Abstract
Histidine decarboxylase gene (HDC) encodes histidine decarboxylase which is the crucial enzyme for the biosynthesis of histidine. Studies have shown that histamine is likely to be involved in the regulation of reproduction system. To find the possible correlation between HDC gene and AANM (age at natural menopause), we selected 265 postmenopausal women from 131 nuclear families and performed a transmission disequilibrium test. Significant within-family associations with AANM for SNP rs854163 and SNP rs854158 of HDC gene were observed (P values = 0.0018 and 0.0197, respectively). After 1000 permutations, SNP rs854163 still remained significant within-family association with AANM. Consistently, we also detected a significant within-family association between haplotype block 2 (defined by SNP rs854163 and rs860526) and AANM in the haplotype analyses (P value = 0.0397). Our results suggest that the HDC gene polymorphisms are significantly associated with AANM in Caucasian women.
Keywords: HDC, Association, Age at natural menopause, SNP
Menopause status is one important anthropological variable influencing the overall health of women, especially those in advanced ages. Abnormal low age at natural menopause (AANM) correlates with female infertility due to ovarian aging [1]. Early menopause was also reported to be associated with the increased risks of fracture [2,3], coronary heart disease [4,5], and disorders of the central nervous system [6,7]. In addition, the Caucasian women with menopause ages of about 40–44 years have 4% higher risk of mortality than those with menopause ages of about 50–54 years [8].
There is a wide variation in AANM, varying between 40 and 60 years. Extensive studies have been conducted to search for the predictors of AANM. Some lifestyle factors were reported to be associated with AANM, such as obesity, smoking, alcohol consumption, and breastfeeding [9–11]. However, these lifestyle factors just accounted for a small part of the large variation in AANM [12]. Additionally, genetic factors were found to play an important role in the variation of AANM. Family study showed that the menopausal ages of daughters were significantly correlated with mothers’ [13]. Heritability estimates of AANM ranged from 0.63 to 0.87, suggesting a strong genetic control [14–16]. To date, the exact genes involved remain unknown, though some candidate genes underlying AANM have been proposed, such as ESR1 (estrogen receptor alpha), CYP1B1 (cytochrome P450, family 1, subfamily B, poly-peptide 1), Factor V Leiden, and FMR1 (fragile X mental retardation 1) [17–20].
Histidine decarboxylase (HDC) gene encodes histidine decarboxylase, which is the crucial enzyme for synthesis of histamine in human body. As an important bioactive substance, histamine is crucial to various physiological activities [21]. It has been found that histamine directly stimulated the secretion of GnRH (gonadotropin-releasing hormone), which is the key molecule in the regulation of gonadal hormone release [22]. Furthermore, study in mice suggested that peripheral histamine was an important regulatory factor of gonadal development during embryogenesis and sex steroid metabolism in adulthood [23]. All these findings support the hypothesis that histamine may play a role in the regulation of reproductive system.
To test the possible association between HDC gene polymorphisms and AANM, we performed a family-based association analyses in postmenopausal Caucasian women.
Material and methods
Subjects
This study was approved by the Creighton University Institutional Review Board. Signed informed-consent documents were obtained from all study participants before they entered the study. The design and sampling procedures have been published before [24]. In brief, all of 1873 participants from 405 nuclear families are US Caucasians of European origin and recruited for various genetic studies. Information about ages at menarche and menopause, number of pregnancies, alcohol consumption, use of hormone before menopause (hormone replacement therapy and oral contraceptives) and surgical history of reproductive system (hysterectomy and ovariectomy) was recorded for each subject by nurse-administered questionnaires. Menopausal status was defined as the date of last menses followed by 12 months of no menses. Specifically for present genetic association studies of AANM, women with surgical menopause or a history of hormone replacement therapy before menopause were excluded. Finally, data on AANM for 265 postmenopausal women from 131 nuclear families were available.
Genotyping
Genomic DNA was extracted from whole blood using a commercial isolation kit (Gentra Systems, Minneapolis, MN, USA) following the procedure detailed in the kit. DNA concentration was assessed by a DU530 UV/VIS Spectrophotometer (Beckman Coulter, Inc, Fullerton, CA, USA). A total of 14 SNPs in and around HDC gene were selected on the basis of the following criteria: (1) validation status, especially in Caucasians, (2) an average density of 1 SNP per 3 kb, (3) degree of heterozygosity, i.e., minor allele frequencies (MAF) > 0.05, (4) functional relevance and importance, (5) reported to dbSNP by various sources. 14 SNPs were successfully genotyped using the high-throughput BeadArray SNP genotyping technology of Illumina Inc. (San Diego, CA, USA) and 11 were analyzed subsequently (three rare SNPs with MAF < 0.05 were discarded in the analyses due to insufficient statistical power). The average rate of missing genotype data was reported by Illumina to be ~0.05%. The average genotyping error rate estimated through blind duplicating was reported to be less than ~0.01%. The analyzed SNPs covered the full transcript length of the HDC gene, with the intermarker distance of ~2.1 kb apart on average.
Statistical analyses
PedCheck (O’Connell and Weeks) was used to check Mendelian consistency of SNP genotype data and any inconsistent genotypes were removed. Then the error checking option embedded in Merlin [25] was run to identify and disregard the genotypes flanking excessive recombinants, thus further reducing genotyping errors. Allele frequencies for each SNP were calculated by allele counting, and the Hardy–Weinberg equilibrium was tested using the PEDSTATS procedure embedded in Merlin. Population haplotypes and their frequencies were inferred for HDC gene using PHASE v2.1.1 software among unrelated parents. LD structure was defined, using GOLD (http://www.sph.umich.edu/csg/abecasis/GOLD/), to chart pairwise D’ statistics derived from haplotype data. HaploBlockFinder (http://cgi.uc.edu/cgi-bin/kzhang/haploBlockFinder.cgi/) was used to identify block structures and select haplotype-tagging SNPs (htSNPs) of HDC gene. To infer haplotypes defined by the tagging SNPs within each block of HDC gene for all of the subjects, we adopted the algorithm of integer linear programming (ILP) implemented in PedPhase V2.0 (http://www.cs.ucr.edu/~jili/haplotyping.html), which is based on LD assumption and able to recover phase information at each marker locus with great speed and accuracy even in the presence of 20% missing data [26]. The quantitative transmission disequilibrium test (QTDT) software (http://www.sph.umich.edu/csg/abecasis/QTDT/) was used to test each SNP and haplotype with estimated frequencies greater than 5% for association with AANM variation. We adopted the orthogonal model implemented in QTDT for our analyses [27].The orthogonal tests were carried out in a variance component framework decomposing the genotype score into orthogonal between-(βb) and within-family (βw) components. The between-family component is sensitive to population admixture, while the within-family component is significant only in the presence of linkage disequilibrium (LD) and robust to population stratification/admixture. In the absence of population stratification, total association is more powerful to detect association compared with within-family association method. Linear regression and two tailed t tests were performed to detect potential confounders of AANM, including alcohol consumption, BMI and menarche.
Results
The basic characteristics of 265 study subjects are presented in Table 1. The mean AANM of these subjects is 49.28 years (standard deviation, SD = 3.78), ranging from 38 to 58 years. In this study, no significant difference in AANM was observed for all the chosen subjects stratified by alcohol consumption, BMI and menarche age (P values > 0.05).
Table 1.
Characteristics of study subjects included in this study
| Characteristics | Mean | SD |
|---|---|---|
| Age at menarche (years) | 13.10 | 1.37 |
| Age at menopause (years) | 49.28 | 3.78 |
| Height (cm) | 162.98 | 6.12 |
| Weight (kg) | 72.00 | 15.43 |
| BMI (kg/m2) | 27.67 | 5.56 |
Abbreviations: BMI, body mass index; SD, standard deviation.
The information of 11 SNPs of the HDC gene is shown in Table 2. All of them were in Hardy-Weinberg equilibrium (P values > 0.05). MAFs ranged from 0.17 to 0.42 in our sample. Five blocks were identified within the HDC gene (Table 3). As shown in Fig. 1, block 1 is located in the 5′-promoter region, block 2 extends from intron 1 to intron 2, block 3 lies in intron 2, block 4, the largest one, spans from intron 2 to intron 9, block 5 is localized in the 3′-UTR.
Table 2.
Information of the SNPs studied for HDC gene
| SNP | dbSNP | Polymorphism | MAF (%) | Distancea (bp) | Position |
|---|---|---|---|---|---|
| SNP 1 | rs2114447 | A/G | 0.36 | — | Promoter |
| SNP 2 | rs12901373 | A/C | 0.36 | 1936 | Promoter |
| SNP 3 | rs2187576 | A/G | 0.40 | 3277 | Promoter |
| SNP 4 | rs854163 | A/G | 0.26 | 4825 | Intron 1 |
| SNP 5 | rs860526 | A/G | 0.42 | 2727 | Intron 2 |
| SNP 6 | rs854158 | A/G | 0.32 | 1934 | Intron 2 |
| SNP 7 | rs8029889 | A/G | 0.21 | 2177 | Intron 2 |
| SNP 8 | rs2853766 | A/G | 0.23 | 2657 | Intron 4 |
| SNP 9 | rs854150 | C/G | 0.37 | 6084 | Intron 9 |
| SNP 10 | rs1802536 | A/C | 0.17 | 1262 | 3′-UTR |
| SNP 11 | rs10519263 | A/G | 0.17 | 3544 | 3′-UTR |
Note. Distance
denotes the distance to previous SNP.
Table 3.
The information of all the studied haplotypes of HDC gene
| Haplotype | Component SNPs | Frequency (%) |
|---|---|---|
| Hap1 | SNPs 1–3 | |
| AAA | 63.63 | |
| AAG | 36.37 | |
| Hap2 | SNPs 4–5 | |
| AA | 23.92 | |
| AG | 34.14 | |
| GA | 1.39 | |
| GG | 40.55 | |
| Hap3 | SNP 6 | |
| A | 68.1 | |
| G | 31.9 | |
| Hap4 | SNPs 7–9 | |
| AAA | 0.26 | |
| AAG | 1.13 | |
| AGC | 19.52 | |
| AGG | 41.22 | |
| GAC | 0.44 | |
| GAG | 21.06 | |
| GGC | 0.67 | |
| GGG | 15.7 | |
| Hap5 | SNPs 10–11 | |
| AA | 82.63 | |
| AG | 17.37 |
Significant evidence of within-family association between HDC and AANM was observed for both single-SNP markers and haplotypes. Table 4 summarized the association analyses results. As shown by the data, both SNP 4 and SNP 6 showed significant within-family associations with AANM (P values = 0.0018 and 0.0197, respectively). A suggestive within-family association was observed for SNP 5 (P value = 0.0677). After 1000 permutations by QTDT, SNP 4 still remained the significant within- family association with AANM, giving the P value of 0.0150 (to reach the experiment-wide significance level of α = 0.05, the single test threshold P value = 0.0170). Significant within-family association was also observed between HDC gene block 2 containing SNPs 4–5 and AANM (P value = 0.0397). All these suggest that the association analyses results of SNP 4 and block 2 were consistent. We further assessed the effect of different alleles of SNP 4 on AANM. The mean AANM for subjects carrying the T allele of SNP 4 is 49.39 years (SD = 3.72), and for the non-carriers it is 47.81 years (SD = 4.54).
Table 4.
Summary of QTDT analyses results
| dbSNP | Population stratification (P value) | Total association (P value) | Within-family association (P value) | |
|---|---|---|---|---|
| SNP 1 | rs2114447 | NS | 0.4405 | — |
| SNP 2 | rs12901373 | NS | 0.4405 | — |
| SNP 3 | rs2187576 | NS | 0.7255 | — |
| SNP 4 | rs854163 | 0.0059 | 0.1038 | 0.0018 |
| SNP 5 | rs860526 | NS | 0.2350 | 0.0677 |
| SNP 6 | rs854158 | 0.0530 | 0.1070 | 0.0197 |
| SNP 7 | rs8029889 | NS | 0.2019 | — |
| SNP 8 | rs2853766 | NS | 0.2353 | — |
| SNP 9 | rs854150 | NS | 0.2756 | 0.0947 |
| SNP 10 | rs1802536 | NS | 0.5263 | — |
| SNP 11 | rs10519263 | NS | 0.5263 | — |
| Hap2 | ||||
| AA | rs854163 (SNP 4) | |||
| rs860526 (SNP 5) | NS | 0.1308 | 0.0397 | |
Note. 1. Five haplotype blocks within the HDC gene were tested but only the significant results were reported here.
2. Significant P values (P < 0.05) are highlighted in bold.
Discussion
We utilized the transmission disequilibrium test to find the possible correlation between HDC gene polymorphisms and AANM. In our study population, HDC gene demonstrates significant association with AANM.
Our most impressive finding is the significant association between the SNP 4 of HDC gene and AANM. SNP 4 is located at the boundary between exon 1 and intron 1 of the HDC gene. In our association analyses, SNP 4 and the related haplotype block 2 presented significant within-family association with AANM. The association with AANM for SNP 4 remained significant even after 1000 permutations implemented in QTDT. However, we did not detect significant total association for SNP 4, which could be explained by the existence of population stratification at the SNP 4 locus (P value = 0.0059). We also found that the subjects carrying T allele of SNP 4 were 3.30% later in AANM than the non-carriers on average. These results suggest that the SNP 4 locus could be in strong linkage dis-equilibrium with the casual genetic variants influencing the AANM variation. Further study with denser markers is required to locate the exact causal loci within the HDC gene.
Twin study has suggested that more than 60% of the AANM variation could be explained by the genetic factors [14–16]. However, very few genes have been reported to affect AANM [17–20]. To our knowledge, the possible effect of HDC gene on AANM variation has never been reported. Yet, biological studies focusing on the function of HDC gene in vivo may provide some clues in this regard.
In females, the follicle storage established during the fetal stage and the rate of follicles atresia in later life determine the number of mature follicle, which is believed to be the major determinant of the onset of menopause [28]. Different kinds of hormone, growth factors, and cytokine are involved in these processes [29–32]. Bodis J’s study found that histamine could directly stimulate the steroid production of ovarial granulosa cells [33]. Histamine is also recognized as an autocrine growth factor [34,35], which influences the secretion of various growth factors and cytokines in human body [36–38]. What is more interesting, histamine could induce ovulation in the isolated perfused ovary [39]. In addition, histamine is an important neuro-modulator in the central nervous system which also exerts a role in the regulation of AANM [40]. Some studies focusing on the biological function of histamine at hypothalamus suggested that histamine influenced the secretion of GnRH (gonadotropin-releasing hormone), GTH (gonadotropic hormone), and even estrogen [22,41,42], which are the key molecules for the regulation of female reproduction system. Taken together, those biological and physiological studies all lent support to our finding that the HDC gene polymorphisms influence the variation of AANM.
In conclusion, we provide the evidence that the HDC gene polymorphisms are associated with AANM in Caucasians for the first time. To confirm our results, replication studies with denser markers and larger sample size will be needed. Moreover, further molecular genetics studies may be necessary to find the exact causal alleles within HDC gene that influence the AANM variation.
Fig. 1.
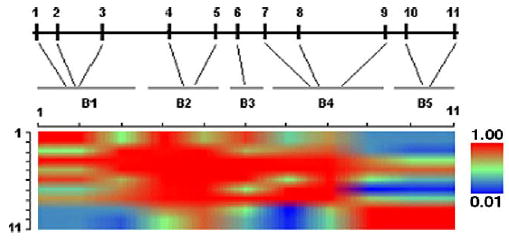
LD intensity measured by |D′| across loci for HDC gene. Pairwise LD for SNPs were calculated by GOLD. Five haplotype blocks (B1-B5) were identified within HDC gene.
Acknowledgments
The study was partially supported by grants from National Institute of Health, the State of Nebraska. This study also benefited from 211 State Key Research Fund by Xi’an Jiaotong University.
References
- 1.te Velde ER, Dorland M, Broekmans FJ. Age at menopause as a marker of reproductive ageing. Maturitas. 1998;30(2):119–125. [PubMed] [Google Scholar]
- 2.van d V, van Der Weijer PH, Barentsen R. Early menopause: increased fracture risk at older age. Osteoporos Int. 2003;14(6):525–530. doi: 10.1007/s00198-003-1408-1. [DOI] [PubMed] [Google Scholar]
- 3.van der KM, de Laet CE, McCloskey EV, et al. Risk factors for incident vertebral fractures in men and women: the Rotterdam Study. J Bone Miner Res. 2004;19(7):1172–1180. doi: 10.1359/JBMR.040215. [DOI] [PubMed] [Google Scholar]
- 4.Hu FB, Grodstein F, Hennekens CH, et al. Age at natural menopause and risk of cardiovascular disease. Arch Intern Med. 1999;159(10):1061–1066. doi: 10.1001/archinte.159.10.1061. [DOI] [PubMed] [Google Scholar]
- 5.van der Schouw YT, van der GY, Steyerberg EW, Eijkemans JC, Banga JD. Age at menopause as a risk factor for cardiovascular mortality. Lancet. 1996;347(9003):714–718. doi: 10.1016/s0140-6736(96)90075-6. [DOI] [PubMed] [Google Scholar]
- 6.Henderson VW. Menopause and disorders of the central nervous system. Minerva Ginecol. 2005;57(6):579–592. [PubMed] [Google Scholar]
- 7.Genazzani AR, Spinetti A, Gallo R, Bernardi F. Menopause and the central nervous system: intervention options. Maturitas. 1999;31(2):103–110. doi: 10.1016/s0378-5122(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 8.Mondul AM, Rodriguez C, Jacobs EJ, Calle EE. Age at natural menopause and cause-specific mortality. Am J Epidemiol. 2005;162(11):1089–1097. doi: 10.1093/aje/kwi324. [DOI] [PubMed] [Google Scholar]
- 9.Kinney A, Kline J, Levin B. Alcohol, caffeine and smoking in relation to age at menopause. Maturitas. 2006;54(1):27–38. doi: 10.1016/j.maturitas.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Riener EK, Keck C, Worda C, Hefler LA, Tempfer CB. Body mass index but not a polymorphism of the interleukin-1 receptor antagonist (IL-1 RA) gene is associated with age at natural menopause. Gynecol Obstet Invest. 2004;58(2):117–120. doi: 10.1159/000078941. [DOI] [PubMed] [Google Scholar]
- 11.Dvornyk V, Long JR, Liu PY, et al. Predictive factors for age at menopause in Caucasian females. Maturitas. 2006;54(1):19–26. doi: 10.1016/j.maturitas.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 12.van Noord PA, Dubas JS, Dorland M, Boersma H, te VE. Age at natural menopause in a population-based screening cohort: the role of menarche, fecundity, and lifestyle factors. Fertil Steril. 1997;68(1):95–102. doi: 10.1016/s0015-0282(97)81482-3. [DOI] [PubMed] [Google Scholar]
- 13.Torgerson DJ, Thomas RE, Reid DM. Mothers and daughters menopausal ages: is there a link? Eur J Obstet Gynecol Reprod Biol. 1997;74(1):63–66. doi: 10.1016/s0301-2115(97)00085-7. [DOI] [PubMed] [Google Scholar]
- 14.de Bruin JP, Bovenhuis H, van Noord PA, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16(9):2014–2018. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 15.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman’s reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83(6):1875–1880. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 16.Treloar SA, Do KA, Martin NG. Genetic influences on the age at menopause. Lancet. 1998;352(9134):1084–1085. doi: 10.1016/S0140-6736(05)79753-1. [DOI] [PubMed] [Google Scholar]
- 17.Weel AE, Uitterlinden AG, Westendorp IC, et al. Estrogen receptor polymorphism predicts the onset of natural and surgical menopause. J Clin Endocrinol Metab. 1999;84(9):3146–3150. doi: 10.1210/jcem.84.9.5981. [DOI] [PubMed] [Google Scholar]
- 18.Hefler LA, Grimm C, Heinze G, et al. Estrogen-metabolizing gene polymorphisms and age at natural menopause in Caucasian women. Hum Reprod. 2005;20(5):1422–1427. doi: 10.1093/humrep/deh848. [DOI] [PubMed] [Google Scholar]
- 19.van Asselt KM, Kok HS, Peeters PH, et al. Factor V Leiden mutation accelerates the onset of natural menopause. Menopause. 2003;10(5):477–481. doi: 10.1097/01.GME.0000056040.51813.1A. [DOI] [PubMed] [Google Scholar]
- 20.Mallolas J, Duran M, Sanchez A, et al. Implications of the FMR1 gene in menopause: study of 147 Spanish women. Menopause. 2001;8(2):106–110. doi: 10.1097/00042192-200103000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S. [Physiological function mediated by histamine synthesis] Yakugaku Zasshi. 2003;123(7):547–559. doi: 10.1248/yakushi.123.547. [DOI] [PubMed] [Google Scholar]
- 22.Noris G, Hol D, Clapp C, Martinez de la EG. Histamine directly stimulates gonadotropin-releasing hormone secretion from GT1-1 cells via H1 receptors coupled to phosphoinositide hydrolysis. Endocrinology. 1995;136(7):2967–2974. doi: 10.1210/endo.136.7.7789322. [DOI] [PubMed] [Google Scholar]
- 23.Pap E, Racz K, Kovacs JK, et al. Histidine decarboxylase deficiency in gene knockout mice elevates male sex steroid production. J Endocrinol. 2002;175(1):193–199. doi: 10.1677/joe.0.1750193. [DOI] [PubMed] [Google Scholar]
- 24.Xiong DH, Shen H, Xiao P, et al. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res. 2006;21(3):424–437. doi: 10.1359/JBMR.051202. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Jiang T. Computing the minimum recombinant haplotype configuration from incomplete genotype data on a pedigree by integer linear programming. J Comput Biol. 2005;12(6):719–739. doi: 10.1089/cmb.2005.12.719. [DOI] [PubMed] [Google Scholar]
- 27.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson SJ, Nelson JF. Follicular depletion during the menopausal transition. Ann N Y Acad Sci. 1990;592:13–20. doi: 10.1111/j.1749-6632.1990.tb30312.x. [DOI] [PubMed] [Google Scholar]
- 29.Behl R, Kaul R. Insulin like growth factor 1 and regulation of ovarian function in mammals. Indian J Exp Biol. 2002;40(1):25–30. [PubMed] [Google Scholar]
- 30.Quirk SM, Cowan RG, Harman RM, Hu CL, Porter DA. Ovarian follicular growth and atresia: the relationship between cell proliferation and survival. J Anim Sci. 2004;82(ESuppl):E40–E52. doi: 10.2527/2004.8213_supplE40x. [DOI] [PubMed] [Google Scholar]
- 31.Bornstein SR, Rutkowski H, Vrezas I. Cytokines and steroidogenesis. Mol Cell Endocrinol. 2004;215(1–2):135–141. doi: 10.1016/j.mce.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Tamura H, Kumasaka K, Miyajima A, Suga T, Kogo H. Ovarian immune cells express granulocyte-macrophage colony- stimulating factor (GM-CSF) during follicular growth and luteinization in gonadotropin-primed immature rodents. Mol Cell Endocrinol. 1998;142(1–2):153–163. doi: 10.1016/s0303-7207(98)00109-9. [DOI] [PubMed] [Google Scholar]
- 33.Bodis J, Tinneberg HR, Schwarz H, Papenfuss F, Torok A, Hanf V. The effect of histamine on progesterone and estradiol secretion of human granulosa cells in serum-free culture. Gynecol Endocrinol. 1993;7(4):235–239. doi: 10.3109/09513599309152507. [DOI] [PubMed] [Google Scholar]
- 34.Rivera ES, Cricco GP, Engel NI, Fitzsimons CP, Martin GA, Bergoc RM. Histamine as an autocrine growth factor: an unusual role for a widespread mediator. Semin Cancer Biol. 2000;10(1):15–23. doi: 10.1006/scbi.2000.0303. [DOI] [PubMed] [Google Scholar]
- 35.Cricco GP, Davio CA, Martin G, et al. Histamine as an autocrine growth factor in experimental mammary carcinomas. Agents Actions. 1994;43(1–2):17–20. doi: 10.1007/BF02005757. [DOI] [PubMed] [Google Scholar]
- 36.van d V, van Buul-Offers SC, Gloudemans T, Roholl PJ, Sussenbach JS, Den OW. Histamine-stimulated expression of insulin-like growth factors in human glioma cells. Br J Cancer. 1997;75(8):1091–1097. doi: 10.1038/bjc.1997.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niisato N, Ogata Y, Furuyama S, Sugiya H. Histamine H1 receptor-induced Ca2+ mobilization and prostaglandin E2 release in human gingival fibroblasts. Possible role of receptor-operated Ca2+ influx. Biochem Pharmacol. 1996;52(7):1015–1023. doi: 10.1016/0006-2952(96)00417-0. [DOI] [PubMed] [Google Scholar]
- 38.Leonardi A, DeFranchis G, De PM, Fregona I, Plebani M, Secchi A. Histamine-induced cytokine production and ICAM-1 expression in human conjunctival fibroblasts. Curr Eye Res. 2002;25(3):189–196. doi: 10.1076/ceyr.25.3.189.13479. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt G, Owman C, Sjoberg NO. Histamine induces ovulation in the isolated perfused rat ovary. J Reprod Fertil. 1986;78(1):159–166. doi: 10.1530/jrf.0.0780159. [DOI] [PubMed] [Google Scholar]
- 40.Wise PM, Smith MJ, Dubal DB, et al. Neuroendocrine modulation and repercussions of female reproductive aging. Recent Prog Horm Res. 2002;57:235–256. doi: 10.1210/rp.57.1.235. [DOI] [PubMed] [Google Scholar]
- 41.Ohtsuka S, Nishizaki T, Tasaka K, et al. Estrogen stimulates gonadotropin-releasing hormone release from rat hypothalamus independently through catecholamine and histamine in vitro. Acta Endocrinol (Copenh) 1989;120(5):644–648. doi: 10.1530/acta.0.1200644. [DOI] [PubMed] [Google Scholar]
- 42.Van Kirk EA, Halterman SD, Moss GE, Rose JD, Murdoch WJ. Possible role of histamine in the regulation of secretion of luteinizing hormone in the ewe. J Anim Sci. 1989;67(4):1006–1012. doi: 10.2527/jas1989.6741006x. [DOI] [PubMed] [Google Scholar]


