Abstract
Agrobacterium T-DNAs were used to deliver transposable Dissociation (Ds) elements into the nuclei of potato (Solanum tuberosum) cells. A double-selection system was applied to enrich for plants that only contained a transposed Ds element. This system consisted of a positive selection for the neomycin phosphotransferase (nptII) gene positioned within Ds followed by a negative selection against stable integration of the cytosine deaminase (codA) gene-containing T-DNA. Sixteen of 29 transgenic plants were found to contain a transposed element while lacking any superfluous T-DNA sequences. The occurrence of this genotype indicates that Ds elements can transpose from relatively short extrachromosomal DNA molecules into the plant genome. The frequency of single-copy Ds transformation was determined at 0.3%, which is only about 2.5-fold lower than the potato transformation frequency for backbone-free and single-copy T-DNAs. Because of the generally high expression levels of genes positioned within transposed elements, the new transformation method may find broad applicability to crops that are accessible to Agrobacterium T-DNA transfer.
Agrobacterium-mediated plant transformation is based on the transfer of single-stranded plasmid DNA segments (T-DNAs) from the infecting pathogen to plant cell nuclei (Tinland et al., 1994; Yusibov et al., 1994). Although the virE2-coated linear DNA is temporarily protected from nuclease attack (Rossi et al., 1996), only about 1% of transferred T-DNAs escape eventual degradation by integrating into double-stranded chromosome breaks (Bravo-Angel et al., 1999; Chilton and Que, 2003; Rommens et al., 2004). The expression level of T-DNA-residing transgenes is often affected by position-dependent gene-silencing effects, which can be overcome by linking the transgenes to matrix-associated DNA regions (Peach and Velten, 1991; Mlynarova et al., 2003).
In contrast to the apparently passive integration of T-DNAs, mobile elements, such as maize (Zea mays) Activator (Ac), are generally believed to transpose via cut-and-paste DNA recombination. Excision is initiated by the assembly of an active synaptic complex in which the two ends of the element are paired and held together by bound Ac-transposase (Tpa) subunits (Gorbunova and Levy, 2000). In analogy with other transposition systems, reinsertion may occur when the 3′ hydroxyl at each end of the excised element performs a nucleophilic attack on the host DNA (Reznikoff, 2003). In the final stage of the transposition process, noncomplementary ends of the broken donor DNA molecule are processed, rejoined, and filled. This repair process generates both a small excision site footprint, often comprising a few base pair of transposon-end sequences and characteristic duplication of the target octonucleotide insertion site.
The Ac element encodes an 807-amino acid Tpa that mediates the initiation of transposition by specifically binding to multiple motifs positioned near the terminus of the element (Kunze and Starlinger, 1989). Separation of the two functions of Ac creates a two-component transposition system. An expression cassette for the Tpa gene represents the first component, whereas the second component consists of a nonautonomous Dissociation (Ds) element that contains the terminal sequences required for transposition (Rommens et al., 1992). Such two-component systems have been introduced into a variety of plant species by transforming plants with T-DNAs carrying both Ds and the Ac-Tpa gene (Hehl and Baker, 1989). As opposed to the unpredictability of T-DNA integration, Ds elements frequently transpose from stably integrated T-DNAs into gene-associated sequences that are adjacent to predicted matrix-associated DNA regions (Meissner et al., 2000; Koprek et al., 2001; Ito et al., 2002; Kim et al., 2004; Pan et al., 2005; Zhao et al., 2006). The site preference for Ds integration has been linked to generally high expression levels of genes positioned within the element (Koprek et al., 2001). Upon transposition, gene expression levels need to be stabilized by segregating the Tpa source away from Ds in progeny plants. This requirement may complicate breeding efforts, especially if the transposable element and its activator are tightly linked. Furthermore, the necessity to either self or cross fertilize transgenic plants sets hurdles to applications of this method in crops that are vegetatively propagated and suffer from in-breeding depression, such as potato (Solanum tuberosum).
The need to employ T-DNA transformation procedures for the introduction of transposons into plants can be circumvented by promoting the transposition of elements from extrachromosomal DNAs into the plant genome. Earlier methods were based on the use of either plasmids or geminiviruses that contain a Ds element. These methods yielded very low transformation frequencies for wild tobacco (Nicotiana plumbaginifolia) protoplasts (Houba-Herin et al., 1994) and rice (Oryza sativa; Sugimoto et al., 1994). They may be difficult to apply to plant species that are not as accessible to protoplast transformation and regeneration. Here, we describe a new method that employs Ds-containing T-DNA. Transformed cells were selected for the presence of Ds and against preservation of T-DNA. The majority of regenerated plants contained a single Ds insertion without additional foreign DNA. The frequency of single-copy and backbone-free transformation events was only about 2.5-fold lower than that determined for conventional T-DNA transformation of potato. Importantly, our studies indicate that transposable elements can excise from nonreplicating and relatively short linear DNA segments for efficient integration into the plant genome.
RESULTS
Production of Transformed Plants
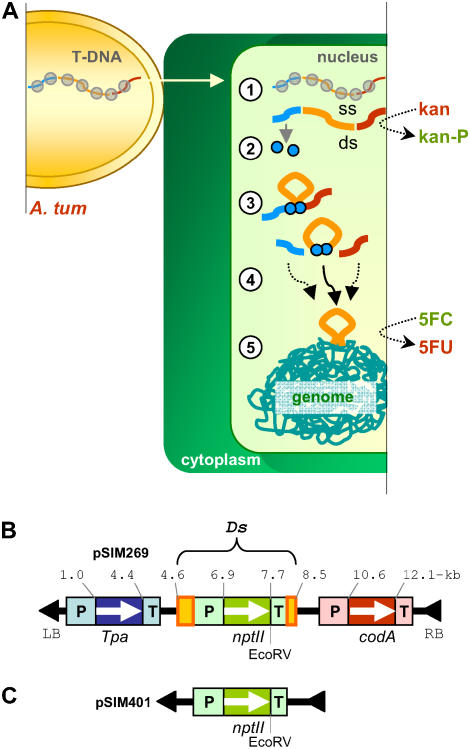
Previous studies indicated that T-DNAs often degrade within days upon transfer to plant cells (Bravo-Angel et al., 1999; Rommens et al., 2004). As modeled in Figure 1A, we hypothesized that such linear DNA molecules could function as transient vehicles for nuclear delivery of transposable Ds elements. Early transposition events might coincide with loss of T-DNA, thus resulting in the regeneration of plants only containing Ds. This theory was tested by employing the T-DNA shown in Figure 1B. The Ds element of this T-DNA consisted of an expression cassette for the neomycin phosphotransferase (nptII) gene inserted between the minimal Ac-derived sequences that are required for nonautonomous transposition (Weil and Kunze, 2000). The element was positioned next to the Ac-Tpa gene fused to the asparagus (Asparagus officinalis) Pr1 (AoPr1) promoter. By rapidly responding to both wounding and infection (Warner et al., 1993), the AoPr1 promoter was expected to drive early Tpa gene expression and allow Ds excision prior to T-DNA degradation. To select against stable T-DNA integration, an expression cassette for the negative selectable marker gene cytosine deaminase (codA) was placed on the other side of Ds. The protein encoded by this gene converts 5-fluorocytosine (5FC) into toxic 5-fluorouracil (Perera et al., 1993). A binary vector carrying Ds flanked by Tpa and codA gene expression cassettes, designated pSIM269, was introduced into Agrobacterium LBA4404, and the resulting strain was used to infect three groups of 1,500 potato stem explants. Control transformations were performed with a binary vector carrying the nptII gene inserted between T-DNA borders (Fig. 1C).
Figure 1.
Transposon-based transformation. A, Model of Ds transposition from an extrachromosomal T-DNA into the genome of a plant cell. The T-DNA contains a Tpa gene expression cassette (blue), a Ds element carrying an expression cassette for the nptII gene inserted between the ends of the maize element Ac (orange), and a codA gene inserted between regulatory sequences (red). Plant transformation is initiated by the transfer of single-stranded (ss) and virE2-coated (gray circles) T-DNA from Agrobacterium (A. tum) to the plant cell nucleus (step 1). After obtaining its double-stranded (ds) form, expression of the Tpa and codA genes will result in the production of Tpa (blue circles) and the conversion of 5FC to toxic 5-fluorouracil, respectively (step 2). Tpa will bind to the Ac-derived sequences and catalyze excision of Ds (steps 3 and 4). The excised element, possibly together with broken T-DNA fragments, may integrate into the plant cell genome (step 5). Application of a sequential selection on kanamycin and 5FC was predicted to yield transformants that contained a Ds element and lacked T-DNA. B, Diagram of the T-DNA of pSIM269 and pSIM401. Black triangles represent the left and right border of the T-DNA.
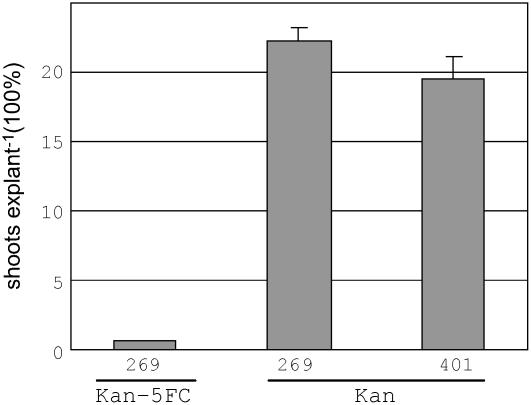
After a 2-d cocultivation period, infected explants were subjected to kanamycin to select for proliferating cells that contained Ds. A subsequent transfer to medium supplemented with 5FC allowed selection against stable integration events of codA-carrying T-DNA. Figure 2 shows that this double-selection procedure yielded 0.65% ± 0.02% shoots explant−1. Much higher frequencies were obtained by either omitting the 5FC step (22.2% ± 1.1% shoots explant−1) or employing the conventional T-DNA vector pSIM401 for standard transformation and selection (19.5% ± 1.6% shoots explant−1; Fig. 2). These results indicate that approximately 97% of kanamycin-resistant cells were eliminated during the negative selection procedure, apparently because these cells contained stably integrated T-DNAs that expressed the codA gene.
Figure 2.
Potato transformation frequencies (transformed shoots explant−1) × 100% for Agrobacterium strains containing pSIM269 and pSIM401, respectively. After cocultivation, explants were placed on medium containing kanamycin (kan). After 20 d, one group of explants infected with Agrobacterium:pSIM269 was transferred to 5FC (Kan-5FC). Data represent the average ± se of three experiments.
Plants Transformed with Ds Only
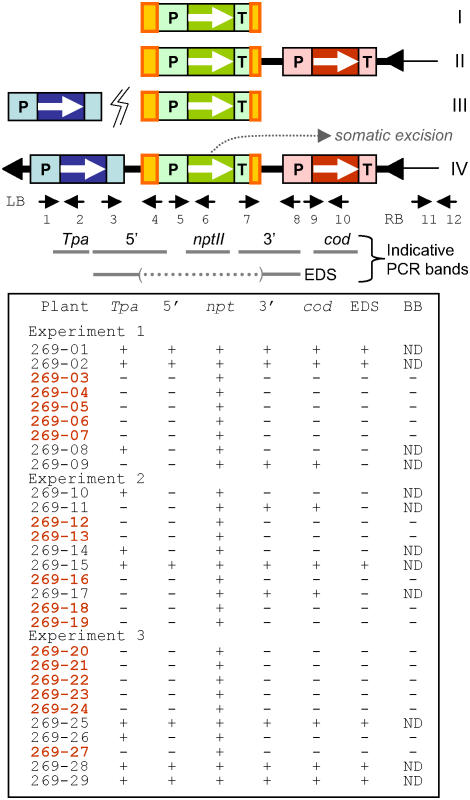
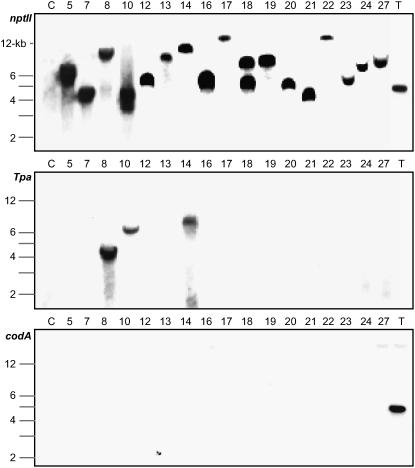
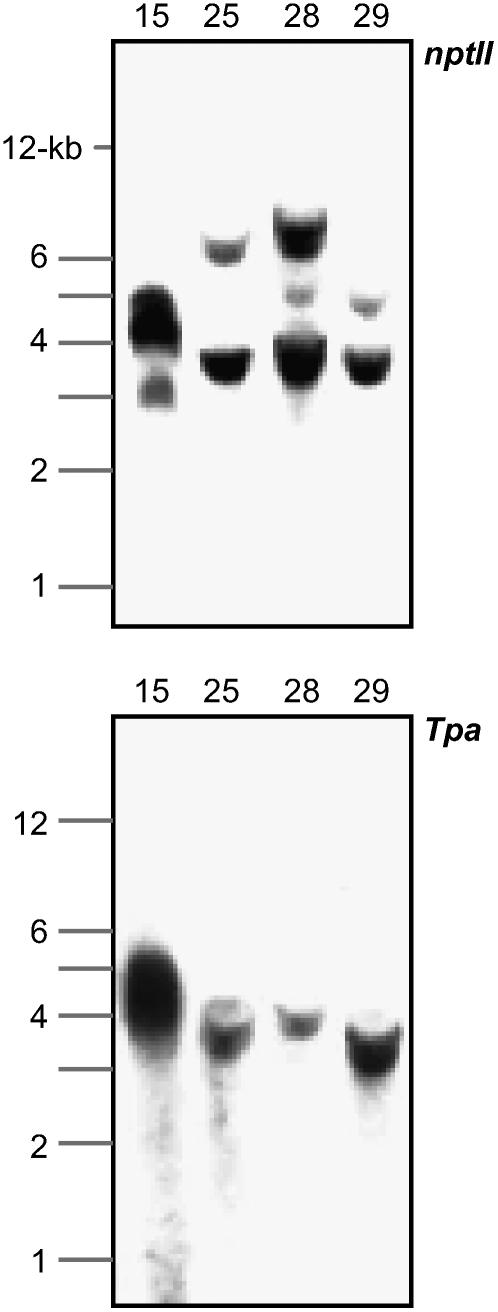
The 5FC-insensitive plants obtained from infections with the pSIM269 strain were genotyped by carrying out PCR on each of at least three leaves per plant. As shown in Figure 3, this analysis identified four different genotypes. The first and largest genotypic group consisted of 16 plants that were positive for Ds, but lacked other parts of the T-DNA and backbone of pSIM269 (plants 269-3, 4, 5, 6, 7, 12, 13, 16, 18, 19, 20, 21, 22, 23, 24, and 27). The consistency of PCR results indicated that the group I (Ds) plants represented stably transformed rather than chimeric events. Further evidence for the genetic uniformity of these plants was obtained from DNA gel-blot analysis. This experiment demonstrated that the majority of transformants, 15 of 16, contained a single Ds-hybridizing band that displayed a similarly strong signal intensity to that of a control pSIM368 T-DNA plant, which contains expression cassettes for the nptII and codA genes (Fig. 4; data not shown). Hybridization of stripped DNA filters with probes derived from Tpa and codA genes confirmed the absence of T-DNA sequences that had originally flanked Ds (Fig. 4; data not shown). Collectively, our results support the model that Ds elements can transpose from nonintegrating T-DNAs into the progenitor cells of transformed plants (Fig. 1A).
Figure 3.
PCR-based genotyping of transgenic plants. Primers are indicated as arrows with sequences shown in “Materials and Methods.” Roman numerals indicate the four different genotypes that were identified. 5′, Junction of upstream T-DNA and flanking 5′ end of Ds; 3′, junction of the 3′ end of Ds and flanking downstream T-DNA; cod, codA gene; BB, pSIM269 plasmid backbone.
Figure 4.
DNA gel-blot analysis of group I and group III plants. Lanes contain 10 μg each of EcoRV-digested plant DNA. Filters were hybridized with internal fragments of nptII (top) and Tpa (bottom). Plant DNA fragments hybridizing with nptII represent independent insertions of Ds. These fragments have a size of at least 3.1 kb, which is the distance from one end of Ds to the internal EcoRV site. C, Untransformed controls; T, pSIM368 T-DNA plant.
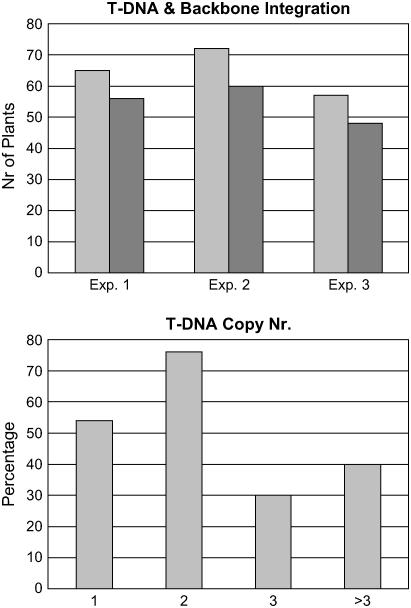
The recovery of 15 Ds-only plants from 4,500 explants indicates a Ds transformation frequency of 0.3% ± 0.02%. This frequency is much lower than the above-described 19.5% for conventional pSIM401 transformation. In this latter case, however, PCR analysis found 84% ± 1% of plants contained backbone DNA (Fig. 5A), whereas approximately 73% of plants represented multiple T-DNA integration events (Fig. 5B). Transgenic plants carrying such superfluous DNA are more difficult to characterize molecularly and are often discarded early on during the commercial line selection process. Thus, 0.8% of control pSIM401 plants contained backbone-free and single-copy T-DNA insertions. This frequency is only 2.5-fold higher than that for single-Ds transformation.
Figure 5.
Frequencies for backbone DNA integration and multiple-copy T-DNA integration for conventional potato transformation. A, Three groups of kanamycin-resistant transgenic plants (light gray bars) were PCR analyzed for the presence of backbone DNA (dark gray bars). B, Copy numbers were defined by hybridizing EcoRV-digested DNA with an nptII gene-derived probe.
Employment of the pSIM269 Strain Can Give Rise to Aberrant Ds Transposition Events
Three transformed plants contained the Ds element together with (1) a junction fragment comprising Ds and downstream DNA; and (2) the codA gene (269-9, 11, and 17; Fig. 6). The absence of T-DNA sequences upstream from Ds in these group II plants can be explained most easily by assuming that the element dissociated from its T-DNA at the 5′ side only. The same hypothesis was used before to explain the frequently occurring deletion of transposon-flanking DNA sequences in both plants and bacteria (Dooner et al., 1988; Roberts et al., 1991; Jilk et al., 1993). Indeed, sequence analysis of plant 269-9 demonstrated that this plant contained new sequences upstream from the transposable element (data not shown).
Figure 6.
DNA gel-blot analysis of pSIM269 T-DNA (group IV) plants. See the legend of Figure 3 for details.
The genotype of four additional plants, 269-8, 10, 14, and 26, resembled that of the second group, but, interestingly, lacked the junction of Ds and the downstream T-DNA (Fig. 3). These group III plants may have originated from extrachromosomal excision followed by independent integration of both Ds and one of the broken T-DNA fragments. Evidence for this hypothesis came from DNA gel-blot analyses, which demonstrated that the four plants contained the nptII and Tpa genes on separate EcoRV fragments (Fig. 4; data not shown). For instance, plant 269-8 contained the nptII gene on an 11-kb fragment, whereas the Tpa gene was located on a fragment with a size of 5 kb. An alternative explanation for the molecular organization of this third group of plants is that a transposed Ds element cointegrated with independently transferred and severely truncated T-DNA. Such a course of events is unlikely, however, because truncated T-DNAs usually contain the 5′ part of the T-DNA rather than the 3′ part (Young and Nester, 1988). All Ds bands displayed high signal hybridization intensities, which are indicative of initial integration events. The absence of weak Ds bands suggests that the Ac-Tpa gene of group III plants was not functionally active (Fig. 4). This inactivity may be due to either position-integration effects or truncation of the Tpa gene expression cassette.
Some Plants Derived from Transformation with the pSIM269 Strain Represent Conventional T-DNA Integration Events
The last group of plants (269-1, 2, 15, 25, 28, and 29) were positive for internal fragments of not only the nptII and Tpa genes, but also the codA gene. Position integration effects may, in these exceptional cases, have limited expression of the negative selectable marker gene, allowing the plants to survive subjection to 5FC. The six group IV plants also contained both upstream and downstream junction fragments for Ds and flanking T-DNA sequences. Such junctions are designated here the 5′ and 3′ full donor site (FDS) and indicate the presence of Ds at its original position in the T-DNA. However, the elements were not entirely dormant because primers located about 0.2 kb upstream and downstream, respectively, from the original site of Ds (Pr3 and 8) could in all cases be used successfully to amplify an 0.4-kb empty donor site (EDS) fragment (Fig. 3). Further evidence for this somatic excision of Ds in some, but not all, plant cells came from DNA gel-blot analysis. In addition to bands that hybridized strongly to both Tpa and nptII (Ds) genes, all plants of this group contained additional minor bands that hybridized only to nptII (Fig. 6; data not shown). This activity of Ds has been described before for a variety of plant species transformed with Ds-containing T-DNAs, including potato (Haring et al., 1989; Pavingerova et al., 2001).
Expression Levels of the Ds-Residing nptII Gene
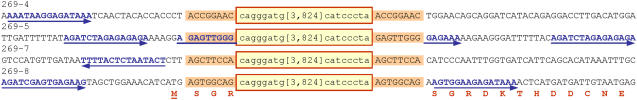
The integration sites of Ds elements that had transposed from extrachromosomal T-DNAs were characterized by carrying out inverse PCRs for the randomly chosen group I and group III plants 269-4, 5, 7, and 8. Sequence analyses of recovered junction fragments demonstrated that the elements had maintained their integrity during transposition from extrachromosomal T-DNAs into the plant genome. Furthermore, the integration events were in all cases unique and associated with typical 8-bp target site duplications (Fig. 7). In plant 269-4, the insertion is 1.5 kb downstream of the 1,3-β-glucan glucanhydrolase gene AJ586738. Plant 269-5 contained an insertion in the 5′-untranslated leader sequence of a putative CCCH-type zinc finger protein (GenBank accession no. BG889316), and the insertion of plant 269-8 was positioned immediately downstream of the start codon of an unknown potato gene (DV627343). These integration events provide new examples for the previously observed tendency of transposable elements to often insert near the 5′ end of genes (Pan et al., 2005). In contrast, plant 269-7 contained the element positioned in the terminator region of an unknown gene (CV498058) at a distance of 438 bp downstream from the 3′-transcript cleavage site. We also found that sequences within approximately 200 bp from the integration site consisted of AT-rich (64%) DNA comprising at least one putative motif with the purine-rich consensus sequence 5′-AGATNTAGAGAGAAA (Fig. 7; data not shown). Collectively, our results demonstrate that Ds elements can excise from extrachromosomal T-DNAs to subsequently integrate into plant genomes.
Figure 7.
Ds integration sites. Intact transposable elements are boxed with terminal octonucleotides (shown in lowercase) and omitted nucleotides (shown between brackets). Target site duplications are highlighted. Sequences resembling the 5′-AGATCTAGAGAGAAA consensus are shown in blue with arrows indicating their orientation. Amino acids encoded by the gene that is disrupted in plant 269-8 are shown below the DNA sequence in red and bold with the first Met (M) underlined.
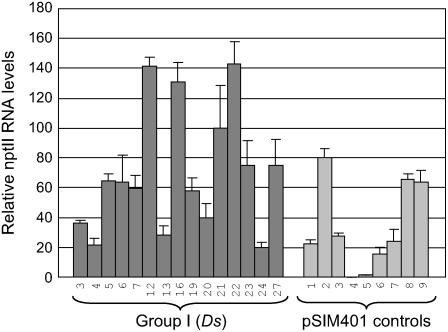
To test our initial hypothesis that transposon-based transformation would support higher frequencies of transgene expression than conventional T-DNA transformation, we determined the nptII RNA levels in both Ds plants and pSIM401 controls by real-time quantitative reverse transcription (RT)-PCR. Figure 8 shows that expression levels of the 15 Ds plants were, on average, twice as high as those of nine pSIM401 controls (P = 0.03). This difference was due, in part, to very low expression levels in plants pSIM401-4 and 5. Apparently, these nptII+ plants had undergone transgene silencing after surviving kanamycin selection. Three Ds plants that contained unusually high amounts of RNA also contributed to the generally higher gene expression levels of transposed nptII genes. Although further studies are needed to elucidate this link, our data indicate that the nptII gene is generally expressed at higher levels if positioned within a transposed Ds element than if part of an integrated T-DNA. Our results also demonstrate that Ds elements can excise from extrachromosomal T-DNAs for subsequent integration into regions that often promote high levels of gene expression.
Figure 8.
Expression levels of the nptII gene. Data represent the mean ± se of three independent experiments as determined by quantitative real-time RT-PCR.
DISCUSSION
We have shown that T-DNAs containing both a Ds element and Ac-Tpa gene can be used as vehicles for transposition-based plant transformation. Excision from nonintegrating T-DNAs is followed by Ds integration into the plant genome, often without cointegration of T-DNA sequences. The resulting Ds plants are not chimeric, indicating that transposition occurred from nonintegrating T-DNAs into the genomes of progenitor cells. Previous studies demonstrated that Ds elements can excise from chromosomes, plasmids, and geminiviruses. Our results now also indicate that linear and short (approximately 4.5-kb) flanking DNA segments are sufficient to support cut-and-paste recombination processes of Ds. Such excision events are likely to have occurred after the originally single-stranded and virE2-coated T-DNAs converted into their double-stranded form (Tzfira et al., 2003) prior to T-DNA degradation.
Given the relatively low frequency of early transposition, it is not surprising that almost all group I plants contained a single copy of the Ds element. However, our results contradict earlier findings on Ds transposition from plasmid DNA into the genome of wild tobacco (Houba-Herin et al., 1994). Although these earlier studies indicated very low frequency of Ds transformation (approximately 0.001%), more than one-third of plants obtained through this method contained three copies of Ds. This discrepancy between plasmid- and T-DNA-based delivery systems may be explained by the assumption that the copy number of plasmids in the nuclei of polyethylene glycol-treated protoplasts is higher than that of T-DNAs in the individual plant cells of Agrobacterium-infected explants. The abundance of plasmids may have given rise to multiple simultaneous transposition events.
Ds elements that excised from integrated T-DNAs were often shown to integrate into DNA regions that support high gene expression levels (Koprek et al., 2001). We found this tendency also to be linked to transposition from nonintegrating T-DNAs. Thus, transposable elements may be used as efficient vehicles for the high-level expression of genes of interest. Further studies need to be performed to determine whether such high gene expression levels can also be obtained with plant genes, which may be more easily silenced (Garcia-Perez et al., 2004). Infrequently, Ds integration may result in the inactivation of agronomically important genes. Such mutations are not likely to cause mutant phenotypes because potato suffers from inbreeding depression and is generally not self fertilized to produce homozygotes.
It may be possible to further increase Ds transformation frequencies by modifying either the construct or the selection procedures. For instance, the timing of Tpa gene expression may be optimized by employing promoters, such as the 35S promoter of the Cauliflower mosaic virus, that are known to trigger early Ds excision if fused to the Tpa gene (Scofield et al., 1992). Alternatively, identification of plants only containing Ds may be facilitated by employing different kinds of selection strategies. Insertion of Ds between a promoter and the nptII gene would make it possible to apply transient selection procedures that were developed to generate marker-free transformed plants (Rommens et al., 2004). Proliferating cells that survive this treatment would contain excised elements. A negative selection system can then be used to eliminate most T-DNA integration events, whereas a visual marker within Ds would facilitate the identification of regenerated shoots that contain a transposed element.
Plant 269-18 is a unique group I plant in that it contains two Ds elements. These elements may have independently transposed from two cotransferred and nonintegrating T-DNAs. However, it is also possible that copy number was doubled by the occurrence of a second transposition event from replicated into unreplicated DNA in a similar manner as shown before for Ds transposition in maize (Chen et al., 1992; Duval-Valentin et al., 2004). In that case, the similar intensity of the two visualized Ds bands would indicate that the second transposition occurred very early during cell proliferation.
Because Ac-Tpa sources have only been identified in maize, Ds elements are likely to remain stably integrated at their original positions in the potato genome. In fact, there have been no reports on Ac-Tpa-independent transposition of Ds in any heterologous plant system. Expression levels of genes that reside within the transposed Ds elements are also expected to remain stable. One argument for this assumption comes from the finding that transposon methylation has only been associated with nontransposed elements that reside in T-DNAs located within highly repetitive genomic regions (Koprek et al., 2001). Furthermore, studies on the progeny of transgenic rice plants containing transposed Ds elements indicate that expression levels are not likely to be affected by a gradual methylation of Ds (Szeverenyi et al., 2006).
Our studies also identified transformation events that were associated with aberrant transposition. Group II plants contained Ds, codA, and the intermediary 3′ FDS, but lacked the 5′ FDS and EDS. Most likely, this genotype was generated by insertion of truncated T-DNAs. Alternatively, the absence of upstream sequences was a consequence of Ds excision attempts. Such activities would result in adjacent deletions that have been reported for both plant and bacterial transposons (Dooner et al., 1988; Roberts et al., 1991; Jilk et al., 1993). The third group consists of plants that contained both an intact Ds element and at least part of the Ac-Tpa gene. However, these plants lacked the junction between the two elements. They may have arisen from the independent integrations of an excised Ds and a resulting broken T-DNA fragment. Further studies will be required to elucidate the precise molecular basis of both the Ds 5′ FDS codA and Tpa-Ds genotypes.
In contrast to the early transpositions that yielded group I to group III plants, transpositions from integrated T-DNAs resulted in the development of chimeric group IV plants. Even if such late transpositions would be followed by rare recombination events that triggered T-DNA deletion, the multiple resulting cell lines would be extremely unlikely to eventually give rise to plants resembling any of the other groups of plants.
The two-component Ds-Ac-Tpa system described here may not represent the only tool kit for transposon-based transformation. Various plant species have been shown to contain elements that belong to the Ac-Ds family. Such elements include Tip100 of common morning glory (Ipomoea purpurea; Ishikawa et al., 2002), Pac1 of pearl millet (Pennisetum glaucum; MacRae et al., 1994), and various elements in sugar cane (Saccharum officinarum; de Araujo et al., 2005). Furthermore, it may be possible to employ other transposable element systems, such as Arabidopsis (Arabidopsis thaliana) Tag1 (Frank et al., 1997) and maize En-Spm (Kumar et al., 2005).
The two different protoplast-based Ds transformation systems that were described more than a decade ago have found little or no utility in commercial crop transformation programs. Drawbacks of these early methods include low transformation frequencies and cumbersome procedures for the proliferation and regeneration of protoplasts from crop plants. The use of T-DNAs as Ds delivery systems makes it possible now to use explants rather than protoplasts for transposition-based transformation. The ease of use and broad applicability of an explant transformation/regeneration system may encourage broader exploitation of transposable elements as tools for plant transformation. Transposition-based plant transformation provides new means of genetically modifying plants without inserting bacterial DNA into their genomes. A previously developed method employs plant-derived transfer DNAs as alternatives to Agrobacterium T-DNAs (Rommens et al., 2004, 2005). Application of any of these two methods may alleviate some of the public concerns about the permanent introduction of bacterial, viral, and synthetic DNA into the genomes of food crops (Rommens, 2004).
MATERIALS AND METHODS
Plant Transformation
Ten-fold dilutions of overnight-grown Agrobacterium LBA4404 cultures were grown for 5 to 6 h, precipitated for 15 min at 2,800 rpm, washed with Murashige and Skoog liquid medium (PhytoTechnology Laboratories) supplemented with Suc (3%, pH 5.7), and resuspended in the same medium to 0.2 OD600 nm. The resuspended cells were mixed and used to infect 0.4- to 0.6-mm internodal segments of the potato (Solanum tuberosum) variety Ranger Russet. Infected stems were incubated for 2 d on coculture medium (one-tenth Murashige and Skoog salts, 3% Suc, pH 5.7) containing 6 g/L agar A111 (Phytotechnology) at 22°C in a Percival growth chamber (16 h light) and subsequently transferred to callus induction medium (Murashige and Skoog medium supplemented with 3% Suc, 2.5 mg/L of zeatin riboside, 0.1 mg/L of naphthalene acetic acid, and 6 g/L of agar) containing timentin (150 mg/L) and kanamycin (100 mg/L). After about 3 weeks, proliferating explants would start to develop shoots. This shoot formation was then enhanced by transfer, 1 week later, to shoot induction medium (Murashige and Skoog medium supplemented with 3% Suc, 2.5 mg/L of zeatin riboside, 0.3 mg/L of GA3, and 6 g/L of agar) containing timentin, kanamycin, and, for most explants infected with Agrobacterium:pSIM269, 5FC (500 mg/L). The resulting shoots were harvested 1 month after transfer and placed on Murashige and Skoog medium with 3% Suc, 6 g/L of agar, and timentin (150 mg/L) for root development.
Plant Genotyping
Plant DNA was isolated by using a robust and reliable method described previously (Rommens et al., 2004). The primer pairs Pr1-Pr2 were used to visualize a junction fragment for the AoPR1 promoter and the linked Tpa gene, Pr3-Pr4 for 5′ FDS, Pr5-Pr6 for the promoter-nptII gene fusion within Ds, Pr7-Pr8 for 3′ FDS, Pr9-Pr10 for promoter-codA gene junction, and primers Pr3-Pr8 for EDS. The primer pair used to determine the presence of vector backbone sequences was Pr11-Pr12 (Table I).
Table I.
Primers
| Name | Sequence |
|---|---|
| Pr1 | 5′-CGTTCTTGAATCAAAGTGACCTGTAC |
| Pr2 | 5′-CTCTTGTGGTAGACATCAACTTGGC |
| Pr3 | 5′-GCATGCTAAGTGATCCAGATG |
| Pr4 | 5′-CTGCAGTCATCCCGAATTAG |
| Pr5 | 5′-AGGAAGGAATTCCCCCGGATCAGC |
| Pr6 | 5′-AGGAGCAAGGTGAGATGACAGG |
| Pr7 | 5′-GGAATTCGCGTAGACTTATATGGC |
| Pr8 | 5′-TGATGACCAAAATCTTGTCATCCTC |
| Pr9 | 5′-GAATCAGCTAATCAGGGAGTGTG |
| Pr10 | 5′-GCCATGCGCGTTGTTTCACATCG |
| Pr11 | 5′-CACGCTAAGTGCCGGCCGTCCGAG |
| Pr12 | 5′-TCCTAATCGACGGCGCACCGGCTG |
| Pr13 | 5′-TGGGTGGAGAGGCTATTCGGCTATGA |
| Pr14 | 5′-ATGTCCTGATAGCGGTCCGCCACA |
| Pr15 | 5′-CGTTCTTGAATCAAAGTGACCTGTAC |
| Pr16 | 5′-CTCTTGTGGTAGACATCAACTTGGC |
| Pr17 | 5′-ACGAAAACGGAACGGAAACGGGATATAC |
| Pr18 | 5′-ATCGGTTATACGATAACGGTCGGTACG |
| Pr19 | 5′-GTAGAGCTAGTTTCCCGACCGTTTCAC |
| Pr20 | 5′-GATTTTCCCATCCTACTTTCATCCCTG |
| Pr21 | 5′-CGACCGGATCGTATCGGTTTTCGATTAC |
| Pr22 | 5′-GCACATAAGTGATAAGTCTTGGGCTC |
| Pr23 | 5′-CGGTAGAGGTATTTTACCGACCGTTAC |
| Pr24 | 5′-CTAACATAAGAAGCCATATAAGTCTAC |
| Pr25 | 5′-CGTCGCATTCCAGATTATCCA |
| Pr26 | 5′-ACGAAGAAACGACAAATCCCAAC |
| Pr27 | 5′-TGCAGGACGAGGCAGCGCGGCTAT |
| Pr28 | 5′-AGGAGCAAGGTGAGATGACAGG |
DNA Gel-Blot Analysis
Plant DNA isolation was conducted using the DNeasy kit (Qiagen). EcoRV digests were run on gel, blotted, and hybridized with radioactively labeled probes as described previously (Rommens et al., 1992). Primers for amplification of fragments that were used to generate probes to visualize the presence of the nptII (Ds) and Ac-Tpa genes (remaining T-DNA) were Pr13-Pr14 and Pr15-Pr16, respectively.
Isolation of Target Sites
To amplify the junctions of Ds and upstream flanking plant DNA, inverse PCR was carried out using PstI, XbaI, and AsnI digests (2 μg). After 1 h of DNA circularization, DNA ligase treatment, and removal of salts using the QIAquick PCR purification kit (Qiagen), DNA digests were used as templates for PCR (30 cycles) with primers Pr17 and Pr18. The amplified products (1 μL) were used as a template for subsequent PCR (35 cycles) with a set of nested primers (Pr19 and Pr20). The Ds junctions with downstream flanking DNA were obtained by using EcoRI, AflIII, and EcoRV digests with Pr21-Pr22 for the first PCR, and Pr23-Pr24 for the second PCR. DNA fragments were separated by agarose gel electrophoresis, isolated using Qiagen's gel isolation kit, cloned into plasmid pGEMT-Easy (Invitrogen), and sequence analyzed using Big Dye Terminator, version 3.0 sequencing chemistry (Applied Biosystems) and an ABI 3730 DNA sequencer.
RNA Analysis
For quantitative real-time RT-PCR, RNA was isolated from pSIM269 and pSIM401 tobacco (Nicotiana tabacum) leaves and treated with RNAse-free DNAseI (Invitrogen). Reactions were performed using 25 ng RNA and the QuantiTech SYBR Green RT-PCR kit (Qiagen) according to the manufacturer's instructions. Internal control primers (Pr25 and Pr26) were designed to amplify the cytochrome oxidase (Cox) gene. Primers for the nptII gene were Pr27 and Pr28.
Acknowledgments
We are grateful to Scott Simplot, Bill Whitacre, and Dr. Kathy Swords for fruitful discussion and support and to Dr. Jaime Menendez-Humara for carrying out the plant transformations.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Caius M. Rommens (crommens@simplot.com).
References
- Bravo-Angel AM, Gloeckler V, Hohn B, Tinland B (1999) Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J Bacteriol 181 5758–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Greenblatt IM, Dellaporta SL (1992) Molecular analysis of Ac transposition and DNA replication. Genetics 130 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton MD, Que Q (2003) Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo PG, Rossi M, de Jesus EM, Saccaro NL Jr, Kajihara D, Massa R, de Felix JM, Drummond RD, Falco MC, Chabregas SM, et al (2005) Transcriptionally active transposable elements in recent hybrid sugarcane. Plant J 44 707–717 [DOI] [PubMed] [Google Scholar]
- Dooner HK, English J, Ralston EJ (1988) The frequency of transposition of the maize element activator is not affected by an adjacent deletion. Mol Gen Genet 211 485–491 [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G, Marty-Cointin B, Chandler M (2004) Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J 23 3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Liu D, Tsay YF, Ustach C, Crawford NM (1997) Tag1 is an autonomous transposable element that shows somatic excision in both Arabidopsis and tobacco. Plant Cell 9 1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez RD, Houdt HV, Depicker A (2004) Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38 594–602 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA (2000) Analysis of extrachromosomal Ac/Ds transposable elements. Genetics 155 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring MA, Gao J, Volbeda T, Rommens CM, Nijkamp HJ, Hille J (1989) A comparative study of Tam3 and Ac transposition in transgenic tobacco and petunia plants. Plant Mol Biol 13 189–201 [DOI] [PubMed] [Google Scholar]
- Hehl R, Baker B (1989) Induced transposition of Ds by a stable Ac in crosses of transgenic tobacco plants. Mol Gen Genet 217 53–59 [DOI] [PubMed] [Google Scholar]
- Houba-Herin N, Domin M, Pedron J (1994) Transposition of a Ds element from a plasmid into the plant genome in Nicotiana plumbaginifolia protoplast-derived cells. Plant J 6 55–66 [DOI] [PubMed] [Google Scholar]
- Ishikawa N, Johzuka-Hisatomi Y, Sugita K, Ebinuma H, Iida S (2002) The transposon Tip100 from the common morning glory is an autonomous element that can transpose in tobacco plants. Mol Genet Genomics 266 732–739 [DOI] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Mizukado S, Sakurai T, Kanahara H, Seki M, Shinozaki K (2002) A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol 129 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilk RA, Makris JC, Borchardt L, Reznikoff WS (1993) Implications of Tn5-associated adjacent deletions. J Bacteriol 175 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun B, Park SH, Park JY, Lee EJ, Kim MJ, et al (2004) Rapid, large-scale generation of Ds transposant lines and analysis of the Ds insertion sites in rice. Plant J 39 252–263 [DOI] [PubMed] [Google Scholar]
- Koprek T, Rangel S, McElroy D, Louwerse JD, Williams-Carrier RE, Lemaux PG (2001) Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol 125 1354–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CS, Wing RA, Sundaresan V (2005) Insertional mutagenesis in rice using the maize En/Spm elements. Plant J 44 879–892 [DOI] [PubMed] [Google Scholar]
- Kunze R, Starlinger P (1989) The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J 8 3177–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae AF, Huttley GA, Clegg M (1994) Molecular evolutionary characterization of an activator (Ac)-like transposable element sequence from pearl millet (Pennisetum glaucum) (Poaceae). Genetica 92 77–89 [DOI] [PubMed] [Google Scholar]
- Meissner R, Chague V, Zhu Q, Emmanuel E, Elkind Y, Levy AA (2000) Technical advance: a high throughput system for transposon tagging and promoter trapping in tomato. Plant J 22 265–274 [DOI] [PubMed] [Google Scholar]
- Mlynarova L, Hricova A, Loonen A, Nap JP (2003) The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell 15 2203–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Li Y, Stein L (2005) Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol 137 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavingerova D, Briza J, Niedermeierova H (2001) Timing of transposition of Ac mobile element in potato. Biol Plant 44 347–353 [Google Scholar]
- Peach C, Velten J (1991) Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol 17 49–60 [DOI] [PubMed] [Google Scholar]
- Perera RJ, Linard CG, Signer ER (1993) Cytosine deaminase as a negative selective marker for Arabidopsis. Plant Mol Biol 23 793–799 [DOI] [PubMed] [Google Scholar]
- Reznikoff WS (2003) Tn5 as a model for understanding DNA transposition. Mol Microbiol 47 1199–1206 [DOI] [PubMed] [Google Scholar]
- Roberts DE, Ascherman D, Kleckner N (1991) IS10 promotes adjacent deletions at low frequency. Genetics 128 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens CM (2004) All-native DNA transformation: a new approach to plant genetic engineering. Trends Plant Sci 9 457–464 [DOI] [PubMed] [Google Scholar]
- Rommens CM, Bougri O, Yan H, Humara JM, Owen J, Swords K, Ye J (2005) Plant-derived transfer DNAs. Plant Physiol 139 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens CM, Humara JM, Ye J, Yan H, Richael C, Zhang L, Perry R, Swords K (2004) Crop improvement through modification of the plant's own genome. Plant Physiol 135 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rommens CM, van Haaren MJ, Buchel AS, Mol JN, van Tunen AJ, Nijkamp HJ, Hille J (1992) Transactivation of Ds by Ac-transposase gene fusions in tobacco. Mol Gen Genet 231 433–441 [DOI] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B (1996) Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 93 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Harrison K, Nurrish SJ, Jones JD (1992) Promoter fusions to the Activator transposase gene cause distinct patterns of Dissociation excision in tobacco cotyledons. Plant Cell 4 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Otsuki Y, Saji S, Hirochika H (1994) Transposition of the maize Ds element from a viral vector to the rice genome. Plant J 5 863–871 [DOI] [PubMed] [Google Scholar]
- Szeverenyi I, Ramamoorthy R, Teo ZW, Luan HF, Ma ZG, Ramachandran S (2006) Large-scale systematic study on stability of the Ds element and timing of transposition in rice. Plant Cell Physiol 47 84–95 [DOI] [PubMed] [Google Scholar]
- Tinland B, Hohn B, Puchta H (1994) Agrobacterium tumefaciens transfers single-stranded transferred DNA (T-DNA) into the plant cell nucleus. Proc Natl Acad Sci USA 91 8000–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Frankman LR, Vaidya M, Citovsky V (2003) Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol 133 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner SA, Scott R, Draper J (1993) Isolation of an asparagus intracellular PR gene (AoPR1) wound-responsive promoter by the inverse polymerase chain reaction and its characterization in transgenic tobacco. Plant J 3 191–201 [DOI] [PubMed] [Google Scholar]
- Weil CF, Kunze R (2000) Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat Genet 26 187–190 [DOI] [PubMed] [Google Scholar]
- Young C, Nester EW (1988) Association of the virD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J Bacteriol 170 3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov VM, Steck TR, Gupta V, Gelvin SB (1994) Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc Natl Acad Sci USA 91 2994–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Palotta M, Langridge P, Prasad M, Graner A, Schulze-Lefert P, Koprek T (2006) Mapped Ds/T-DNA launch pads for functional genomics in barley. Plant J 47 811–826 [DOI] [PubMed] [Google Scholar]