]
- Jambunathan N, McNellis TW (2003) Regulation of Arabidopsis COPINE 1 gene expression in response to pathogens and abiotic stimuli. Plant Physiol 132 1370–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Okresz L, Bogre L, Hirt H (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5 415–424 [DOI] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK Jr, Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF (2004) Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact 17 511–520 [DOI] [PubMed] [Google Scholar]
- Klement Z, Farkas GL, Lovrekovich L (1964) Hypersensitive reaction induced by phytopathogenic bacteria in the tobacco leaf. Phytopathology 54 474–477 [Google Scholar]
- Lee J, Rudd JJ, Macioszek VK, Scheel D (2004) Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem 279 22440–22448 [DOI] [PubMed] [Google Scholar]
- Ligterink W, Kroj T, zur Nieden U, Hirt H, Scheel D (1997) Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276 2054–2057 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK-Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7 301–308 [DOI] [PubMed] [Google Scholar]
- Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18 1027–1034 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Ichimura K, Irie K, Morris P, Giraudat J, Matsumoto K, Shinozaki K (1998) Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett 437 56–60 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi-Shinozaki K, Matsumoto K, Shinozaki K (1996) A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 93 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10 339–346 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant Microbe Interact 17 696–706 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Bent AF (2003) Deciphering host resistance and pathogen virulence: the Arabidopsis/Pseudomonas interaction as a model. Mol Plant Pathol 4 517–530 [DOI] [PubMed] [Google Scholar]
- Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF (2006) Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18 764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4 392–400 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hua J (2004) A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, Sun Q, Hua J (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45 166–179 [DOI] [PubMed] [Google Scholar]
- Yu IC, Parker J, Bent AF (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.