Abstract
Formal description of plant phenotypes and standardized annotation of gene expression and protein localization data require uniform terminology that accurately describes plant anatomy and morphology. This facilitates cross species comparative studies and quantitative comparison of phenotypes and expression patterns. A major drawback is variable terminology that is used to describe plant anatomy and morphology in publications and genomic databases for different species. The same terms are sometimes applied to different plant structures in different taxonomic groups. Conversely, similar structures are named by their species-specific terms. To address this problem, we created the Plant Structure Ontology (PSO), the first generic ontological representation of anatomy and morphology of a flowering plant. The PSO is intended for a broad plant research community, including bench scientists, curators in genomic databases, and bioinformaticians. The initial releases of the PSO integrated existing ontologies for Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and rice (Oryza sativa); more recent versions of the ontology encompass terms relevant to Fabaceae, Solanaceae, additional cereal crops, and poplar (Populus spp.). Databases such as The Arabidopsis Information Resource, Nottingham Arabidopsis Stock Centre, Gramene, MaizeGDB, and SOL Genomics Network are using the PSO to describe expression patterns of genes and phenotypes of mutants and natural variants and are regularly contributing new annotations to the Plant Ontology database. The PSO is also used in specialized public databases, such as BRENDA, GENEVESTIGATOR, NASCArrays, and others. Over 10,000 gene annotations and phenotype descriptions from participating databases can be queried and retrieved using the Plant Ontology browser. The PSO, as well as contributed gene associations, can be obtained at www.plantontology.org.
Angiosperms are one of the most diverse groups of plants that vary greatly in morphology, size, habitat, and longevity. Agriculture is almost entirely dependent on angiosperms. Besides providing food and fiber, angiosperms are important sources for pharmaceuticals, lumber, paper, and biofuel. Understanding the origins, mechanisms, and functions of morphological diversity in flowering plants is one of the fundamental questions in plant biology. Modern approaches to studying plant development integrate classical knowledge in plant anatomy and development with molecular genetics and genomics tools. Among powerful tools, analyses of mutants that affect developmental processes have shed new light on our understanding of the complexity of plant development. More recently, high-throughput, genome-wide phenomic screens in Arabidopsis (Arabidopsis thaliana; for review, see Alonso and Ecker, 2006), and large-scale gene expression-profiling technologies (for review, see Rensink and Buell, 2005) generated a huge amount of data in plant science. These tools and resources have the potential to contribute to efforts to link genes with developmental morphology (i.e. genotype with phenotype) and make an impact on our understanding of functions of genes involved in plant development. However, an accurate interpretation of the function of genes that control various aspects of plant development must be embedded in detailed knowledge of the anatomy and morphology of a plant. Explicitly, the structural features of plant cells, tissues, and organs need to be correctly understood and uniformly described. Accurate and standardized nomenclature for plant anatomy and morphology is also required for comparative purposes (i.e. for comparisons of genes involved in plant development among related or evolutionarily distant taxa). Semantic perplexity presents a major obstacle for conducting such comparative studies in plants; similar plant structures are described by their species-specific terms. For example, in scientific publications, fruit is often referred to as silique in Arabidopsis, grain or caryopsis in rice (Oryza sativa), and kernel in maize (Zea mays). Conversely, the inherent ambiguity of some plant anatomical terms led to the same or similar terms being applied to different structures (e.g. cork cell in the epidermis of grasses and cork cell in the periderm in all other angiosperms).
Standard vocabulary for describing anatomy and developmental stages was developed for several plant species at major plant genomic databases, such as Arabidopsis at The Arabidopsis Information Resource (TAIR; Berardini et al., 2004), rice and other cereals at Gramene (Yamazaki and Jaiswal, 2005), and maize at MaizeGDB (Vincent et al., 2003). These vocabularies have been used to describe gene expression data and mutant or natural variant phenotypes in several plant databases. However, they were developed independently of each other and were based on different principles and rules. In addition, variation in nomenclature used for different taxonomic groups in angiosperms presented obstacles for conducting queries in more than one plant database and retrieving meaningful results. For the purpose of comparative genomics, diverse terminology needed to be organized into a standardized language that could be shared among individual databases and used for accurate description of phenotypes and gene expression data.
To address these problems, the Plant Ontology Consortium (POC; Jaiswal et al., 2005) has developed a simple and extensible controlled vocabulary that describes anatomy, morphology, and growth and developmental stages of a generic flowering plant. In addition, the POC has established a database through which the data curated using this vocabulary can be accessed in a one-stop manner. Here, we describe the first representation of anatomy and morphology of a generic flowering plant, the Plant Structure Ontology (PSO). This ontology represents the morphological-anatomical aspect of the Plant Ontology (PO); the temporal aspect and the Plant Growth and Developmental Stages Ontology have been described elsewhere (Pujar et al., 2006). We also discuss the guiding principles and rationale for the development and maintenance of the PSO and its importance for describing phenotypes and large-scale gene expression data in reference plants and crop species.
DEVELOPMENT OF THE PSO: RATIONALE
An ontology is a concise and unambiguous description of relevant entities and their explicit relations to each other (Schulze-Kremer, 2002). Entities (terms) are linked by specific relationships, with paths from more specific to more general terms upward in the ontology tree. Thus, the information from one hierarchical level can be propagated up to the next level, allowing users to make inferences and to perform queries at different levels. Each term in the ontology has a textual definition, an accession number (identifier or ontology ID), and a specific relationship to at least one parental term (Bard and Rhee, 2004). The unique identifier and relationships in the ontology are interpretable by a computer, which makes possible computational processing and retrieval of information associated with each term. For example, lists of genes annotated to the terms in an ontology can be compared and terms that are overrepresented in one list over another can be determined using statistical tests. This underscores their main appeal in the field of biology and, to a large extent, explains the recent increase in the number of bioontologies (Blake, 2004).
The best known bioontology, the Gene Ontology (GO), was the first to offer a practical solution for describing gene products in a human- and computer-comprehensible manner spanning diverse taxonomic groups (Gene Ontology Consortium, 2006; http://www.geneontology.org). The GO consists of three mutually independent ontologies; each describes cellular components, biological processes, or molecular functions that occur in organisms. Over the years, the GO has become a standard for describing functional aspects of gene products in a consistent way in various genomic databases. Following the GO paradigm and embracing the idea of generic, standardized terminology that can be used across diverse taxonomic groups, the POC has largely adopted the ontology design model and rules established by the GO consortium. However, the PSO is conceptually different and is governed independently from the GO. Some important differences between the PO and GO are discussed in more detail below.
The PSO is the first multispecies ontology of plant anatomy and morphology. Its main purpose is to provide a standardized set of terms describing plant structures—a tool for annotation of gene expression patterns and phenotypes of germplasms across angiosperms. Hence, this vocabulary is intended for a broad plant research community, including curators in genomic databases, bioinformaticians, and bench scientists. The PSO initially integrated existing species-specific ontologies for Arabidopsis, maize, and rice; however, it is not intended only for a few model plant organisms. Rather, we envision it as a continuously expanding ontology that will gradually encompass crop species and woody species. Recently, the ontology has been expanded to include terms for Fabaceae, Solanaceae, additional cereal crops (wheat [Triticum aestivum], oat [Avena sativa], barley [Hordeum vulgare]), and poplar (Populus spp.), a model plant organism for woody species.
A common set of criteria was established to ensure that the PSO would be biologically accurate and adequately meet practical requirements for annotation. Analysis of the three original species-specific plant ontologies—predecessors of the PSO—greatly influenced our decisions on the rationale and design for the PSO. Foremost, we defined the scope of this ontology to be limited to anatomical and morphological structures pertinent to flowering plants during their normal course of development. Botanical terms, from the cellular to the whole organism level, are entities (i.e. terms [in italics in this article]) in the PSO. Besides this main criterion for creating a term, in some cases (following annotation requirements), we have considered derivation (i.e. origin of plant parts and cell lineages, as well as spatial/positional organization of tissues, organs, and organ systems of a flowering plant (e.g. leaf abaxial epidermis and leaf adaxial epidermis).
We established general rules for deciding when not to add terms to the ontology. To a great extent, qualifiers (or attributes) of the terms are avoided, and the ontology makes only very limited use of attributes. Thus, the term corolla is included, but the terms “sympetalous corolla” and “apopetalous corolla” are not. Attributes that are specific for describing mutant plants (e.g. wrinkled seed) are also excluded. Because it does not include attributes, the PSO is insufficient as, nor is it intended to be, a taxonomic vocabulary on its own and does not address phylogeny of angiosperms. Moreover, the most granular terminology in the PSO is at the cell-type level. Therefore, terms for subcellular compartments are not included in the PSO. These terms are handled by the GO Cellular Component ontology. In addition, temporal landmarks (i.e. morphological and anatomical changes that occur via developmental progression of organs and organ systems) are excluded from the PSO; this aspect is a part of the Plant Growth and Developmental Stages Ontology (Pujar et al., 2006). Nonetheless, some temporal aspects are indirectly present in the PSO. Unlike in animal systems, most plant organs are developed in the postembryonic phase of the life cycle. Many plant structures develop continually, whereas others exist only temporarily; that is, at a particular time during the life cycle. Structures that exist even in a very short period of time, such as a leaf primordium, are included as terms in the PSO. For example, terms such as apical hook (defined as a hook-like structure that develops at the apical part of the hypocotyl in dark-grown seedlings in dicots) and leaf primordium (defined as an organized group of cells that will differentiate into a leaf that emerges as an outgrowth in the shoot apex) exist in the PSO. A leaf primordium is merely the first visible appearance of a leaf and, therefore, both terms, leaf and leaf primordium, describe the same entity (leaf) at different time points in development. There are genes that are expressed in organ primordia, such as JAGGED and FILAMENTOUS FLOWER genes in Arabidopsis (both expressed in leaf, sepal, petal, stamen, and carpel primordia) with expression levels declining in the developing or adult organs (Dinneny et al., 2004). To accurately annotate expression patterns of such genes, we created separate terms for each primordium structure. Currently, the PSO has 11 such terms.
To integrate terms from different species, we extensively used synonymy wherever feasible. This allows users to search existing plant databases using either a generic term or its taxon-specific synonyms. For example, silique, caryopsis, and kernel are listed as synonyms of the term fruit. Therefore, a search for fruit in the PO database would retrieve all genes expressed in the silique of Arabidopsis, caryopsis of rice, and kernel of maize. In reality, silique, caryopsis, and kernel are types (classes) of fruit, rather than strict synonyms. However, for the purpose of this ontology, specific types of a few high-level terms (e.g. fruit, inflorescence, and stem) are included only as synonyms. Thus, we intentionally overlooked an enormous morphological diversity of flowering plants in favor of cross species comparisons, generic searches, and intuitive ontology browsing. Therefore, synonyms in the PSO can be taxon-specific morphological forms of a generic structure. Also, an entity in the PSO can either be a term or a synonym, but not both. In a few cases where synonymy was not a suitable option, we created new terms as specific classes. Typical examples are the terms tassel and ear, staminate and pistilate inflorescences specific to the genus Zea, respectively. In addition to the synonyms described above, the PSO contains a number of terms that have authentic (exact) synonyms. Examples include the terms male gametophyte (synonym: pollen grain), female gametophyte (synonym: embryo sac), or perisperm (synonym: seed nucellus). Extensive use of synonymy in the PSO resulted in reduced granularity (i.e. the degree of detail in the ontology) and emphasized generic aspects of the ontology. As a rule, a high level of granularity was limited in the PSO because we strove to keep the ontology relatively simple, yet sufficiently broad and generic to encompass a number of flowering plants.
CONTENT OF THE PSO
A term (also called a node) in the PSO is an entity that represents a component of plant structure, such as cell, tissue, organ, and organ system. Each plant structure in the PSO has a term name, a unique numerical identifier (accession no.), a definition, and a specified relationship to at least one other term. An accession number always starts with the PO prefix followed by seven digits (e.g. PO:0009011). Once assigned to the PO term, the accession number never changes or gets reassigned to another term. Users should always cite an ontology term by its exact name and a complete accession number, including the prefix. Similar to the GO, the PSO is organized into a hierarchical network called the Directed Acyclic Graph (for definition, see http://www.nist.gov/dads/HTML/directAcycGraph.html). Three types of parent-child relationships are used in the PSO to specify the type of association between two terms: is_a, part_of, and develops_from (described in more detail in Jaiswal et al., 2005).
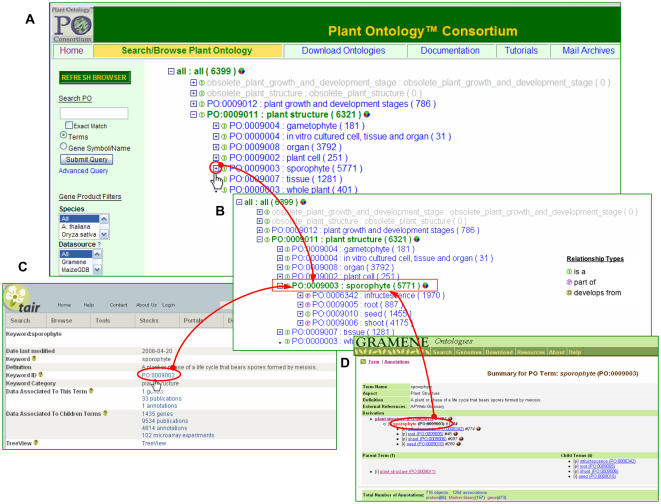
The term plant structure (PO:0009011) is the highest level of the PSO. Each term immediately below plant structure represents high-level structures (broadly defined entities) that contain specific classes or types, positioned in the hierarchy as their direct descendants, called children terms. There are five direct children of plant structure: plant cell, tissue, organ, gametophyte, and sporophyte (Fig. 1). The remaining two nodes, in vitro cultured cell, tissue, and organ and whole plant, were originally included in all three plant species-specific anatomical ontologies that preceded the PSO. Because these terms were used in annotations by all three databases, we included them as top-level nodes. The latter node, whole plant, is conceptually inconsistent (not a botanical term) from the rest of the terms in the PSO and is intentionally left without children terms. We recommend that this term be used as a last option—only when precise annotation to any other term in the PSO is not possible. Sporophyte and gametophyte exist as separate terms because they represent diploid and haploid generations of the plant life cycle, respectively. The largest node, sporophyte, includes seed, root, shoot, and infructescence as direct children nodes. The term shoot is broadly defined as part of the sporophyte composed of the stems and leaves and includes shoot apical meristems. It has phylome, stem, and inflorescence as part_of children terms, and rhizome, shoot borne shoot, root borne shoot, stolon, and tuber as specific types of a shoot. The term embryo (part_of seed) consists of a number of terms that are applicable for both eudicots and monocots (particularly members of Poaceae). Compared to eudicots, embryo development in grasses is more advanced; a fully developed embryo has body parts, such as coleoptile, coleorhizae, and scutellum, which are nonhomologous or absent in eudicots. Because no plant embryo has all body parts that are designated as part_of embryo in the PSO, we adopted a nonrestrictive part_of relationship type; the child must be a part_of the parent to exist in the ontology. However, a parent structure does not have to be composed of all of its part_of children. For example, scutellum is necessarily part_of embryo; that is, wherever scutellum exists, it is always a part of an embryo. However, not all embryos have scutellum (only embryos in Poaceae do). The high-level term infructescence was created to accommodate terms that describe both simple fruits, formed from a single ovary (e.g. grape [Vitis vinifera]), and compound fruits, formed from multiple ovaries (e.g. pineapple [Ananas comosus] or mulberry [Morus]). Currently, this node has only one direct descendant, fruit, which refers to a simple fruit. Terms specifically describing compound fruit will be included at a later time. Similar to the embryo node, the fruit node contains several part_of children and not every fruit type necessarily has all part_of descendants. Overlapping subsets of part_of terms can be created, each applicable to siliques of Arabidopsis and other Brassicaceae, caryopsis in cereals, and berry, a fleshy type of fruit, in tomato (Solanum lycopersicum) and other Solanaceae.
Figure 1.
A, A screen shot of the ontology browser; top nodes of the PSO. Clicking on the [+] or [−] sign in front of a term vertically expands or collapses the ontology tree, respectively. B, Expanded sporophyte node. A mouse click on a term itself opens a term detail page (data not shown). Numbers in parentheses next to the term name indicate the number of the annotations for unique object types associated with a term (including annotations to all children terms). Legends for relationship-type icons are shown on the right. Term detail pages for the PSO term sporophyte at TAIR (C) and Gramene (D) databases. Each database has hyperlinks back to the PSO, allowing for quick access to the POC browser and database. At species-specific databases, additional annotations to the term sporophyte can be found.
Recently, terms relevant for the Solanaceae and Fabaceae families, perennials, and woody species were added to the PSO. For example, terms such as tuber (and its children terms subterranean tuber and aerial tuber) and root nodule (with children terms adventitious root nodule, determinate nodule, and indeterminate nodule) were added to accommodate annotations to genes and germplasms in the Solanaceae and Fabaceae families. In addition, the first attempt to add terms relevant for perennials and woody species was made (such as epicomic shoot, defined as a shoot developing from a trunk), with more terms still to be incorporated. A number of terms for secondary growth were also added, grouped under secondary xylem (such as heartwood, sapwood, growth ring, growth ring boundary, and others), secondary phloem (such as bark, libriform fiber, septate fiber, and phloem fiber), and vascular cambium (such as ray initial and fusiform initial), including several cell-type terms under parenchyma cell, such as wood parenchyma cell, with direct descendants, axial wood parenchyma cell, and ray wood parenchyma cell (with additional children terms underneath).
At the very top level of the hierarchy is the node obsolete. As in the GO, a term that has been removed from the ontology is never permanently deleted. Instead, the term and its assigned identifier are kept in the ontology file for the record. The definition is appended with OBSOLETE and an explanation is provided as to why a term was removed. The note in the definition or comment field might also contain suggested terms for searching and annotating. Obsoleted terms are not intended for use. Consequently, obsoleted terms do not have any annotations associated with them. In many cases, terms in the obsolete node are valid botanical terms (such as tunica and corpus); they are simply no longer in use in the PSO, mainly to avoid having duplicated terms that describe a similar plant structure. Instead of using the outdated concept of tunica and corpus, shoot apex organization is described by the following terms: central zone, peripheral zone, and rib zone. Other examples include terms depicting plant-specific subcellular structures (e.g. filiform apparatus), all of which were made obsolete in the PSO to avoid overlap with the GO. Users are advised to use cellular component terms in the GO instead.
COMPARISON OF THE PSO TO OTHER BIOLOGICAL ONTOLOGIES
Currently, the PSO is the only available morphological-anatomical ontology that is pertinent to more than one organism. Original Arabidopsis Anatomy Ontology from TAIR and Cereal Plant Anatomy Ontology from Gramene were retired, whereas maize vocabulary is still in use at the MaizeGDB (together with the PSO). Species-specific vocabularies describing anatomical features and developmental stages have been developed for animals, among others, fruit fly (Drosophila melanogaster; FlyBase Consortium, 2002), zebrafish (Sprague et al., 2003), and mouse (Burger et al., 2004; Hayamizu et al., 2005), and for human (Hunter et al., 2003), see Open Biomedical Ontologies (OBO; http://obo.sourceforge.net/browse.html). Unlike the PSO, anatomical vocabularies for fruit fly and vertebrates have developmental components (i.e. stages) and are much larger. For example, the Drosophila anatomy ontology has over 6,000 terms, whereas the mouse adult anatomy ontology has 2,700 terms.
OBO stipulate that large overlaps between bioontologies should be avoided, with an idea that terms from orthogonal (mutually independent) ontologies can be combined to make more complex ontologies. We attempt to minimize and, ultimately, anticipating software implementation, completely eliminate overlaps of the PSO with other bioontologies under the OBO umbrella. Because the GO includes terms for subcellular structures, the PSO excludes them. Several terms describing plant subcellular structures were made obsolete in the PSO and were introduced to the GO. Subsequently, GO IDs were added in the comment section for these terms to properly inform the users. Examples include several terms describing pollen wall components and also the term filiform apparatus. Inevitably, an apparent overlap between the plant cell node (PO:0009002) in the PSO and the plant_cell (CL:0000610) in the Cell Ontology (Bard et al., 2005) still exists; we are looking for the best solutions to eliminate this overlap.
ANALYSIS OF THE PSO AND DISTRIBUTION OF ANNOTATIONS
Compared to the GO and other anatomical ontologies, the PSO is a rather small ontology. The top-level term (also called root node), plant structure (PO:0009011), has 726 children terms (release PO_0906; Table I), of which 384 (or 53%) are leaf terms, also called terminal nodes (the most specific terms with no children terms below), and 342 (47%) interior nodes (terms with children). In addition, the PSO currently has 304 synonyms assigned to 149 terms. The relatively small size of the PSO reflects the generic nature of the ontology; often, the most granular terms are specific to taxonomic groups and are included only when necessary (i.e. to retain biological accuracy and to comply with annotation requirements). Having reached a balance between broadness and granularity, the PSO is a stable and inclusive vocabulary. All of the top nodes, with the exception of the infructescence, are populated with necessary terms to describe the phenotypes and gene expression data in angiosperms that are currently being annotated.
Table I.
Statistics of PSO terms and annotations, as of August 31, 2006, release PO_0906
n.a., Not applicable.
| No. | % Total | |
|---|---|---|
| Terms | 727 | 100 |
| Terminal or leaf terms | 384 | 53 |
| Interior terms | 342 | 47 |
| Terms with synonyms | 149 | 20 |
| Total no. of synonyms | 304 | n.a. |
| Annotated terms | 423 | 58 |
| Directly annotated terms | 321 | 44 |
| Annotated leaf terms | 156 | 21 |
| Total annotations | 10,146 | 100 |
| Total annotations to leaf terms | 1,480 | 15 |
| Total leaf term annotations excluding whole plant | 1,075 | 11 |
| Annotations to interior terms | 8,666 | 85 |
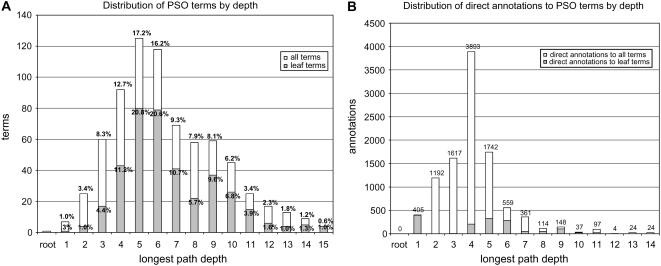
We analyzed the structure of the PSO and the distribution of annotations to the PSO terms to assess the breadth, depth, and current usage of the ontology. The depth of a term was defined as the number of nodes in the longest path from the root to that term. Distribution of the depths of the terms in the PSO is shown in Figure 2A. The mean and mode of the depth in the ontology was 6.5 and 5, respectively, indicating that the majority of the terms were fairly granular. The longest depth was 15, with the majority of the leaf terms (86%) having the depth between three and 10 (Fig. 2A). To some extent, this variability is due to the nature of the domain that the PSO describes (i.e. anatomy and morphology of an angiosperm). Certain morphological structures of an angiosperm are more complex, resulting in deeper depths (such as flower or leaf), whereas others are much simpler (such as male gametophyte and female gametophyte). The pattern of distribution for terminal terms was similar to that for interior terms.
Figure 2.
Analysis of the ontology and annotations in the PSO. A, Distribution of all PSO terms (white) and leaf terms (gray) as a function of longest path depth from the root. B, The number and distribution of direct annotations as a function of longest path depth from the root.
The number and distribution of the annotations at different depths of the ontology are a measure of the usage of the ontology, indicating how adequate the depth of the ontology is for the annotations of gene expression data and phenotypic descriptions. Because annotation to the most granular terms is the ultimate curation goal, we analyzed the current distribution of direct annotations across the PSO and distribution of annotation to leaf terms (Fig. 2B). The majority of direct annotations (83%) are made to nodes with a depth between two and five nodes, indicating that terms with more granularity (with a path depth of seven or more nodes) are less frequently used for direct annotations. Direct annotations to leaf terms are distributed between terms of depth between four and 11, with the exception of 405 annotations to the top-level term whole plant (Fig. 2B). Because this term does not have any children, it appears as a terminal term in the PSO at the first node. However, it is not a granular term and is excluded from further analysis. Only 155 leaf nodes, or 41% of total leaf nodes (excluding whole plant node), have direct annotations (1,075 annotations), counting for 11% of total annotations to the PSO terms (Table I). Close to 90% of the annotations are made to nonleaf terms and the majority of the leaf terms are not currently used in annotations. This suggests that the granularity of the ontology seems to be sufficient for the majority of the branches in the ontology. These data may also be indicative of the extent of knowledge of gene expression and phenotype characterization and could be further analyzed to determine which aspects of the ontology are less well studied than others. It is also possible that the distribution of the annotation reflects the extent of curation efforts in contributing databases and could be used to strategize directions in curation efforts. Finally, it may also reflect the current state of the technology used for gene expression data. Commonly available technology for measuring gene expression data (e.g. microarray technology, northern blots, reverse transcription-PCR) are most frequently applied to organs and organ systems, which are high-level terms in the ontology. This is not necessarily true for in-depth analyses of mutant phenotypes, even though a large number of phenotypic descriptions are generated in greenhouses or in the field, where observations are made using limited tools. As new technologies become more available for plant researchers, such as laser-capture microdissection, which allows for the procurement of specific cells of nearly any plant tissue, more granular terms in the PSO will likely be used for annotations.
APPLICATIONS OF THE PSO AND AVAILABILITY OF ANNOTATIONS
Several plant databases now use the PSO as the main ontology for annotating gene expression data and for describing phenotypes. TAIR and Gramene retired their species-specific anatomical ontologies for Arabidopsis and cereals, respectively, and have been using the PSO exclusively. In MaizeGDB, the original maize vocabulary (Vincent et al., 2003) has been partially integrated with PSO terms, and the goal is to complete this integration in the near future. At this time, both sets of terms are used for annotations, which can then be queried using PSO or maize-specific term names. The PSO is currently implemented in several genomic databases and is displayed at TAIR (www.arabidopsis.org), Gramene (www.gramene.org), Nottingham Arabidopsis Stock Centre (NASC), the European Arabidopsis center (www.arabidopsis.info), BRENDA, the comprehensive enzyme information system (www.brenda.uni-koeln.de), and the MaizeGDB (www.maizegdb.org).
The POC database is set up as a portal through which the data curated using PO for different plant organisms, such as Arabidopsis, rice, and maize, can be easily accessed at one site. Information from one hierarchical level in the ontology is propagated up to the next level (i.e. annotation to any given term with is_a or part_of relationship type implies automatic annotation to all ancestors of that term). Therefore, users can make inferences and perform queries at different levels in the PSO. For example, all Arabidopsis, rice, and maize genes expressed in the flower and phenotypes with altered floral development can be retrieved not only by a search using the term flower, but also by a search using the term inflorescence, of which the flower is a part. Also, a search with the term flower should retrieve all genes expressed in stamens, pistils, petals, or sepals. To elucidate the primary application of the PO, the annotation process in contributing databases is described below, followed by specific examples of how scientists can efficiently use the PSO in their research.
Annotations to the PSO in Participating Databases
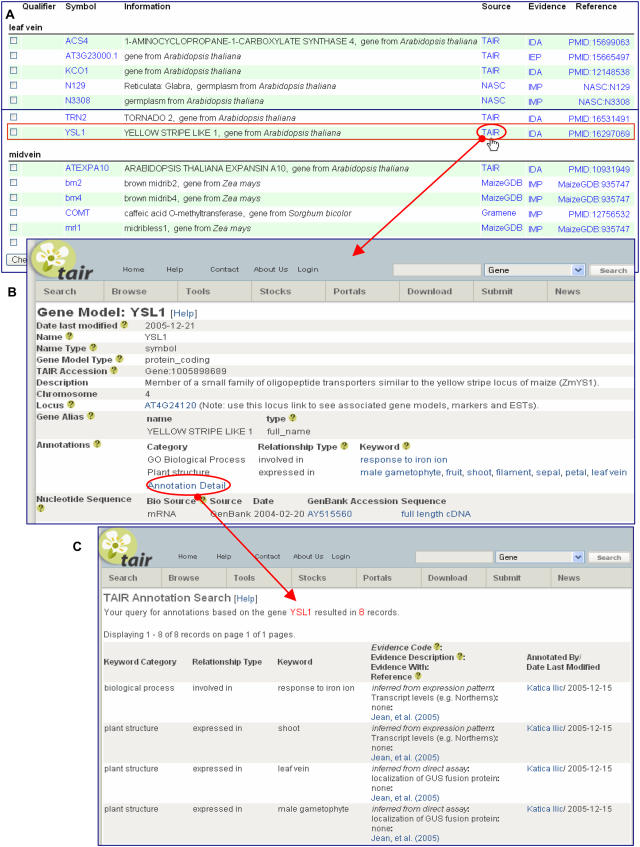
A user interested in genes involved in leaf vascular development can query the PSO by entering an appropriate term, for example, leaf vein, in the PO browser and retrieve all annotations to this term and its children terms (midvein and secondary vein) in Arabidopsis, rice, and maize. The list includes genes that are expressed in leaf veins as well as phenotypes with altered leaf vein development. Annotations to this term are contributed by TAIR, NASC, and Gramene (Fig. 3A). The user can obtain more information about each gene or germplasm by clicking on the name of the contributing database (Source), as shown for the Arabidopsis YELLOW STRIPE LIKE 1 (YSL1) gene, annotated by TAIR (Fig. 3B).
Figure 3.
A, Annotations to the term leaf vein (PO:0020138) in the POC database. Hyperlinks in the Source column take the user to the gene page in the database where the annotation was created. Encircled is a hyperlink to TAIR database for the annotation of Arabidopsis YSL1 gene. B, Gene detail page at TAIR for the YSL1 gene, where additional information about the YSL1 gene can be retrieved. C, TAIR annotation detail page for YSL1 gene, where all annotations to the PO and GO are listed with the evidence code and evidence description for each annotation entry.
Functional annotation of a gene, which is an association between a gene and a term in an ontology, summarizes information about its function at the molecular level, its biological roles, protein localization patterns, and spatial/temporal expression patterns (Berardini et al., 2004). Generally, annotation tasks are carried out at genomic databases, by manual or computational methods. All annotations contributed to the POC are composed manually by curators (biologists with an advanced degree) who either extract the information from published literature and generate concise statements by creating gene-to-term associations (Berardini et al., 2004; Clark et al., 2005) or record phenotype descriptions directly by observing plants (natural variants and mutants) in greenhouses or in the field. Literature curation is usually conducted at species-specific genomic databases (TAIR, Gramene, and MaizeGDB). Curators at plant stock centers, such as NASC, Arabidopsis Biological Resource Center, and Maize Genetics Cooperation Stock Center, often combine their in-house description of germplasms, based on greenhouse observations and/or stock donor information, with information available from the literature. Each gene-to-term association is a separate annotation entry and a gene can be annotated with several ontology terms. For instance, the YSL1 gene in Arabidopsis is annotated to multiple PO terms in TAIR (Fig. 3, B and C). YLS1 is expressed in male gametophyte, fruit, shoot, filament, sepal, petal, and leaf vein, with evidence codes inferred from expression pattern (IEP) and inferred from direct assay (IDA), extracted from the publication by Jean et al. (2005). Evidence codes are defined types of evidence, which are used to support the annotation. Most commonly used evidence codes for annotating gene expression data and phenotypes are IDA, IEP, and inferred by mutant phenotype (IMP). In addition to the evidence code, TAIR provides evidence description, which depicts more specific assay types for supporting the annotations. For instance, YSL1 is expressed in the shoot, with evidence code IEP and evidence description transcript levels (e.g. northerns; Fig. 3C). Details on evidence codes and evidence descriptions can also be found online (http://www.plantontology.org/docs/otherdocs/evidence_codes.html). More details on literature curation using controlled vocabulary and components of annotations can be found elsewhere (Berardini et al., 2004; Clark et al., 2005). Each contributing database has developed its own annotation interface and has taken different approaches to displaying gene and phenotype annotations. However, association files contributed to the POC Concurrent Versions System repository are uniformly formatted and are compliant to POC standards.
Use of the PSO in Gene Expression and Protein Localization Experiments
Besides gene annotations, another common application of the PSO is in categorizing experiments and describing biological samples. For example, databases containing large-scale gene expression profiling data, such as GENEVESTIGATOR (Zimmermann et al., 2004) and NASCArrays (Craigon et al., 2004), are using the PSO to show genes that are expressed in certain plant structures and to describe microarray experiments, respectively. The Plant Expression Database (Shen et al., 2005) is currently incorporating PSO terms in their microarray experiment sample description and also in their data submission forms (R. Wise, personal communication). Similarly, ArrayExpress plans to implement PSO terms in the near future (H. Parkinson, personal communication). NASCarrays uses PSO terms to describe tissue sample sources used in microarray experiments (as BioSource Information; Supplemental Fig. S1).
Researchers can, and are encouraged to, use the PSO for describing tissue samples for various transcript analyses (e.g. northern blot/reverse transcription-PCR, β-glucuronidase/green fluorescent protein, in situ mRNA hybridization), protein localization experiments (e.g. immunolabeling, proteomic data), and gene expression assays from microarray experiments or laser-capture microdissection experiments in their publications and Web sites. Descriptions of other expression data, such as expressed sequence tags (ESTs) and cDNA libraries, can be enhanced by using proper botanical terms and accession numbers from the PSO. These datasets are submitted to dbEST at the National Center for Biotechnology Information (NCBI) and consistent use of standardized anatomical terms can greatly improve cross species comparison. For instance, a user interested in finding all ESTs from EST libraries generated from pollen grains across plant taxa could query the NCBI GenBank using the unique ID for the PSO term male gametophyte (synonym: pollen grain), PO:0020091, and retrieve all ESTs generated from pollen tissue samples. Currently, such a query is not feasible at the NCBI; instead, a search for the words pollen AND plant retrieves all EST entries in which both words, pollen and plant, appear anywhere in the text. The Gramene database has already started using the PSO for tissue-type description of 201 EST and cDNA libraries for cereals obtained from dbEST. The list of libraries and the links to the PSO terms can be viewed at http://www.gramene.org/db/ontology/association_report?id=PO:0009011&object_type=Marker%20library.
In summary, the consistent use of PSO terms across different plant species and use of available annotations of gene expression data and phenotype descriptions are valuable aids to bench scientists and can facilitate new discoveries. Researchers involved in large-scale expression profiling projects or those who generated mutant collections and are creating their own databases to store phenotypic data are encouraged to use the PSO. The POC has already been contacted by a number of such laboratories with questions on how to use the ontologies for describing tissue samples in EST collections, laser-capture microdissection experiments, microarray experiments, and mutant phenotype collections. We are continuously making an effort to reach out to our prospective users and to meet the particular annotation needs of the collaborating databases, as well as the needs of the broader plant research community. Users are encouraged to contact the POC to get help, contribute their feedback, and suggest new ontology terms by writing to po-dev@plantontology.org.
Comparison of Gene Expression and Phenotype Data in Arabidopsis, Rice, and Maize
The data curated using the PSO, contributed by participating plant databases, can be easily accessed by performing one-stop queries in the POC database. As of August 31, 2006, the database has over 4,400 unique genes and nearly 1,900 germplasms annotated with PSO terms, with a total of over 10,000 associations, contributed by TAIR, Gramene, MaizeGDB, and NASC. Annotations are displayed and can be queried using the PO browser tool (http://www.plantontology.org/amigo/go.cgi), a modified AmiGO tool (see “Materials and Methods”). A user interested in genes involved in inflorescence development and their comparison between grasses (rice and maize) and Arabidopsis can search for the term inflorescence (PO:0009049) and retrieve all gene annotations and phenotypic descriptions associated with this term. Direct annotations to the PSO term and annotations to all its children terms are displayed on the term detail page. Hyperlinks to the original publications from which annotations were extracted provide quick access to the original experimental data and methodology, which, combined with a direct link to the gene and locus detail pages at contributing databases, leads to quick access to deposited DNA and protein sequences. Also, on the gene detail pages at Gramene and TAIR, functional annotations with GO terms are displayed and hyperlinked to the GO, providing access from the PO to the GO through these links.
The gene expression data available at the POC Web site combined with sequence similarity and phylogenic analysis can facilitate comparative structural and functional studies of related plant genes. Although it is yet to be experimentally verified that the evolutionary conservation among plant genomes is manifested by functional similarity, such as distinct overlapping expression patterns of orthologous genes, available annotations of gene expressions can be used as a starting point in such studies. This approach can be particularly useful for orthologs in maize and rice, considering their evolutionary relatedness (i.e. their monophyletic origin) and, to some degree, also for comparison to their putative orthologs in Arabidopsis. A known example is the study of functional complementation and overlapping expression patterns of the vp1 gene in maize and its Arabidopsis ortholog ABI3, both genes involved in seed maturation and germination (Suzuki et al., 2001). ABI3 is expressed in the Arabidopsis embryo and seed coat (TAIR), whereas germplasm of the maize vp1 mutant is annotated to the PSO term fruit (MaizeGDB). Thus, the query for the term fruit, of which the seed is a part, using species-specific filters for Arabidopsis and maize (available on the PO browser) would retrieve all genes/germplasms annotated in these two species, including vp1 and ABI3. Although the PO database does not yet have tools to address orthology or even sequence similarity in rice, maize, and Arabidopsis, annotation data available at the POC Web site can be used as a starting point for detailed studies of the function and expression of putative orthologous genes in rice and maize and their corresponding homologs in Arabidopsis. Web sites such as InParanoid provide orthology information for sequenced eukaryote genomes (O'Brien et al., 2005) and could be used in combination with the POC to address these questions.
Extended Annotation of Mutant Phenotypes Using Controlled Vocabularies
Describing a phenotype is a complex task; to capture relevant biological information about an entire set of characteristics of an organism, one needs to consider all observable (measurable) traits, qualitative and quantitative, the type of assays, and specific experimental conditions in which interaction of genotype and environment occurs. Traditionally, curators at plant genomic databases have relied on the free-text description (usually as a short summary), often combined with images of mutant phenotypes and natural variants. This approach largely limits data manipulation and searches and prevents easy comparison across species.
PSO is an essential ontology to use to move toward more systematic annotation of phenotypes. However, it depicts the plant structures only during normal development of a plant. It does not include terms that describe morphological variations of cells, tissues, and organs in mutated plants (e.g. fasciated ear) or qualitative and quantitative descriptors (e.g. type of branching, trichome shape, spikelet density). Thus, additional ontologies are required for capturing relevant biological information about phenotypes fully. If used exclusively, the PSO would be insufficient to capture all of the details of a phenotype in a controlled vocabulary format.
Recently, the NASC, Gramene, and MaizeGDB moved toward combining PO terms with other ontologies to annotate mutant phenotypes and natural variants to allow computation and more efficient cross species comparison. At Gramene, PO terms are used in conjunction with Trait Ontology terms (Yamazaki and Jaiswal, 2005) to describe phenotypes. As an example, the phenotype description of the allele cg.1, cigar shape panicle (cg) gene in rice (Seetharaman and Srivastava, 1969; Prasad and Seetharaman, 1991) is shown in Supplemental Figure S2A. This mutation affects the morphology of a panicle, rachis, and grain (see the text description in Supplemental Fig. S2); thus, the annotations were made to PSO terms inflorescence (PO:0009049), stem (PO:0009047), and seed (PO:0009010). In addition to PO terms, curators from the Gramene database chose terms from another ontology, Trait Ontology (Yamazaki and Jaiswal, 2005), to annotate the cg.1 allele in rice: panicle type (TO:0000089), seed length (TO:0000146), seed size (TO:0000391), and stem length (TO:0000576).
A different approach has been taken by the NASC database for describing mutant phenotypes and natural variants in Arabidopsis. In addition to a free-text description, short statements, referred to as an entity, attribute, value (EAV) description, are composed by combining terms from orthogonal (i.e. nonoverlapping) ontologies. This model has been tested in pilot projects at a few model organism databases, namely, ZFIN (Sprague et al., 2003) and FlyBase (FlyBase Consortium, 2002). The EAV model relies on the Phenotype and Trait Ontology (PATO)—a species-independent controlled vocabulary created as a schema in which the qualitative phenotypic data are represented as nouns and phrases (Gkoutos et al., 2005). The core of the PATO is composed of a set of attribute and value terms (such as color, shape, and size; green, serrate, and dwarf), which are recently converted to a single hierarchy of qualities (G. Gkoutos, personal communication). At the NASC database, the allele ckh1-1 (in Landsberg erecta background), a mutation of the CYTOKININ-HYPERSENSITIVE 1 gene in Arabidopsis, is annotated to the PO terms inflorescence (PO:0009049) and to the PATO term ShortHeight-Value (PATO:0000569), creating the following syntax: inflorescence:short:height. An additional annotation to primary root (PO:0020127) is followed by ShortLength-Value (PATO:0000574), creating the syntax primary root:short:length. Thus, multiple controlled vocabulary statements can be created for any germplasm/seed stock.
Presently, the POC database and ontology browser are not set up to display annotations to multiple ontologies. Therefore, controlled vocabulary annotations to ontologies other than the PO can be viewed on gene/germplasm/stock detail pages at contributing databases, which can be accessed by clicking on the appropriate database link (Supplemental Fig. S2B). More details on using the Trait Ontology and the PATO and EAV model can be found at the Gramene and NASC Web sites, respectively. Whereas Trait Ontology is plant specific and was created for the purpose of annotating mutants in rice and other cereal crops, PATO ontology is species independent and intended for description of mutant phenotypes across kingdoms. PATO terms can be used in combination with a wide range of other ontologies that describe entities, such as GO, Cell Ontology, and anatomical and developmental stage ontologies, among others.
CHALLENGES FOR FUTURE DEVELOPMENT OF PLANT ONTOLOGIES
Proliferation of Terms
A major concern for the PSO is the proliferation of terms. The number of terms needs to be large enough for precise annotation of genes and phenotypes, but small enough for curators and end users to navigate the ontology easily. The terminology for describing plants is rich and complex and is often species or family specific. Available visualization and editing software portrays the ontologies as strictly hierarchical, whereas plant structure is not. Rather, it is modular in nature, with a relatively small number of tissue and cell types recurring, often with slight modification, in different organ systems at different times during development. Converting a modular structure to a formal hierarchy requires extensive redundancy in the ontology. For example, a flower might be a part of a cyme, a raceme, or any other inflorescence types. To maintain the appropriate upward flow of information through the hierarchy, we would need to create a term specifying a distinct type of flower within each inflorescence type. Thus, we faced the possibility of creating the terms flower of cyme, flower of raceme, flower of panicle, etc., followed by stamen of flower of cyme, gynoecium of flower of cyme, etc. With inflorescence and fruit types, we solved the problem by placing all of the different inflorescence and fruit types as synonyms of inflorescence and fruit, respectively. This effectively removed one hierarchical level from the ontology at these positions (Supplemental Fig. S3).
Synonymy was not appropriate to account for staminate and pistillate inflorescences of Zea, which are physically separate and morphologically distinct (monoecious) from each other. The two types of inflorescence often have different phenotypes in single-gene mutants and identical genes are often deployed differently in each. Maize geneticists thus often want to be able to distinguish these two. Therefore, the maize ear and tassel are the only two inflorescence types that are treated as a type of inflorescence.
The solution by synonymy does not fully eliminate the problems with proliferation of terms. Users of the ontology will find extensive residual redundancy in some areas. Ultimately, new visualization and ontology editing software and a different approach to creating ontologies will be needed to reflect the modularity of biological reality more precisely and intuitively.
Homology Assessment and Taxon-Specific Forms
The PSO is designed to be a practical tool for annotating genes and germplasms and to be, as far as possible, neutral on questions of homology. Thus, for example, the terms cotyledon and scutellum are not treated as synonyms, even though there is a body of thought that suggests that they might be derived from the same sort of ancestral structure. As our knowledge of plant structure continues to develop, however, some of these terms may be merged.
More problematic, but also perhaps more interesting, are structures that are unique to particular clades of plants. These are currently accommodated by the sensu designation, but as major groups are added, the number of such terms is likely to increase. For example, stipules are considered to have arisen independently in multiple lineages and they may prove to be developmentally and genetically distinct. If true, in addition to a common term stipule (PO:0020041), the PO could be faced with multiple terms such as stipule sensu Rubiaceae, stipule sensu Fabaceae, stipule sensu Brassicaceae, etc. Handling a phylogenetic relationship is beyond the scope of the PSO currently, but it is an important topic to address in the long run. As more genes are annotated from more species, the PSO may help to discover whether similar structures that have evolved independently are produced by very distinct underlying genetic mechanisms.
MATERIALS AND METHODS
Ontology Development and Maintenance
The PSO was based on three species-specific ontologies, TAIR Anatomy Ontology (Berardini et al., 2004), the Cereal Ontology from Gramene, and the maize (Zea mays) Ontology (Vincent et al., 2003). However, most PSO terms and definitions were adopted from a few well-known textbooks and glossaries. Most definitions come from Plant Anatomy (Esau, 1977) and from the Angiosperm Phylogeny Web site created and maintained by Peter Stevens and the Missouri Botanical Garden (http://www.mobot.org/MOBOT/research/APweb). Definitions were sometimes taken verbatim from references or modified for clarity. Original publications are often consulted. In addition, opinions of plant researchers in respective areas of expertise are periodically sought by the POC. The ontologies were created and edited using the GO ontology editing tool, Directed Acyclic Graph Editor, which is freely available from SourceForge (http://sourceforge.net/project/showfiles.php?group_id=36855).
As part of an ongoing effort to actively maintain the plant ontologies, the POC meets on a regular basis to discuss new terms and ontology structure suggestions. Users are encouraged to use the feedback navigation bar menu option on the POC Web site to suggest new ontology terms or send feedback or contact the POC at po-dev@plantontology.org. Ontology and annotation updates are released on the POC Web site the last week of every month. Each release is indicated by the month and year of the release date (i.e. PO_0906), displayed at the left side of the ontology browser header. POC ontology and association files in the Concurrent Versions System are tagged accordingly to connect the respective flat files with the database release. The same files that are used for each database release are also posted at the SourceForge OBO Web site (http://obo.sourceforge.net/cgi-bin/detail.cgi?po_anatomy). Synchronization between POC ontology releases and participating database releases of PO is handled individually by each database. The individual databases regularly update their PO versions either on a monthly (TAIR) or quarterly (Gramene) basis.
Ontology and Annotation Analysis
To generate statistics for the path depth of PSO terms and annotations, we downloaded, installed, and queried the PO MySQL database version 09/06 (http://www.plantontology.org/download/database). Term depths were determined by querying the number of nodes in the longest path length from the root node. This measure of depth was used so that a parent-child relation would never decrease the depth of a term.
Database and Ontology Browser
We used the GO database schema and ontology browsing tool, AmiGO, for storing and displaying the PSO and its annotations, respectively. The AmiGO browser, a Web-based tool for searching ontologies and their associations developed by the GO consortium, is freely available open-source software. We made minor modifications to make it more suitable to the specific requirements of PO. Modifications of AmiGO pertinent to the general ontology community were contributed to GO. The PO browser accesses the MySQL POC database at Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. The structure of the POC database and main features of the Web site have been previously described (Jaiswal et al., 2005).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A, AtGeneExpress microarray experiment description with tissue sample description using the PSO term cotyledon (PO:0020030) indicated as the tissue source at NASCarrays. B, The PO accession number for the term cotyledon is hyperlinked to the NASC term detail page where a user can find all other annotations to the term cotyledon displayed at the NASC database.
Supplemental Figure S2. Annotations to the PSO term inflorescence.
Supplemental Figure S3. Ontology browser term detail view for the term inflorescence (PO:0009049).
Supplementary Material
Acknowledgments
We acknowledge the Gene Ontology Consortium for software infrastructure and technical support. We thank our two industry collaborators, Monsanto and their Genome Knowledge Enhancement Program, and Pioneer Hi-Bred International, Inc., for contributing their species-specific ontologies. We also thank Quentin Cronk, Rex Nelson, Naama Menda, Victoria Carollo, and William Friedman for their participation in ontology development, as well as numerous researchers and curators who have reviewed the plant ontologies and are listed individually online (http://www.plantontology.org/docs/otherdocs/acknowledgment_list.html).
This work was supported by the National Science Foundation (grant no. 0321666 to the Plant Ontology Consortium).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Seung Y. Rhee (rhee@acoma.stanford.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Ecker JR (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7 524–536 [DOI] [PubMed] [Google Scholar]
- Bard JB, Rhee SY (2004) Ontologies in biology: design, applications and future challenges. Nat Rev Genet 5 213–222 [DOI] [PubMed] [Google Scholar]
- Bard JB, Rhee SY, Ashburner M (2005) An ontology for cell types. Genome Biol 6 R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J (2004) Bio-ontologies-fast and furious. Nat Biotechnol 22 773–774 [DOI] [PubMed] [Google Scholar]
- Burger A, Davidson D, Baldock R (2004) Formalization of mouse embryo anatomy. Bioinformatics 20 259–267 [DOI] [PubMed] [Google Scholar]
- Clark JI, Brooksbank C, Lomax J (2005) It's all GO for plant scientists. Plant Physiol 138 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D (2004) The role of JAGGED in shaping lateral organs. Development 131 1101–1110 [DOI] [PubMed] [Google Scholar]
- Esau K (1977) Anatomy of Seed Plants, Ed 2. Wiley & Sons, New York
- FlyBase Consortium (2002) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res 30 106–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2006) The Gene Ontology (GO) project in 2006. Nucleic Acids Res 34 D322–D326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D (2005) Using ontologies to describe mouse phenotypes. Genome Biol 6 R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M (2005) The adult mouse anatomical dictionary: a tool for annotating and integrating data. Genome Biol 6 R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A, Kaufman MH, McKay A, Baldock R, Simmen MW, Bard JB (2003) An ontology of human developmental anatomy. J Anat 203 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, et al (2005) Plant ontology: A controlled vocabulary of plant structures and growth stages. Comp Funct Genom 6 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean ML, Schikora A, Mari S, Briat JF, Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44 769–782 [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer ELL (2005) Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33 D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Seetharaman R (1991) Inheritance of grain size and its interrelationship in rice. Indian J Genet Plant Breed 51 352–356 [Google Scholar]
- Pujar A, Jaiswal P, Kellogg EA, Ilic K, Vincent L, Avraham S, Stevens P, Zapata Z, Reiser L, Rhee SY, et al (2006) Whole plant growth stage ontology for angiosperms and its application in plant biology. Plant Physiol 142 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink AW, Buell RC (2005) Microarray expression profiling resources for plant genomics. Trends Plant Sci 10 603–610 [DOI] [PubMed] [Google Scholar]
- Schulze-Kremer S (2002) Ontologies for molecular biology and bioinformatics. In Silico Biol 2 179–193 [PubMed] [Google Scholar]
- Seetharaman R, Srivastava DP (1969) Inheritance of semi-dwarf stature and cigar shaped panicle in rice. Indian J Genet Plant Breed 29 220–226 [Google Scholar]
- Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA (2005) BarleyBase—an expression profiling database for plant genomics. Nucleic Acids Res 33 D614–D618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Clements D, Conlin T, Edwards P, Frazer K, Schaper K, Segerdell E, Song P, Sprunger B, Westerfield M (2003) The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res 31 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, Cocciolone S, McCarty DR (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28 409–418 [DOI] [PubMed] [Google Scholar]
- Vincent PL, Coe EH, Polacco ML (2003) Zea mays ontology—a database of international terms. Trends Plant Sci 8 517–520 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Jaiswal P (2005) Biological ontologies in rice databases: an introduction to the activities in Gramene and Oryzabase. Plant Cell Physiol 46 63–68 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.