Abstract
The advent of high-throughput phenotyping technologies has created a deluge of information that is difficult to deal with without the appropriate data management tools. These data management tools should integrate defined workflow controls for genomic-scale data acquisition and validation, data storage and retrieval, and data analysis, indexed around the genomic information of the organism of interest. To maximize the impact of these large datasets, it is critical that they are rapidly disseminated to the broader research community, allowing open access for data mining and discovery. We describe here a system that incorporates such functionalities developed around the Purdue University high-throughput ionomics phenotyping platform. The Purdue Ionomics Information Management System (PiiMS) provides integrated workflow control, data storage, and analysis to facilitate high-throughput data acquisition, along with integrated tools for data search, retrieval, and visualization for hypothesis development. PiiMS is deployed as a World Wide Web-enabled system, allowing for integration of distributed workflow processes and open access to raw data for analysis by numerous laboratories. PiiMS currently contains data on shoot concentrations of P, Ca, K, Mg, Cu, Fe, Zn, Mn, Co, Ni, B, Se, Mo, Na, As, and Cd in over 60,000 shoot tissue samples of Arabidopsis (Arabidopsis thaliana), including ethyl methanesulfonate, fast-neutron and defined T-DNA mutants, and natural accession and populations of recombinant inbred lines from over 800 separate experiments, representing over 1,000,000 fully quantitative elemental concentrations. PiiMS is accessible at www.purdue.edu/dp/ionomics.
The genome within all living systems acts through the transcriptome, proteome, metabolome, and ionome to direct all aspects of an organism's diverse functions. The dynamic response and interaction of these biochemical omes defines how a living system functions and its study, functional genomics, is now one of the biggest challenges in the life sciences. To address this challenge, quantitative and qualitative high-throughput molecular phenotyping tools have been developed, which, when coupled to similarly high-throughput genotyping tools, allow gene-to-function connections to be made rapidly. Such tools and resources include DNA microarray transcript profiling and genotyping, proteomic, metabolomic, and ionomic profiling, catalogued saturation collections of insertional mutants, and numerous mapping populations. However, the advent of these high-throughput technologies has created a deluge of information that is challenging to deal with, not only because of its sheer volume, but also because of the difficulties in interpreting the various measurements in the context of different genotypes and organismal physiologies. To maximize the value of such large datasets, it is also critical that they are rapidly disseminated to the broader research community, allowing for multiple community approaches to data mining and discovery.
This expansion of high-throughput functional genomics projects has led to an exciting proliferation of Web-based data and resource-cataloging sites, as well as numerous open access data analysis engines (for review, see Rhee and Crosby, 2005). There are also several projects under way (e.g. Arabidopsis [Arabidopsis thaliana] Web services [http://bioinfo.mpiz-koeln.mpg.de/araws]) in which multiple databases are federated within a common framework using a network of standardized Web service providers. However, what is currently lacking in this large set of community genomics tools is a discovery environment that integrates defined workflow controls for genomic-scale data acquisition and validation, data storage and retrieval, and data analysis, indexed around the genomic information of the organism of interest. Such a system should be open access and facilitate rapid generation of new knowledge tying genes to functions.
The Purdue Ionomics Information Management System (PiiMS) described here is founded on our National Science Foundation Plant Genome and Arabidopsis 2010 funded ionomics project. The ionome is a new conceptual framework for thinking about plant mineral nutrition and was described by Salt and colleagues (for review, see Salt, 2004) to include all the metals, metalloids, and nonmetals present in an organism (Lahner et al., 2003), extending the term metallome (Outten and O'Halloran, 2001; Williams, 2001; Szpunar, 2004) to include biologically significant nonmetals, such as N, P, S, Se, Cl, and I. PiiMS couples flexible data storage and search tools with our high-throughput inductively coupled plasma (ICP)-mass spectrometry (MS) phenotyping platform and allows Web-based open access to our large ionomics dataset. PiiMS can be accessed at www.purdue.edu/dp/ionomics.
In PiiMS, we have developed workflow controls and data storage systems to facilitate the high-throughput data acquisition required by our ionomics projects. PiiMS models the physical workflow in the laboratory and divides it into stages based on the physical activities and the information and data generated throughout the experiment life cycle. In this way, it can provide workflow management support as well as the capability to capture contextual information (metadata) necessary to fully describe the experiment. This system allows us to control and capture, through a series of input portals, the flow of information in our high-throughput ionomics system. Information mapping the position of the plants in the experimental unit to the sample analysis vial is captured, helping to reduce errors in sample identification after analysis. Information relating to the plant's genotype and genealogy, date planted and harvested, environmental, and ICP-MS analytical conditions are also captured. After ICP-MS analysis is completed, all analytical data are uploaded into PiiMS via a Web-based upload portal. All experimental metadata and ICP-MS analytical data are associated in the database. Tools are also implemented in the system to manage various aspects of the PiiMS environment, including creation of new sample maps, addition of new genetic and genealogy information, and management of users and instruments. Within our Ionomics project, PiiMS is used daily to manage a continuous functional genomics pipeline containing over 3,000 active samples. PiiMS manages plant growth, harvest, sample preparation, and ICP-MS analysis and is used daily to analyze and store over 2,000 fully quantified elemental concentrations. PiiMS also provides on demand data reanalysis and open access search and retrieval capability across the complete ionomics dataset. Similar ideas for the development of supportive data collection tools for plant metabolomics have recently been proposed (Jenkins et al., 2005), but to our knowledge PiiMS is the first true implementation of these ideas integrated into an existing high-throughput analytical phenotyping platform.
Most functional genomics approaches, including transcriptomics, proteomics, metabolomics, and ionomics, consist of a similar experimental design, which includes sample generation, preparation, and analysis followed by data processing, storage, and retrieval. Because of these commonalities, the overall architecture of PiiMS is adaptable across various functional genomics platforms, providing a good model for the management of functional genomics approaches in general. Currently, we are also in the process of designing middleware systems, software that sits in between the user interface and the database to allow integrated data analysis across multiple functional genomics databases.
GENERAL APPLICATIONS OF THE PIIMS DATASETS
PiiMS currently contains data on the concentrations of P, Ca, K, Mg, Cu, Fe, Zn, Mn, Co, Ni, B, Se, Mo, Na, As, and Cd in over 60,000 shoot tissue samples of Arabidopsis from over 800 separate experiments, representing more than 1,000,000 fully quantitative elemental concentrations. Data are being added to the database through 2008 and possibly after that, if funding permits. The dataset can be divided into two main types that can be queried separately or as a whole, depending on the user's interest.
Forward-Genetics Mutant Screens
There are currently three forward-genetic screens with the first round of screening completed in the database. These include a screen of fast-neutron and ethyl methanesulfonate (EMS)-mutagenized Columbia (Col-0) populations (Lehle Seed) grown in soil under low-Fe conditions and an EMS population in low-P conditions. Further nutrient conditional screens are currently under way. All putative M2 generation mutants identified in the low-Fe screens have been grown to seed, the M3 plants have been reanalyzed under the same conditions, and the data are available in the database. For a full description of the results of the fast-neutron screen, which includes the identification of 51 confirmed mutants, see Lahner et al. (2003). All confirmed mutants from the fast-neutron screen have been submitted to the Arabidopsis Biological Resource Center (ABRC). For the low-P screen, M2 putative mutants are in the process of being grown to seed for reanalysis and the data will be uploaded to the public database as it is generated.
The concept of open access has long been a part of science because scientists share their reagents, mutants, and protocols. However, usually researchers only share the final products of their research. For example, researchers who conduct genetic screens generally only share the lines that they identified as mutants. The complete dataset from the screens is usually not shared. Taking advantage of the ability of PiiMS to display data for every sample that we have run, we have designed a screening protocol that enables anyone to view the primary data from our screens and rerun lines that were not initially selected. This enables open access to the process, which we hope will result in better mutant identification. The two EMS screens were conducted using the open access mutant identification protocol. Of the approximately 1,600 M2 plants screened under each condition, we identified putative mutants by visual inspection of z-score plots (see section on data visualization) and collected seed from selected individual plants (178 plants in the Fe screen and 233 in the P screen). All other M2 plants were grown and harvested in rows so the seed from six to 12 plants is bulked into a single tube. If users identify additional putative mutants using data in PiiMS, either by visual inspection of an ionomic phenotype of interest or through new bioinformatics algorithms, these pools can be easily screened to reidentify the putative mutant. Of the 178 EMS Fe putative mutants, 137 produced seed and were reanalyzed. From visual inspection of z-score plots, we identified 56 lines as confirmed mutants that will be deposited in the ABRC. Several confirmed mutants have also been backcrossed to Col-0 and outcrossed to Landsberg erecta (Ler-0) for mapping and the data from F1 and F2 populations are also contained in the database.
All confirmed mutants are available to the public via the ABRC and can be identified by searching PiiMS. Once obtained, researchers are free to use these mutants in their own laboratories for mapping studies to identify genes involved in regulating the shoot elemental composition, or ionome.
Another resource for genetic variation that can be used to determine gene function is the large number of natural accessions of Arabidopsis that have been collected and deposited in the various Arabidopsis stock centers. The database contains ionomic data from over 110 different natural accessions, including the 96 accessions previously genotyped by Nordborg et al. (2005). Accessions identified as high or low in particular elements of interest can be crossed and the F2 population used to map genes of interest. The database already contains data on F1 and F2 populations made from several different accessions. Such an approach was recently used by Rus et al. (2006) to identify HKT1 as driving the natural variation in shoot Na observed in Ts-1 and Tsu-1. Such data can also be used for genome-wide evolutionary studies.
The database also contains ionomic data on various genotyped recombinant inbred line (RIL) populations, including 162 lines of the Cape Verde Islands-1 × Ler-2 (Alonso-Blanco et al., 1998) RIL population run under sufficient and low-soil Fe conditions, 166 lines from the Bay-0 × Shahdara (Loudet et al. 2002) RIL population grown in sufficient Fe soil conditions, and 98 lines of the Col-4 × Ler-0 (Lister and Dean, 1993) and Col-0 × Van-0 RIL populations grown in sufficient Fe soil conditions. Users can download these data and identify quantitative trait loci (QTL) for their element or elements of interest using their preferred algorithms. The recent publication of DNA microarray-based high-density genotyping on the Col-4 × Ler-0 RIL set (Singer et al., 2006) provides the capability for very-high-resolution mapping of ionomic QTLs in this population. Furthermore, recent publication of transcript profile data for 148 RILs from the Bay-0 × Shahdara population (Kliebenstein et al., 2006), of which 59 are included in PiiMS, provides the unique opportunity for identification of QTLs controlling ionomic gene expression networks.
Reverse-Genetics Mutant Screens
The database contains data on homozygous sequence-indexed T-DNA lines in over 1,000 unique genes. The lines include knockouts in transporters and kinases selected by the Arabidopsis 2010 Ionomics group (http://www.cbs.umn.edu/Arabidopsis/ionome), as well as lines sent to us by other users interested in the ionomics phenotype of knockouts in their genes of interest. Of the 806 experiments in the search section of the database, 737 are available in the public search, whereas 71 are still private, waiting for the user's permission to publish or the automatic 6-month release date. As part of the Arabidopsis 2010 project, we anticipate having data on over 2,000 homozygous T-DNA lines in the database by Fall 2008, including a large number of genes of unknown function, along with lines provided by the plant biology community with mutations in genes of interest.
The datasets within PiiMS are an important resource for researchers interested in plant mineral nutrition. However, without the underlying metadata that define the samples, such large datasets become impossible to interpret. When dealing with large data generation pipelines, there is a high risk of typographical data errors. Additionally, most biological molecules, including elements, metabolites, proteins, and transcripts, are highly variable in response to the environment. It is therefore essential that procedures are in place to check that the correct lines are entered and environmental variables under which the experiment is conducted recorded, and that these details are made available to the end user.
CHALLENGES OF FUNCTIONAL GENOMICS DATA MANAGEMENT
As large-scale phenotyping projects progress and datasets grow larger, several features become essential for integrating the large volumes of data into information management systems from which users can manage the data and extract meaningful information.
Standardized Nomenclature
The issue of gene/allele naming, which has long been a concern in biology, is even more important for databases, where users may search with a variety of names for genes or alleles. The Arabidopsis community has already developed a standard genetic and gene nomenclature system (Meinke and Koornneef, 1997; Arabidopsis Genome Initiative, 2000) and has large collections of lines available in publicly searchable databases. By integrating the Arabidopsis Genome Annotation (version 5) and the insertional mutant catalogs curated in the Salk Institute Genomic Analysis Laboratory (SIGnAL) database into PiiMS, we can ensure that the standard line and gene IDs are associated with each sample. Association of the ionomics information in PiiMS with standard gene identifiers allows for the retrieval of ionomic information based on specific gene identifiers. Such indexing of data to standard gene identifiers also facilitates the future interrogation of PiiMS by external Web services, such as Arabidopsis Web services (http://bioinfo.mpiz-koeln.mpg.de/araws), providing a mechanism for cross-platform integration of diverse datasets. Furthermore, the standard gene identifiers will also allow access to the Gene Ontology Consortium set of ontologies describing biological processes, molecular function, and cellular components, and will play an essential role in integrating genomic and functional data in the future. However, ontologies describing high-throughput phenotyping platforms, such as the ionomics platform managed by PiiMS, are lacking. To help address this problem, we have incorporated defined vocabularies into PiiMS beyond those used to define genes. These defined vocabularies help systematize much of the metadata acquisition process and facilitate data retrieval by limit searching on free text fields. For example, growth media types, tissue types, types of acid used in sample digestion, and types of instruments used for sample analysis are predefined in the system and are presented as choices to the user. Work is ongoing in our laboratory to develop a more complete ontology describing high-throughput data acquisition processes that will be used in the future to generalize the workflow to allow PiiMS to be easily adapted to other high-throughput phenotyping platforms.
Quality Control
In comparison to single bench experiments, the amount of attention and time devoted to each sample is greatly reduced in genomic-scale phenotyping projects. It is therefore critical to ensure that the quality of the data is not compromised. To achieve this systematic error checking, PiiMS formalizes data input and provides systems that facilitate application of expert knowledge. In PiiMS, we have integrated automatic checks for consistency between stock center line ID, gene ID, and background accession, followed by human review of lines that do not meet this consistency check. Such checks ensure that line metadata are consistently entered. Tools are also in place in PiiMS to facilitate internal review of data quality by an analytical chemist before the data are released into the database.
Metadata Capture
In addition to the actual data, a large amount of metadata, such as line IDs, growth conditions, and planting/harvesting dates, are also collected and these data are automatically associated with the analytical data in the database. Because these parameters will change over time, this information is critical for future interpretation and reinterpretation of the data. In PiiMS, we have created portals for every step in the workflow process (described below), allowing for standardization and control of metadata collection.
Workflow Formalization
A key component of developing an integrated information management system for a high-throughput functional genomics platform is to formalize the process workflow so that it can be represented in the system. One of the key components of this formalization is to define the way in which samples move through the process from customer submission of lines, planting, harvesting, and sample preparation to sample analysis and analytical data collection and processing. This is not only critical for integrating the database with the functional genomics pipeline, but also for reducing the chances of human error during the process. This is highly preferable to tagging or bar coding individual samples for two reasons. First, tags have to be printed and applied to each sample at each step and then read as the sample transitions from one stage to the next, a time-consuming task with a large potential for introducing error. Second, the metadata associated with each sample have to be assigned to each tag individually, another time-consuming, and possibly error-inducing, task. By formalizing the tray maps and tube configurations, samples belonging to the same line are already grouped and information for the line only has to be entered into the database once, where it is assigned to all lines in the group by the database. In PiiMS, we have created a mechanism that allows the association of sample location in the experimental environment, sample tube, and analytical and metadata in a single integrated database. PiiMS provides tools that allow for the definition and storage of these associations, providing both flexibility of experimental design with a robust mechanism to associate metadata and analytical data.
Data Security and Release
Open access to large, publicly funded functional genomics datasets is not only an obligation of the funding agency, but also critical if the datasets are going to be efficiently mined for valuable biological information. The advantages that open source code has brought to the computing arena, where a diverse set of developers generate robust code, such as Linux and Mozilla, could also be achieved in biology if large functional genomic datasets are open access. This would allow multiple investigators to carefully analyze data from many different perspectives, helping to develop and refine robust biological knowledge. A similar approach has already been taken in the astronomy and social science communities, with resources such as SkyView (http://skyview.gsfc.nasa.gov) and the Inter-University Consortium for Political and Social Research (http://www.icpsr.umich.edu). However, such open access to data raises issues of data security and propriety, particularly because data dissemination is currently best achieved via the World Wide Web.
PiiMS leverages the security features of the industry standard IBM UDB DB2 Enterprise Server upon which it is currently based. Access control also is enforced within PiiMS by assigning different users with different access privileges to the different functionalities within the system. To balance the divergent demands of open access and propriety (prepublished data), we have initiated both a private and a public side of the database. Access to data in the private part of the database is privileged; however, private data is made public 6 months after it is loaded into PiiMS. We feel that this provides sufficient time for publication and also balances the need for public access with data propriety.
OVERALL STRUCTURE OF PIIMS
The user interface for PiiMS divides the PiiMS functionalities into eLaboratory, eManagement, Data Search/Advanced Search, and Order Form. The eLaboratory portal provides tools to both control the workflow in the ionomics pipeline and capture metadata and data produced by the process. The eManagement portal provides tools for the configuration and customization of the various PiiMS portlets for metadata and data acquisition, as well as tools to define and edit information describing the line, tray, and tube configurations. Reporting tools to monitor ordering activity and provide statistics on samples run are also included in the eManagement portal. The Order Form portal provides a controlled mechanism for customers to submit samples for analysis via the ionomics pipeline at Purdue. The Search portals provide tools for searching and visualizing data within PiiMS.
WORKFLOW AND DATA CAPTURE
Customer Submission
Customers wishing to submit lines for ionomic analysis within the pipeline must first create an account using the Sign Up tool. This account provides users with access to a data input portal for the entry of critical information defining each line to be submitted, including line name, other line aliases (AKA), genetic structure, mutation type, background accession, and gene mutated. To enforce standardized nomenclature, we have downloaded and stored locally in PiiMS the information defining the T-DNA insertional mutant collection curated at SIGnAL and stored at http://signal.salk.edu/data and the Arabidopsis accessions at the ABRC. We have also created a customizable list defining other mutants not included in the SIGnAL and ABRC resource lists. To prevent typographic or transposition errors in entering these lines, the system checks the user's submission against these resource lists, alerts the user if there is a discrepancy, and suggests lines from the locally stored catalog that have fields that match those submitted by the user. For example, if a user submits Salk_012345 as a homozygous stock, the database will suggest Salk_012345:genetic structure = segregating and Salk_012345_homo:genetic structure = homozygous. The user may then correct the line, select one of the database suggestions, or ask the system to submit the line as a custom line, which is the classification for lines that are not in the stock centers; for example, lines created in the customer's laboratory. The choice of background line is especially important because it affects the choice of controls that go into each experiment and the background against which the data are normalized. Different accessions can have widely different ionomics profiles. Wassilewskija (Ws-0), for example, accumulates approximately 70% less Mo than Ws-2 and Ws-4. Each order is assigned a number by the system. The customer uses this order number to label seeds for the line being submitted and the seeds are mailed to the ionomics facility at Purdue University. This system allows PiiMS to track all lines submitted to the system individually.
Order Approval
Submitted customer orders are tracked and reviewed in the eManagement portal by the PiiMS system administrator, who checks that submitted lines are appropriate for the system and that the submitted metadata are correctly formatted. The system administrator may alter, accept, or reject the lines. Stock center lines, which have already been checked by the database, are highlighted and can be accepted as a batch, whereas custom lines require individual attention. This process not only prevents mistakes from entering into the metadata, but allows the system administrator to keep track of submitted lines and prioritize them for entry into the ionomics pipeline.
eLaboratory Portal
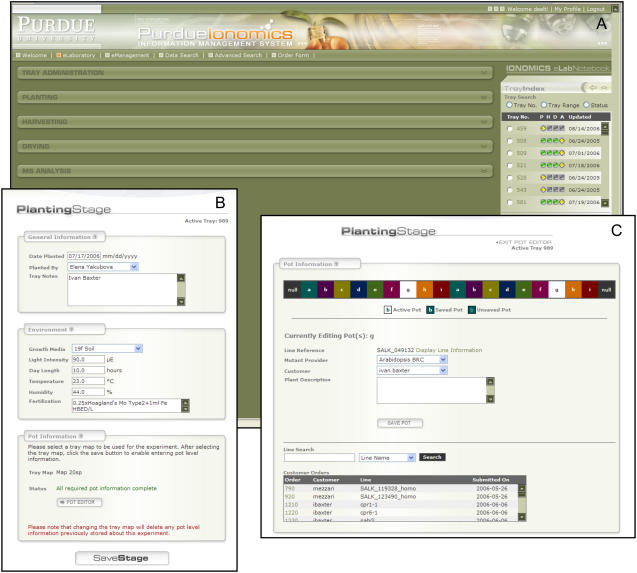
Within the eLaboratory portal, PiiMS divides the Purdue Ionomics pipeline into four process stages defined as Planting, Harvesting, Drying, and MS Analysis (Fig. 1A). These processes can be generalized as experimental subject, sample acquisition, sample preparation, and sample analysis. The physical workflow and metadata for each experiment in the ionomics pipeline flow sequentially through these processes, allowing PiiMS to capture critical metadata at each stage in the process within the portlets. To the right of the process portlets in the PiiMS interface is a list of experiments within PiiMS, with tools that allow experiments to be browsed based on their stage in the process or directly via experiment number. This tool also provides a visual representation of where a given experiment is in the PiiMS workflow. Using this browsing tool, any experiment can be loaded into the eLaboratory portal and all properties associated with that experiment edited via the four workflow-specific process portlets. Once saved, changes are propagated through PiiMS in real time. Tools are also available to create and copy new experiments.
Figure 1.
eLaboratory portal used to control workflow in PiiMS (A), with examples of modules accessed through this portal that are used to collect metadata during the planting stage. These include modules to collect experiment level information (B) and information that defines each line and its physical location in the planting tray (C).
Planting Stage
At this stage, seeds are planted and the appropriate information about the lines being planted is associated with a predefined tray map in the system (Fig. 1B). The physical entity that flows through the system at this stage is a tray composed of several pots. The system tray map displays a visual representation that resembles the physical tray, with pots that will be planted with the same line being colored and labeled the same (Fig. 1C). By clicking on each pot within this virtual tray map, the technician can select orders or internal lines and assign them to that pot. The technician is prompted to enter tray-specific metadata, such as soil batch, day length, light intensity, humidity, temperature, watering solution, date planted, customer identity, and any miscellaneous planting notes (Fig. 1, B and C).
Harvesting Stage
At this stage, leaf samples are harvested from the plants and placed into digestion tubes determined by the selected tube configuration, with specific parameters being captured in the Harvesting Stage portlet (Supplemental Fig. S1A). Tube configurations are predefined within the eManagement portal using a flexible definitions tool (Supplemental Fig. S1B). Each tube configuration is associated with a particular tray map, providing a logical connection within the system between sample position in a plant growth tray and the tube in which the sample is digested and analyzed. Such fixed relationships are then used by the system to merge the metadata and analytical data for each line within the system, allowing systematic indexing of all analytical data using metadata elements collected for each line. The harvesting technician also records which sample tubes are empty and makes notes on the visual appearance of the plants that are recorded for each sample using the Tube Editor function within the system (Supplemental Fig. S1A).
Sample Drying and ICP-MS Analysis Stages
At these stages, the samples are first prepared for ICP-MS analysis by drying and digestion in acid. The concentrations of various elements in the sample are then quantified using ICP-MS. Within the eManagement portal, there are tools to define the list of elements to be analyzed, providing full flexibility in the analysis. The Drying Stage portal requests information defining the drying processes, sample weights, and digestion conditions (Supplemental Fig. S2, A and B). For ICP-MS analysis, the workflow portal requests general information about the ICP-MS instrument, analysis parameters, and operator (Supplemental Fig. S3A). Fields are also provided to make miscellaneous notes on the digested samples; for example, sample is yellow or contains flocculate, within the Tube Editor function (Supplemental Fig. S3B). Finally, tools are provided for data upload to the system (Supplemental Fig. S3A). ICP-MS data can be uploaded as unnormalized solution concentrations and normalized to weight and background lines within the system. Or, alternatively, data can be normalized off line and then solution concentrations, weight-normalized, and background-normalized data uploaded individually. The mathematical justification for weight normalization and background normalization was previously published (Lahner et al., 2003) and the algorithm implemented in PiiMS developed by B. Lahner (B. Lahner, unpublished data). This algorithm is used to derive elemental concentrations per weight of shoot material (μg g−1) and z-score values (sds away from the mean of the background line). During online data normalization, using the Analysis function, graphic tools are provided to allow the ICP-MS analyst to evaluate the data and the normalization process and correct errors when required. Analysis is divided into four stages (Supplemental Fig. S4). In stage 1, a percentage relative sd cutoff is established to select the elements to be used for the weight calculation (Lahner et al., 2003) and options for automatic or manual iteration of the calculation are provided (Supplemental Fig. S4A). Stage 2 calculates weights and presents summary tables for calculated weights, weight-normalized data, and correlations between calculated weights and weighed weights (Supplemental Fig. S4B). If these calculations are acceptable, the process progresses to stage 3, where z-scores are calculated (Lahner et al., 2003). Stage 3 requires the ICP analyst to define how the background mean and sd will be calculated (Supplemental Fig. S4C). Calculations are based on either all plants in a tray if the experiment is a forward-genetics screen or a single background line if the experiment is designed to compare a mutant to its wild type. If line analysis is chosen, then the system determines the appropriate background line based on the accession designation for each line. The analyst is also given the choice of altering which background line will be used (Supplemental Fig. S4C). Once the data normalization method has been chosen, the system calculates the background-normalized data (z-scores) and provides summaries of the calculated values, including z-score plots, percentage change from the background, along with a table of the z-score values (Supplemental Fig. S4, D and E). The system also provides a tool to remove outliers from the calculation (Supplemental Fig. S4E). At each stage of this data normalization process, the analyst can go back and forth through each stage to alter or correct previous decisions. Once the ICP-MS analyst is satisfied with the quality of the data, it is released into the database for general searching and visualization.
DATA SEARCH, DISPLAY, AND DOWNLOAD
PiiMS supports two querying modes of the database, the Data Search and Advanced Search portals. Public users have open access to the Data Search mode. The Data Search mode is accessible on the PiiMS menu bar and also through the magnifying glass icon on the Welcome page. This search mode provides functionalities to search the database based on ionomic phenotype (forward-genetic screen), gene (reverse-genetic screen), line, experiment number, or order number. This mode also allows customers to view the data from their completed sample submissions. Once data of interest has been retrieved, PiiMS provides tools to view data summaries and plots, along with formatting data files for download to local machines. The Advanced Search mode allows the construction of extensive Boolean queries using multiple indexes, including Mutant Type, Ecotype, Gene Name, Gene ID, and Experiment Number and Range. Optional data filters are also available to further refine a search. In the Advanced Search, returned data are formatted for download onto a local machine as a comma-delimited text file. Currently, the Advanced Search is only accessible to expert users who have log-on privileges. Users interested in obtaining access to this portal should contact the PiiMS systems administrators.
Forward-Genetic Search
Currently, PiiMS contains shoot ionomic data on over 7,500 unique Arabidopsis lines, including fast-neutron, EMS, and T-DNA-mutagenized lines, natural accessions, and RILs, of which approximately 1,500 are available in the ABRC and SIGnAL collections. The PiiMS Data Search tool allows users to search this large ionomic dataset to identify lines with defined ionomic differences. For example, a user can search for lines that show reduced Ca and elevated K in shoots (Fig. 2A). Searches are performed on median values from the background-normalized dataset (lines are excluded if n < 4), such that lines can be identified that differ from their wild-type background by a predetermined number of sds. Data can be sorted based on experiment number, alphabetically by line name, or filtered so as to only show those lines for which the gene disruption is known. Reported data can be further sorted in ascending or descending order by clicking on the element of interest (Fig. 2B). Search results are browsed and lines of interested are identified from their ionomic profiles (Fig. 2B). By clicking on the tray number, line name, or ATG number for the line of interest, further experiments and datasets relating to the line can be viewed (Fig. 2C). If tray number is selected, then the information describing all the lines analyzed in that tray can be viewed. If line name or ATG number is selected, then a list of all the experiments that contain the line of interest is displayed. Experiments of interest can then be selected and the complete dataset for each experiment viewed. Using this process, Arabidopsis lines with potentially interesting ionomics phenotypes can be identified in the dataset, ordered from the stock center, and used for the basis of a new research project. In essence, this is an in silico forward-genetic screen.
Figure 2.
Modules accessed through the Data Search portal. These include the primary search interface (A), the output of searches based on ionomic phenotype (low Ca, high K; B) or gene name (NHX; D), and the list of experiments the lines returned from the phenotype search (C). [See online article for color version of this figure.]
Reverse-Genetic Search
By using the Data Search tool to retrieve ionomic data through gene ID (ATG number, gene name, or line name; Fig. 2A), it is possible to use PiiMS as a reverse-genetics tool. Currently, PiiMS contains shoot ionomic data on over 1,000 homozygous T-DNA insertional mutants in defined genes, in many cases with multiple alleles, from a selection of genes with a broad array of predicted functions. Ionomic data on new homozygous T-DNA insertional mutants is also being added to PiiMS when new lines are processed through the ionomic pipeline. Once the search has been initiated based on a gene of interest (Fig. 2A), the system returns a choice of existing lines in the database that match the query (Fig. 2D); lines of interest are then selected and the data retrieval process follows that outlined for the forward-genetic search above.
Data Display and Download
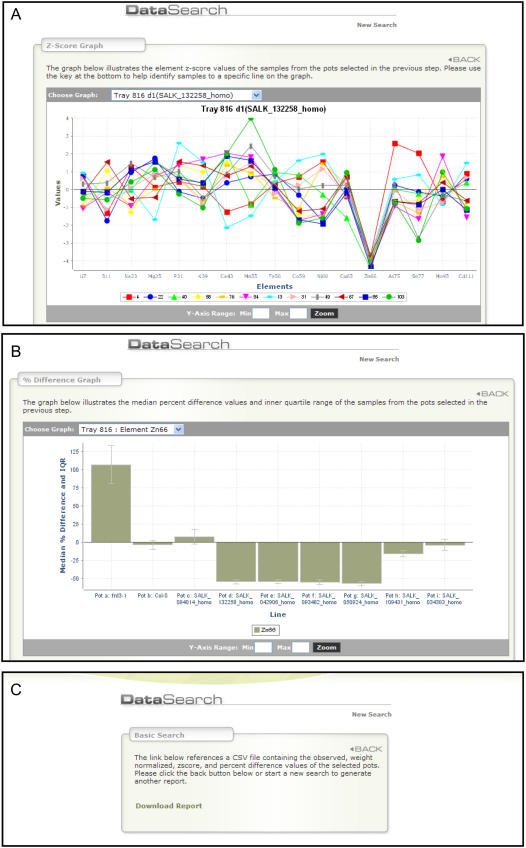
Once the user has recovered data from an experiment of interest, PiiMS provides various tools to summarize, visualize, and download the data. A summary table is provided for the selected line showing the average weight-normalized data along with the wild-type background, percentage difference between line and wild type, and P values to help establish significance differences where they exist. Tools are provided on a menu bar at the bottom of the returned results page to plot the z-score data or percentage difference from wild type for selected lines within the experiment, using drop-down menus to select the line of interest to be displayed (Fig. 3). Background-normalized (z-score, number of sds away from the mean of the background line) data for selected lines can be viewed (Fig. 3A). Data can also be viewed as the percentage change from the median value of the background line for each element, with error bars representing the Interquartile Range (Fig. 3B). By viewing both the z-score and the percentage change plots, it is easy to estimate visually both the magnitude and significance of changes in the ionome for lines of interest. For example, multiple replicate plants (n = 12) of the homozygous line SALK_132258 (hma4) are all approximately 4 sds low in Zn compared to Col-0 (Fig. 3A), and this represents an approximately 50% reduction in total shoot Zn (Fig. 3B). Data formatted as a comma-delimited text file can also be downloaded for further analysis on a local machine (Fig. 3C). A formatted report containing data, summary tables, and plots, in portable document format, can also be downloaded for storage and retrieval on a local machine.
Figure 3.
Visualization of selected ionomic data on the HMA4 mutants as a plot of z-scores (number of sds away from the mean of the wild type grown in the same experiment; A), and median percentage change from the wild type (B) for each element analyzed. Data can also be downloaded as a comma-delimited text file (csv) for further processing on a local machine or summarized in portable document format (pdf; C).
SOFTWARE DESIGN AND AVAILABILITY
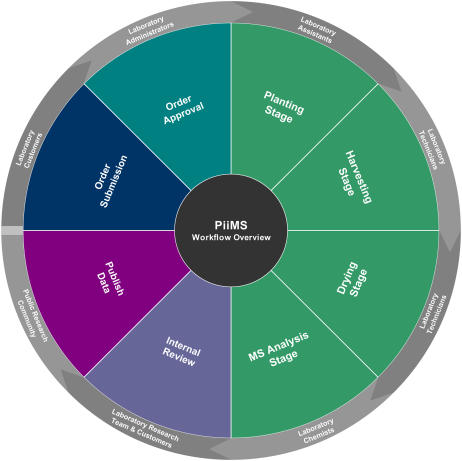
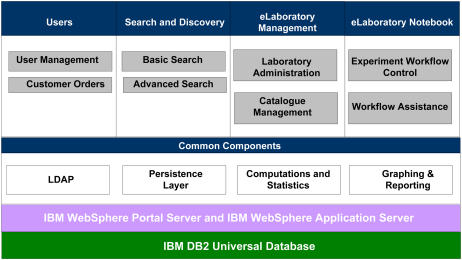
A driving principle in building PiiMS was to be able to support the workflow defined by the different stages that take place in the ionomics laboratory. These different stages correspond to order submission, order approval, planting, harvesting, drying, MS analysis, internal review, and, finally, data release (Fig. 4). At any given point in time, concurrent users are provided with different tools to carry out those tasks and check their status. Based on this requirement, we opted for Portal Technology that splits the user interface into different sections, called portlets, where different content can be displayed independently from other sections while allowing communication of updates between them (Fig. 5). When a PiiMS user signs in, based on the user's membership to specific user groups, the content and functionality, which are permitted to the user groups, are rendered and displayed. This is similar to how a desktop manages the access to software in a multiuser environment. Each portlet on the Web interface corresponds to a tailored use similar to software in a desktop environment. For example, only administrators can access the management portlets in PiiMS where new options can be defined. Hence, each stage of the PiiMS structure is implemented as a single or a combination of portlets coded in Java. The PiiMS Web application has been designed to support multiple users and high traffic for data entry, analysis, and reporting. The infrastructure used for PiiMS deployment reflects this premise. The Web application has been written in Java and deployed on the IBM WebSphere Portal Server and the IBM WebSphere Application Server, which may be operated in a clustered environment when required. These applications were chosen for two primary reasons. First, WebSphere has simple and robust support for portal technology and portlets. Second, our programming team at Purdue has strong expertise in WebSphere. However, our long-term goal is to convert these applications to open-source software to allow complete open exchange.
Figure 4.
Graphic representation of the PiiMS workflow stages and the type of user that interacts at each stage. [See online article for color version of this figure.]
Figure 5.
Graphic representation of PiiMS software architecture and key features. The top layer represents the functionalities of the four major components of PiiMS. The middle layer corresponds to features and functionalities shared by all components. The bottom layer presents the major application and database server technologies used by the current PiiMS implementation. [See online article for color version of this figure.]
Data entering the system undergo multiple levels of translation and validation before finally being stored in a relational format currently using the IBM DB2 database management system. This database captures all necessary data and metadata, including laboratory management data, customer orders, experiments, including ICP-MS data, and analysis data. Although we used IBM DB2, any relational database could be used. Currently, we are moving the PiiMS database to an open-source database management system. The components for data entry, analysis, and reporting are deployed on the portal server as coarse-grained self-contained portlet applications. Each component has a defined workflow, which contains multiple portlets capable of interacting with each other via messaging. The portal server may be used to assign user privileges that span complete portlet applications or limit access to parts of the workflow within the portlet applications. All portlet applications are tied to the relational database via a light-weight transaction manager that provides a common gateway for access and connection monitoring. The transaction manager is responsible for converting objects to relational form and vice versa. Furthermore, PiiMS support for Lightweight Directory Access Protocol integration enables users to fully synchronize their PiiMS account with Purdue University accounts (also known as single sign on). Non-Purdue University community users can also register themselves directly with PiiMS.
We will provide the code to any users willing to purchase the IBM WebSphere software necessary to run it. Furthermore, as part of the continued development of PiiMS, we intend to convert PiiMS to a fully open-source package. As part of this process, we have already completed translation of the database from the IBM DB2 system to the open-source PostgreSQL data management system (http://www.postgresql.org).
FUTURE DEVELOPMENTS
PiiMS is a deployed data management system that successfully integrates workflow control, metadata, and analytical data acquisition with database and data search capabilities. Critically, PiiMS also indexes the acquired data to the Arabidopsis genome allowing the performance of both forward and reverse genetics in silico. By Web-enabling PiiMS, we have also allowed open access to the data necessary for such in silico genetics. However, PiiMS is only the first step in our goal to develop an integrated functional genomics platform. To further enhance our ability to utilize PiiMS to do functional genomics, we are currently developing integrated middleware to interface PiiMS to a suit of data analysis, data reduction, and visualization tools built around predefined and dynamic workflows. These will include tools for data normalization, statistical analysis, clustering, and classification and the evaluation of segregating populations and genetic models. Using Web services, we also plan to integrate the PiiMS ionomics dataset with other Arabidopsis resources. Finally, we are taking the basic architectural principles of PiiMS and generalizing them across other organisms, including rice (Oryza sativa) and yeast (Saccharomyces cerevisiae), as well as other omics technologies, including proteomics and metabolomics.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Harvesting Stage portal.
Supplemental Figure S2. Drying Stage portal.
Supplemental Figure S3. MS analysis portal, first stages.
Supplemental Figure S4. MS analysis portal, later stages.
Supplementary Material
Acknowledgments
This project is part of a larger collaborative effort funded by the National Science Foundation Plant Functional Genomics and 2010 programs, which includes Mary Lou Guerinot, David Eide, Jeff Harper, David E. Salt, Julian Schroeder, and John Ward. PiiMS is built on the IBM WebSphere Portal Server Enterprise Edition, DB2 Universal Database Server Enterprise Edition, and WebSphere Studio gifted by IBM. We also extend our gratitude to John Burr, Sumantra Nandi, Fan Zhang, Stephan Rohde, Wenkui Wang, Brett Lahner, and Elena Yakubova for their contributions to the development of PiiMS.
This work was supported by the Indiana 21st Century Research and Technology Fund (grant no. 912010479), the National Science Foundation Plant Genome Research (grant no. DBI 0077378) and Arabidopsis 2010 (grant no. IOB 0419695), the National Institutes of Health (grant no. 5 R33 DK070290–03), and Purdue University Discovery Park (e-Enterprise Center, Bindley Bioscience Center, and Cyber Center).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David E. Salt (dsalt@purdue.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14 259–271 [DOI] [PubMed] [Google Scholar]
- Jenkins H, Johnson H, Kular B, Wang T, Hardy N (2005) Towards supportive data collection tools for plant metabolomics. Plant Physiol 138 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MA, van Leeuwen H, Loudet O, Doerge RW, St Clair DA (2006) Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21 1215–1221 [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4 745–750 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104 1173–1184 [DOI] [PubMed] [Google Scholar]
- Meinke D, Koornneef M (1997) Community standards for Arabidopsis genetics. Plant J 12 247–252 [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292 2488–2492 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Crosby B (2005) Biological databases for plant research. Plant Physiol 138 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE (2006) Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet 2 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE (2004) Update on plant ionomics. Plant Physiol 136 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Fan Y, Chang HS, Zhu T, Hazen SP, Briggs SP (2006) A high-resolution map of Arabidopsis recombinant inbred lines by whole-genome exon array hybridization. PLoS Genet 2 e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpunar J (2004) Metallomics: a new frontier in analytical chemistry. Anal Bioanal Chem 378 54–56 [DOI] [PubMed] [Google Scholar]
- Williams R (2001) Chemical selection of elements by cells. Coordin Chem Rev 216–217 583–595 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.