Abstract
Pseudomonas aeruginosa, of medical, environmental, and industrial importance, depends on inorganic polyphosphate (poly P) for a wide range of functions, especially survival. Mutants of PAO1 lacking poly P kinase 1, PPK1, the enzyme responsible for most poly P synthesis in Escherichia coli and other bacteria, are defective in motility, quorum sensing, biofilm formation, and virulence. We describe here multiple defects in the ppk1 mutant PAOM5, including a striking compaction of the nucleoid, distortion of the cell envelope, lack of planktonic motility and exopolymer production, and susceptibility to the β-lactam antibiotic carbenicillin as well as desiccation. We propose that P. aeruginosa with reduced poly P levels undergoes ultrastructural changes that contribute to profound deficiencies in cellular functions.
Keywords: carbenicillin, exopolymer, motility, desiccation, nucleoid
Polyphosphate (poly P), a linear chain of phosphate residues linked by phosphoanhydride bonds, is present in all cells and was likely present throughout evolution (1). Poly P is synthesized in prokaryotic cells from ATP by poly P kinases (PPKs) for which two families, PPK1 and PPK2, have been identified. PPK1 is responsible principally for the synthesis of poly P in Escherichia coli, Salmonella enterica serovar Typhimurium, Shigella flexneri, Vibrio cholerae, Helicobacter pylori, Bacillus cereus, Myxococcus xanthus, and, as described here, P. aeruginosa. Homologous PPK1 amino acid sequences are in the databases of >40 organisms, including many bacterial pathogens (2). Knockout mutants of ppk1 in several pathogens demonstrate phenotypes including responses to stresses, motility, and virulence (3–7) as well as developmental defects in other species (8, 9). The only apparent eukaryotic homolog is in the social slime mold Dictyostelium discoideum (2), the mutant of which exhibits growth and developmental phenotypes (10).
We describe here the construction and biochemical characterization of a P. aeruginosa ppk1 knockout mutant, PAOM5, for which the motility, biofilm formation, burned-mouse, and ocular virulence phenotypes were reported in refs. 3, 6, and 11 and which, unlike the WT, is susceptible to predation by D. discoideum (10). Electron microscopy studies reveal significant differences in ultrastructure between the WT and mutant, including compaction of the nucleoid, withdrawal of the cytoplasm from the inner membrane, abnormal envelope structure, as well as a failure to produce exopolymer. Video microscopy studies show that the mutant is almost immotile in liquid medium, despite having about the same number of polar flagella. We also demonstrate that the ppk1 mutant exhibits much reduced viability after exposure to a β-lactam antibiotic, carbenicillin, or the hyperosmolarity of desiccation.
Results
PPK1 Activity and poly P Levels Are Reduced but Not Abolished in the ppk1 Mutant.
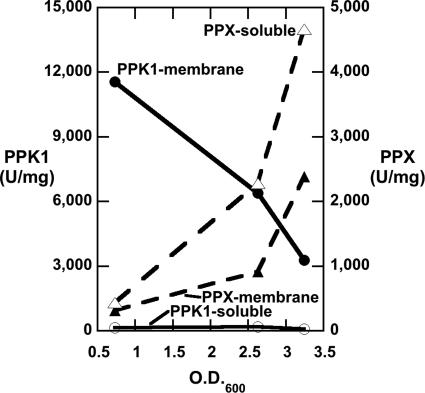
P. aeruginosa PAO1 (WT) grown in rich (LB) medium exhibits high levels of PPK1 activity during mid-exponential phase growth, almost wholly associated with the membrane fraction (Fig. 1). The levels decline as the culture enters stationary phase and continue to decline into stationary phase. ExopolyPase (PPX) activity, mostly solubilized after sonication, is inversely correlated with that of PPK1.
Fig. 1.
WT PPK1 and PPX activities in cells grown in LB medium (see Materials and Methods). The first data point set, at OD600 ≈ 0.6, was taken 3 h after inoculation at mid-exponential phase; the second set, at OD600 ≈ 2.5, was taken 3 h later upon entry into stationary phase; and the third set, at OD600 ≈ 3.2, was taken 4 h after that, well into stationary phase. Enzyme extractions, solubilizations, and assays were performed as described (see Materials and Methods). Data are the arithmetic means of at least duplicate measurements.
Three confirmed ppk1 deletion/Tet insertion (Δppk1::Tet) mutants were selected for enzymatic analysis and poly P determinations (Table 1). PPK1 activities in the membrane fractions of all three were reduced to <5% of the WT, whereas the soluble PPX activities (data not shown) were reduced by no more than 45%. Poly P levels in the WT and the three mutants were determined and checked for susceptibility to the yeast PPX, Saccharomyces cerevisiae PPX1 (12). Levels in the WT are ≈2.8-fold lower than those determined in E. coli under the same conditions (13). All three mutants have considerably higher Poly P levels than the E. coli ppk1 mutant, likely attributable to PPK2 and its homologs, apparently absent in the latter. One of these mutants, PAOM5 (M5), was selected for all further studies because it has intermediate levels of remaining PPK1 activity and poly P.
Table 1.
PPK1 and PPX activities and poly P levels in the WT and mutant
| Strain | Genotype | Enzyme activity*, units/mg of protein |
poly P†, nmol/mg protein |
||||
|---|---|---|---|---|---|---|---|
| PPK1 |
PPX |
scPPX1 digestion in vitro‡ |
|||||
| Membrane | Soluble | Membrane | Soluble | (−) | (+) | ||
| PAO1 | Wild type | 6,400 | 185 | 914 | 2,280 | 16.7 | 0.19 |
| PAOM5 | Δppk1::Tet | 152 | 59 | 303 | 1,255 | 3.4 | 0.03 |
*Strains were grown in LB medium for 6 hr (OD600 = 2.5–2.6); data are the arithmetic means of triplicate assays.
†Strains were grown in poly P-inducing Mops medium (see Materials and Methods) for 2–3 hr (OD600 = 0.3–0.4); data are the arithmetic means of triplicate assays.
‡Extracted poly P was digested with excess scPPX1 (+) or not (−) to determine its susceptibility to the enzyme.
Addition of the amino acid analog serine hydroxamate to WT E. coli grown in minimal salts medium induces a stringent response, resulting in a >6-fold increase in poly P levels to ≈13 nmol/mg (14). The same phenomenon was observed in PAO1 (WT) in the current studies, although the level was significantly higher, near 100 nmol/mg; the mutant had a 10-fold lower level of poly P compared with the WT (data not shown for either strain).
The mutant growth was comparable to that of the WT both in complex and minimal media. However, exponential phase mutant cells consistently were smaller than the WT under all conditions tested. The striking defects in motility, quorum sensing, and biofilm development became manifest only upon entry into stationary phase. When the mutant was complemented with plasmid pHEPAK11 expressing PAO1 ppk1, the PPK1 level increased 25-fold to 170,000 units/mg protein and these defects, as well as other phenotypes including virulence, disappeared (3, 6, 11).
Planktonic Mutant Cells Have Normal Flagella Ultrastructures but Are Immotile.
Phenotypes of the mutant include reduced motilities on solid media compared to WT: 70% reduction in swimming (flagellar-mediated) (15), 90% reduction in swarming (flagellar-mediated) (11), and 35% reduction in twitching (pili-mediated) (11). Regarding the defects in flagellar and pili motilities, scanning electron microscopy showed that both WT and mutant possessed similar numbers and sizes of flagellae and pili grown under the same liquid culture conditions (data not shown), in confirmation of a previous report (11). However, when mutant and WT strains were tested for real-time flagellar motility in liquid medium by video microscopy, the motility of the mutant was <1% that of WT [see supporting information (SI) Movie 1].
Mutant Cells Exhibit Defects in Envelope Ultrastructure and a Compacted Nucleoid.
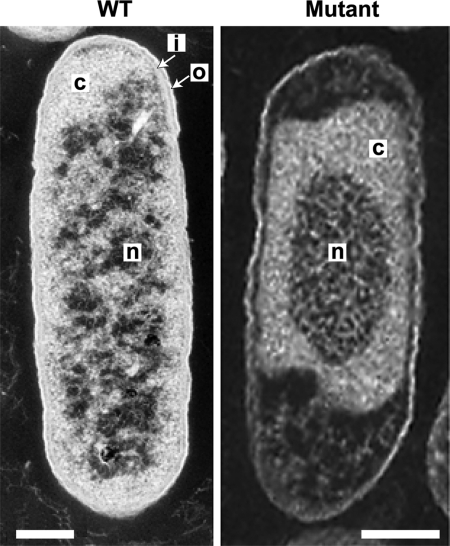
Thin section transmission electron microscopy of all WT cells observed shows a defined outer membrane, periplasm, and inner membrane, collectively the envelope, as well as a uniformly distributed cytoplasm and nucleoid (Fig. 2 Left). Under identical conditions, all mutant cells (Right) observed appear defective in all these features and consistently smaller in size than the WT. There is no clear definition to the cell envelope that appears ruffled and distorted, and the cytoplasm is detached from the poles and borders of the cell in many places. In addition, the nucleoid is significantly compacted relative to the WT. Mutant nucleoids comprised 26 ± 7% of the total cell area (37 cells in five independent fields) compared with 41 ± 11% for WT nucleoids (19 cells in five independent fields). We cannot rule out the possibility that the mutant is more susceptible than the WT to plasmolysis during EM preparation because this was not tested.
Fig. 2.
Transmission electron micrographs show compaction of the nucleoid and disruption of the cell envelope in the mutant. The WT (Left) and mutant (Right) were grown in Mops medium for 3 h (see Materials and Methods). Each cell is representative of the strain and both were included in the relative nucleoid cross-sectional area analyses. Structure designations: n, nucleoid; c, cytoplasm; i, inner membrane; o, outer membrane. (Scale bars: 200 nm.)
Mutant Cells Fail to Produce Exopolymer.
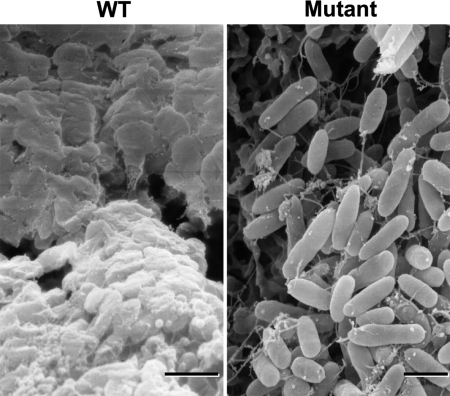
The WT, a nonmucoid species, forms well characterized biofilms, but the mutant fails to produce a comparable biofilm structure as shown by confocal laser microscopy (3); the basis for this defect is not known. Scanning electron microscopy of 3-day-old WT and mutant biofilms demonstrate that both cultures lose their flagella and aggregate on the surface in a stage preceding biofilm formation. However, the mutant fails to produce exopolymer, presumably exopolysaccharides (e.g., alginate), necessary for biofilm formation (Fig. 3); thus, synthesis of exopolymer requires PPK1 activity and/or poly P.
Fig. 3.
Scanning electron micrographs show mutant biofilms fail to produce exopolymer. WT (Left) and mutant (Right) static biofilms were grown on nitrocellulose membranes in LB medium for 3 days (see Materials and Methods). (Scale bars: 1 μm.)
Mutant Cells Fail to Survive Exposure to Carbenicillin.
A feature of P. aeruginosa is its remarkable resistance to a number of antibiotics, particularly β-lactams. The apparent defects in the mutant cell envelope suggested it might be deficient in responses to envelope-related stresses such as a β-lactam antibiotic. Late exponential phase WT and mutant cells were exposed to varying levels of the β-lactam antibiotic, carbenicillin (Cb). Four hours after exposure to 50 μg/ml, WT cells showed no difference in plate counts relative to untreated cells, whereas mutant cells exhibited a 4-fold drop (Table 2). At 100 μg/ml, mutant cells were ≈200-fold more sensitive, a concentration only slightly inhibitory to the WT. This assay is significantly different from that for the minimum inhibitory concentration in that antibiotic is added to late exponential cells not exposed previously to the antibiotic.
Table 2.
Late-log phase mutant cells are sensitive to carbenicillin
| Cb, μg/ml* | Colony forming units, × 107 |
Change, fold | ||
|---|---|---|---|---|
| Strain | Time zero | 4 hours | ||
| 50 | WT† | 73 ± 25 | 220 ± 26 | +3 |
| Mutant | 101 ± 8 | 11 ± 10 | −9 | |
| 100 | WT | 74 | 24 | −3 |
| Mutant | 58 | 0.1 | −580 | |
Drop in colony forming units 4 h after carbenicillin (Cb) addition to late-log phase cultures grown in LB medium. Data for each Cb concentration are obtained from independent experiments; those for 50 μg/ml are the arithmetic mean ± one standard deviation (n = 3).
*Final concentration.
†The growth of WT cells to 4 h after Cb addition was identical to that of untreated cells.
Mutant Cells Fail to Survive Desiccation.
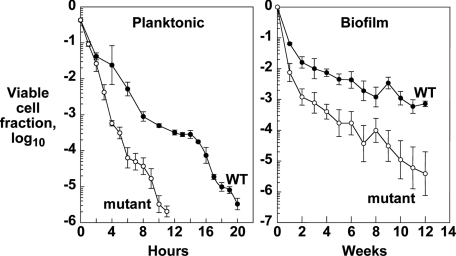
The E. coli ppk1 mutant is far more sensitive than the WT to stresses such as pH shock, oxidative agents, heat shock, and osmotic shock (16). Another environmental stress that all bacteria face in evolution is desiccation. Stationary-phase planktonic cultures of the WT and mutant were grown in LB medium; ≈106 cells in 20 μl were placed on membranes and allowed to dry at room temperature. At time points up to 20 h, replicate samples were assayed for colony forming units (see Materials and Methods). Within 4 h, the mutant exhibits a 10-fold reduced survivability relative to the WT and 100-fold reduction after 11 h (Fig. 4 Left).
Fig. 4.
Planktonic (Left) or biofilm (Right) mutant cells are sensitive to desiccation. Data are geometric means with error bars of 1 standard error of four to six replicate measurements (see Materials and Methods).
Inasmuch as bacteria in a hygroscopic biofilm likely show increased resistance to desiccation, failure of the mutant to produce exopolymer and a mature biofilm suggested it would be more sensitive to desiccation. After 24 h of growth in LB medium to produce static biofilms, cultures were assayed for desiccation survival over a period of weeks. After one week, survival of the mutant decreased 10-fold relative to the WT and declined even more over the next 11 weeks (Fig. 4 Right).
Discussion
P. aeruginosa (PA01) is adapted to natural environments and considered an opportunistic pathogen in humans. The organism is unusual in the number and variety of substrates it can grow on, as well as its intrinsic and developed resistance to antibiotics. Poly P metabolism is crucial for PA01 as shown by null mutants of poly P kinase 1 (ppk1) with deficiencies in motility, quorum sensing, biofilm formation, and virulence (3, 6, 15). We report here that a ppk1 mutant (PAOM5) also fails to produce exopolymer and survive exposure to the β-lactam antibiotic carbenicillin or desiccation. A remarkable compaction of the nucleoid and structural alterations in the cell envelope are further indications of the profound need for ppk1 and/or poly P in PA01.
PPK1 of P. aeruginosa shares some genetic and biochemical features with its homolog in E. coli and also some distinct differences. In E. coli, the operon with the encoding gene and the exopolyPase encoded by ppx are organized in a colinear arrangement, whereas in PA01, the two adjacent genes are convergent and overlap and, thus, do not form an operon; the effects on transcriptional regulation of PAO1 ppk1 are not known. Most significantly, PA01, unlike E. coli, contains a family of three additional genes that encode enzymes for poly P synthesis and utilization that are nonhomologous to PPK1: PPK2, a cryptic PPK2, and PAP, a putative poly P-adenylate phosphotransferase (2), (K. Ishige and A.K., unpublished data). PPK1 of E. coli is a multifunctional enzyme with five demonstrated enzymatic activities, including the synthesis of poly P from ATP (the forward reaction) and the generation of ATP from poly P and ADP (the reverse reaction) (17), both of which have been demonstrated for PAO1 PPK1, although the other three have not. In addition, PAO1 PPK1 forms oligomers, likely octamers, in the presence of short-chain poly P (18). These, in turn, form filaments in the presence of ATP (K. Ishige, D. J. Griffith, and A.K., unpublished data), which is blocked by the addition of phalloidin (19), an inhibitor of actin filament formation.
PPK1 in PA01 is associated almost exclusively with the membrane fraction as in E. coli, and the WT activities in both organisms are similar. Poly P levels in minimal medium are similar in PA01 and E. coli, but under the stringent condition induced by the amino acid analog serine hydroxamate, PA01 levels are considerably higher (14). PPX activity in the growth cycle is inversely related to that of PPK1 in PA01 but directly related in E. coli. Thus, the two organisms, although closely related, have evolved significantly different patterns of poly P metabolism in response to their different habitats.
Three null mutants of PA01 (Δppk1::Tet) showed marked reductions of PPK1 activity as in the E. coli Δppk1ppx::Kan mutant, <4% that of the respective WT (4). However, poly P levels in the PA01 mutants were reduced by 21–73% compared with the parental strain, due presumably to the additional enzymes of poly P synthesis; levels in the E. coli mutant are <5% of the WT. Of the three PA01 mutants isolated, one was selected for the studies presented here.
The PA01 mutant is deficient in all motilities on solid media: swimming, swarming, and twitching. Scanning electron microscopy showed both WT and mutant to have similar numbers of polar flagellae and normal type IV pili, as previously reported (11). The differences between mutant and WT motilities on agar of different densities indicated that the mutant defects might be surface-related and suggested that the flagellar-mediated swimming motility deficiency would be exacerbated under planktonic conditions for lack of a solid surface. Video microscopy showed the virtual lack of swimming motility in the mutant (<1% of WT), possibly due to structural defects in the flagellar motor or its function, although neither have been tested.
Transmission electron microscopy revealed major structural alterations in the nucleoid and envelope of the mutant as well as its smaller size, the first such exponential phase phenotypes observed in any ppk1 mutant. Adaptation to stationary phase begins several generations before its onset (20), suggesting a role for PPK1 in the earliest stages of this process. By inspection, as in Fig. 2, nucleoids in the mutant are strikingly compacted and the cytoplasm is irregularly distributed within all cells. However, the digitally determined mean estimates for WT and mutant relative nucleoid areas in the micrograph cross-sections differ by <50% (41% and 26%, respectively, with considerable variations). This discrepancy is likely due to the difficulty in determining the precise location of the WT nucleoid because it is distributed within the cell with other components. It occupies most of the cell volume so the WT mean value reported is likely an underestimate.
This compaction resembles that observed in E. coli strains that overproduce the histone-like protein H-NS (21). That poly P can readily displace the bound DNA in isolated chromatin (K. Ishige and A.K., unpublished data) indicates the capacity of poly P to influence chromosomal structure and function. Further, DNA microarray data show large, global effects on gene expression in the mutant compared with the WT (K. Ishige and A.K., unpublished data). In the mutant, the arrangement of the inner and outer membranes and the interstitial periplasmic space appear distorted, perhaps as a consequence of the compacted nucleoid and its function. Although the defects in envelope ultrastructure and compacted nucleoid have not been demonstrated in any way to be related to the other observed mutant phenotypes, namely immotility and altered gene expression, respectively, such causal relationships are possible.
Mutant cells fail to produce a WT biofilm (3) for which two explanations are now possible. The lack of planktonic flagellar and twitching motilities, both required for P. aeruginosa biofilm formation (22), may be sufficient. In addition, the failure of the mutant to produce exopolymers, as observed in 3-day-old cultures by scanning electron microscopy, would result in a defective biofilm architecture. Poly P may regulate the expression of alginate, a principal component of Pseudomonad exopolysaccharides (23). A mucoid strain of PA01 has higher levels of poly P than a nonmucoid strain; these levels are greatly reduced in a mutant of algR2, which encodes a master regulator of alginate synthesis. Poly P may be a sensor for alginate production or the source of the GTP required for the synthesis of the GDP-mannose alginate precursor. The coordinated regulation of these two polymers may occur at the transcriptional level. Some Pseudomonas genes, whose products are critical for alginate biosynthesis, exhibit regulation by the alternate sigma factor σE (24). Transcriptional analysis of ppk1 in P. aeruginosa 8830, a stable mucoid strain isolated from cystic fibrosis patient sputum, suggested regulation by a σE-responsive promoter (25); this has not been tested in PAO1.
The defects in the ppk1 mutant described to this point indicate, as with the corresponding E. coli mutant, a failure to respond to a variety of stresses. Among them is the loss of the high level of resistance of PA01 to the β-lactam antibiotic, carbenicillin. This loss might be anticipated from the alterations in envelope structure and the involvement of the sigma factor RpoS, known from E. coli studies to depend on poly P (26), in responses to antibiotics. When assayed for sensitivity to carbenicillin, the mutant showed a several hundredfold decrease in survival at a drug concentration that has little effect on the WT. We are exploring the possibility of changes in the amount and/or reduction in cross-linking of peptidoglycan as cause(s) of the increased sensitivity. Other candidate mechanisms are changes in multidrug efflux pumps or inducible lactamases.
Desiccation is a stress encountered by bacteria throughout evolution. After 10 h of drying, planktonic mutant cells lose viability >100-fold greater than the WT. The sensitivity of the PA01 mutant in static biofilms was 10-fold greater after one week and, by week 12, increased by >400-fold. Increasing hyperosmolarity may be the cause, because the E. coli ppk1 mutant is 10- to 100-fold more sensitive than the WT when exposed to 2.5 M sodium chloride (16). Osmotic pressure induces the E. coli general stress response, reviewed in ref. 27, which is diminished in the E. coli ppk1 mutant by the down-regulation of RpoS function (16, 26). Mutant cells exposed to desiccation thus may be unable to mount an adequate stress response for survival.
The reported studies show poly P (and/or PPK1) to be important for many functions and processes in P. aeruginosa PA01. The ppk1 null mutant exhibits pleiotropic phenotypes including decreased virulence, defective motility and biofilm formation, and a compacted nucleoid. The mutant shares features with the E. coli ppk1 mutant in responses to various stresses. Phenotypes in both organisms are manifest almost entirely upon entry into stationary phase and where tested are eliminated by complementation, indicating similar functions of PPK1 and/or poly P in these two organisms. However, differences between the two suggest that P. aeruginosa in its natural habitat and in infections, human or other, has evolved with special strategies to produce and use poly P. In the capacities that all cells must have to respond and adapt to environmental stresses, physical, chemical, nutritional, and biological, poly P plays a central role. The involvement of poly P in many cellular processes, including development and the fitness of both predator and prey, in all organisms tested demonstrates its widespread biologic importance.
Materials and Methods
Bacterial Strains, Media, and Growth.
WT P. aeruginosa PAO1 was a gift of Daniel W. Martin (East Carolina University, Greenville, NC). P. aeruginosa (WT PAO1 and the derivative Δppk1::Tet mutant, PAOM5) cultures were grown at 37°C with aeration in LB medium or on Pseudomonas Isolation Agar (EM Science, Gibbstown, NJ). Δppk1::Tet derivative strains were selected with 250 μg/ml tetracycline. Strains containing pHEPAK11 were grown in the presence of 300 μg/ml carbenicillin. E. coli DH5α was used for subcloning work.
Growth conditions for inducing poly P accumulations in Mops (3-(N-morpholino)propanesulfonic acid) minimal medium (Sigma, St. Louis, MO) due to phosphate (Pi) and amino acid downshifts were described in ref. 13. Briefly, cells were grown overnight with glucose (4 mg/ml) as a carbon source, 2 mM Pi (as K2HPO4), and 20 μg/ml of all 20 aa followed by at least 1:100 dilution in fresh Mops with glucose (4 mg/ml), 0.1 mM Pi, and 2 μg/ml amino acids. Induction of poly P accumulations by addition of the amino acid analog serine hydroxamate (SHX) to Mops medium to effect amino acid starvation was described in ref. 14. Briefly, cells in medium with 0.4 mM Pi and 40 μg/ml of all 20 aa were grown to an OD540 of ≈0.2, and SHX (0.5 μg/ml) was added.
Construction of Δppk1::Tet Mutant and Complemented Strains.
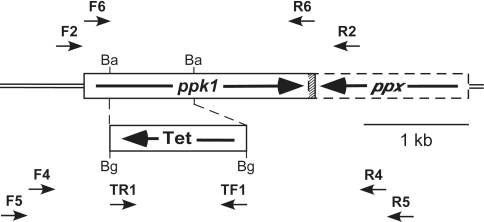
A 2.45-kb DNA fragment containing the chromosomal P. aeruginosa PAO1 ppk1 gene was amplified by PCR with primers F2 and R2 (Fig. 5; for primer information see SI Table 3) and cloned into the HindIII and XbaI sites in pBluescriptII SK(+) to generate pSKPAK1. The PCR fragment starts at 107 bp upstream of the A in the ATG translational start codon (bp 150 in GenBank locus AB007587) and ends 115 bp downstream of the A in the TGA stop codon, not as reported in ref. 11. A 0.8-kb BamHI fragment in the amino terminus half of the ppk1 ORF in pSKPAK1 was replaced with a 1.3-kb BglII fragment containing the tetracycline resistance-encoding gene cassette from pDMB100 (ref. 28; kindly supplied by Daniel W. Martin) in the opposite transcriptional orientation to create pPAK1TC4. WT PAO1 was electroporated with pPAK1TC4, and transformants were selected on LB plates with tetracycline (250 μg/ml). Double cross-over recombination of the mutation was verified first by genomic PCR with combinations of primers F4, F5, R4, R5, TF1, and TR1 and, subsequently, by biochemical determinations of PPK1 activity and poly P levels. Mutant PAOM5 was complemented by transformation with a plasmid overexpressing P. aeruginosa ppk1 under tac promoter control, pHEPAK11, constructed by PCR amplifying the ppk1 gene in plasmid pPPK02F (ref. 29; kindly supplied by Toshikazu Shiba, Fujirebio, Inc., Tokyo, Japan) by using primers F6 and R6 and inserting the resulting product into the HindIII and EcoRV sites in the medium copy number E. coli-P. aeruginosa shuttle vector pMMB66H (30). Transformants were selected on LB agar with 300 μg/ml carbenicillin.
Fig. 5.
Chromosomal organization of ppk1 ppx and construction/verification of the Δppk1::Tet mutation. In PAO1, ppk1 (PA5242; 2,210 bp) and ppx (PA5241; 1,520 bp) are convergently transcribed, and their ORFs overlap by 14 bp at the 3′ ends, denoted by the hatched area. Construction of the mutation is described in Materials and Methods. Tet denotes the tetracycline resistance-encoding gene cassette from pDMB100 (28). Ba and Bg indicate BamHI and BglII endonuclease restriction sites, respectively. The small arrows denote primers (described in SI Table 3) used in cloning ppk1 and in the genetic verification by PCR of the mutation (see Materials and Methods).
Assays for PPK1 and PPX Activities and Poly P Levels.
PPK1 activity was measured as the conversion of [32P-γ]ATP to poly P by thin-layer chromatography analysis as described in ref. 31. PPX activity was measured as the conversion of poly P to Pi as described in ref. 32. Poly P was extracted by the glassmilk method and levels were determined by conversion to ATP by E. coli PPK1 followed by conversion to light in a luciferase reaction and quantification in a TopCount NXT microplate scintillation and luminescence counter (Packard Instruments (PerkinElmer), Boston, MA) as described in ref. 33.
Scanning Electron Microscopy.
Strains were grown in LB medium to an OD600 of 0.6. Cells (109) were seeded onto individual nitrocellulose membranes in 24-well culture plates. The inoculated plates were incubated at 37°C and examined at 12, 24, 48, and 72 h. The membranes were removed from the plate and fixed in 2.5% glutaraldehyde and 0.15% ruthenium red for 3 h at 40°C. They were then rinsed in PBS solution, followed by postfixation in 1% osmium tetroxide/0.1 M sodium cacoldylate (pH 6.5)/0.2% ruthenium red for 2 h at room temperature. The membranes were dehydrated in a series of graded ethanol solutions, dried with a critical-point dryer (Tousimis, Rockville, MD) with carbon dioxide, and mounted on aluminum stubs with silver paste. These were dried for 3 h, and then coated with gold-palladium with a Polaron cool-sputter coater (Quorum Technologies, Newhaven, U.K.). The membranes were examined in a scanning electron microscope (JSM-6300F; JEOL USA, Peabody, MA) at 15 kV.
Transmission Electron Microscopy and Negative Staining.
A suspension of cells grown overnight in LB medium was inoculated into fresh LB medium and grown to an OD600 of 0.6. The log-phase bacteria were fixed in 2.5% glutaraldehyde in PBS, embedded in 1% agar, and postfixed in 1% osmium tetroxide. The fixed samples then were dehydrated through a series of graded ethanol solutions. After further dehydration in propylene oxide, the specimens were embedded in Spurr's low-viscosity embedding resin (Ted Pella, Inc., Redding, CA) and sectioned. For negative staining, uranyl acetate and lead citrate were added and reinforced with evaporated carbon. The specimens were examined with a transmission electron microscope (JEM-1200EXII; JEOL USA) at 80 kV. High-resolution digitized images were used to determine the cross-sectional areas of individual nucleoids relative to those of the corresponding whole cells, expressed as percentages, by using the associated size bar. Areas were calculated by the software program Volocity, version 3.7.0 (Improvision, Ltd., Coventry, U.K.) in the Cell Science Imaging Facility in the Beckman Center at Stanford University. Although Volocity used pixel contrast to define the apparent nucleoid boundaries in all cells, subjective determination was made in some cases, particularly in WT cells. Only whole, undividing cells were used, regardless of orientation, as long as some part of the nucleoid was observable. The values reported are arithmetic means ± 1 standard deviation.
Planktonic Motility Studies.
A suspension of cells grown overnight in LB medium was inoculated into fresh LB medium and grown to an OD600 of 0.6. The cultures were spun down and resuspended in LB medium to a concentration of 1.6 × 108 cells per ml. The bacterial suspension (8 μl) was spotted onto a glass slide, covered with a no. 1.5 (22 × 22 mm) glass coverslip and sealed with clear nail lacquer. The chambered bacteria were inverted and allowed to settle to minimize the influence of convection. Random microscopic fields were selected, and serial video images were captured for 10 sec. Bacteria were visualized with a ×100 phase-contrast oil objective on a Diaphot (Nikon, Tokyo, Japan) inverted microscope. Serial video images were captured at 20 frames per second with a LG-3 frame-grabber (Scion, Frederick, MD), a Dage CCD-72 camera and a Digital Image Processor DSP-2000 (Dage-MTI, Michigan City, IN). The images were captured on a Power Macintosh G3 computer (Apple Computer, Cupertino, CA) using Scion Image 1.62 software (Scion); a freeware version of NIH-Image to view the video can be found at http://rsb.info.nih.gov/nih-image. Estimates of bacterial motility by image analysis, as described in ref. 34, were obtained by using Metamorph software version 4.3 (Universal Imaging, Downingtown, PA). Briefly, serial images of the 200-frame animation sequences were subtracted by using automated Metamorph journals in the following order: the second from the first, the third from the second, to the 200th frame. The subtractions were binarized, and the pixels were counted in every frame of the sequence. Motility in the sequence was expressed as the total number of pixels remaining after the subtraction of two serial video images and then was normalized for bacterial density. The WT is arbitrarily assigned a motility value of 100%.
Carbenicillin-Sensitivity Assays.
Strains were grown overnight in LB and subcultured into replicate flasks of LB medium to a starting OD550 of ≈0.04. Approximately one generation before entry into stationary phase, at an OD550 of ≈0.8, the indicated concentrations of carbenicillin were added to the flasks. Survivability initially was assessed by OD, then by duplicate colony plate counts of serial dilutions in LB medium. The standard error of plating was <20%.
Desiccation Survival Assays.
Strains were grown in LB medium in baffled conical flasks. At 24 h, the ODs were measured and the cultures diluted to 5 × 107 cells per ml. A 45-mm diameter, 0.2-μm pore size, hydrophobic edge, cellulose nitrate membrane (Sartorius, Goettingen, Germany), prewashed in LB medium (10 ml) and dried, was placed in a 9-cm diameter, triple-vent Petri dish. Cells (106) (20 μl) were spotted onto the center of the membrane, which was dried at room temperature (20–24°C). At sampling intervals of every hour for the mutant and every 2 h for the WT up to 12 h, then every hour, the membranes were rehydrated with 100 μl of saline solution and the bacteria were removed with a cell scraper. The scraper was rinsed in 5 ml of saline solution to which the scraped membrane was added and vortexed for 15 sec. Colony counting of the suspensions, either to inoculate the membranes or recovered from them, was performed on dilutions in saline solution plated onto LB agar by using a combination of 20-μl drops and spread plates (200 μl). The plates were incubated at 37°C for 24 h and colonies counted. The fraction of recovered cfu was calculated as the colony counts recovered from the membranes divided by the cfu of the inoculum.
Supplementary Material
Acknowledgments
We thank Drs. Daniel W. Martin and Toshikazu Shiba for plasmids, Kitty Lee at Stanford University's Cell Science Imaging Facility for expert assistance with the nucleoid area analyses, and Drs. A. Dale Kaiser and I. Robert Lehman for helpful comments on the manuscript. This work was supported by the National Institutes of Health (to C.D.F., M.H.R., and A.K.), the U.K. Department of Health (to P.J.W. and M.R.W.B.), and The Ellison Medical Foundation for funding (to A.K.) to support M.R.W.B. during his sabbatical stay in the A.K. laboratory.
Abbreviations
- poly P
polyphosphate
- PPK
polyphosphate kinase
- PPX
exopolyPase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609733104/DC1.
References
- 1.Brown MRW, Kornberg A. Proc Natl Acad Sci USA. 2004;101:16085–16087. doi: 10.1073/pnas.0406909101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Ishige K, Kornberg A. Proc Natl Acad Sci USA. 2002;99:16678–16683. doi: 10.1073/pnas.262655199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. Proc Natl Acad Sci USA. 2000;97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K-S, Rao NN, Fraley CD, Kornberg A. Proc Natl Acad Sci USA. 2002;99:7675–7680. doi: 10.1073/pnas.112210499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price-Carter M, Fazzio TG, Vallbona EI, Roth JR. J Bacteriol. 2005;187:3088–3099. doi: 10.1128/JB.187.9.3088-3099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parks QM, Hobden JA. Invest Ophthalmol Vis Sci. 2005;46:248–251. doi: 10.1167/iovs.04-0340. [DOI] [PubMed] [Google Scholar]
- 7.Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. J Bacteriol. 2005;187:7687–7695. doi: 10.1128/JB.187.22.7687-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi X, Rao NN, Kornberg A. Proc Natl Acad Sci USA. 2004;101:17061–17065. doi: 10.1073/pnas.0407787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Rao NN, Shiba T, Kornberg A. Proc Natl Acad Sci USA. 2005;102:13416–13420. doi: 10.1073/pnas.0506520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Gómez-García MR, Brown MRW, Kornberg A. Proc Natl Acad Sci USA. 2005;102:2731–2735. doi: 10.1073/pnas.0500023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashid MH, Kornberg A. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurst H, Shiba T, Kornberg A. J Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao NN, Liu S, Kornberg A. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda A, Murphy H, Cashel M, Kornberg A. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 15.Rashid MH, Rao NN, Kornberg A. J Bacteriol. 2000;182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao NN, Kornberg A. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzeng C-M, Kornberg A. J Biol Chem. 2000;275:3977–3983. doi: 10.1074/jbc.275.6.3977. [DOI] [PubMed] [Google Scholar]
- 18.Ishige K, Zhang H, Kornberg A. Proc Natl Acad Sci USA. 2002;99:16684–16688. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-García MR, Kornberg A. Proc Natl Acad Sci USA. 2004;101:15876–15880. doi: 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MRW, Williams P. Annu Rev Microbiol. 1985;39:527–556. doi: 10.1146/annurev.mi.39.100185.002523. [DOI] [PubMed] [Google Scholar]
- 21.Spurio R, Durrenberger M, Falconi M, La Teana A, Pon CL, Gualerzi CO. Mol Gen Genet. 1992;231:201–211. doi: 10.1007/BF00279792. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole GA, Kolter R. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim H-Y, Schlictman D, Shankar S, Xie Z, Chakrabarty AM, Kornberg A. Mol Microbiol. 1998;27:717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 24.Shankar S, Ye RW, Schlictman D, Chakrabarty AM. Adv Enzymol Relat Areas Mol Biol. 1995;70:221–255. doi: 10.1002/9780470123164.ch4. [DOI] [PubMed] [Google Scholar]
- 25.Zago A, Chugani S, Chakrabarty AM. Appl Environ Microbiol. 1999;65:2065–2071. doi: 10.1128/aem.65.5.2065-2071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao NN, Kornberg A. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengge-Aronis R. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 28.Martin DW, Schurr MJ, Mudd MH, Deretic V. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 29.Ishige K, Kameda A, Noguchi T, Shiba T. DNA Res. 1998;5:157–162. doi: 10.1093/dnares/5.3.157. [DOI] [PubMed] [Google Scholar]
- 30.Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 31.Ahn K, Kornberg A. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 32.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 33.Ault-Riche D, Fraley CD, Tzeng CM, Kornberg A. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojima M, Tamagawa H, Nagata H, Hanioka T, Shizukuishi S. J Clin Periodontol. 2000;27:405–410. doi: 10.1034/j.1600-051x.2000.027006405.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.