Abstract
Pancreatic endocrine cell differentiation depends on transcription factors that also contribute in adult insulin and glucagon gene expression. Islet cell development was examined in mice lacking MafB, a transcription factor expressed in immature α (glucagon+) and β (insulin+) cells and capable of activating insulin and glucagon expression in vitro. We observed that MafB−/− embryos had reduced numbers of insulin+ and glucagon+ cells throughout development, whereas the total number of endocrine cells was unchanged. Moreover, production of insulin+ cells was delayed until embryonic day (E) 13.5 in mutant mice and coincided with the onset of MafA expression, a MafB-related activator of insulin transcription. MafA expression was only detected in the insulin+ cell population in MafB mutants, whereas many important regulatory proteins continued to be expressed in insulin− β cells. However, Pdx1, Nkx6.1, and GLUT2 were selectively lost in these insulin-deficient cells between E15.5 and E18.5. MafB appears to directly regulate transcription of these genes, because binding was observed within endogenous control region sequences. These results demonstrate that MafB plays a previously uncharacterized role by regulating transcription of key factors during development that are required for the production of mature α and β cells.
Keywords: insulin, MafA, pancreas development
The pancreatic islets of Langerhans are composed of α, β, δ, ε, and pancreatic polypeptide (PP) cells, which produce the hormones glucagon, insulin, somatostatin, ghrelin, and PP, respectively. Collectively, these hormones regulate both fuel and energy metabolism, with insulin and glucagon essential to controlling glucose homeostasis (1). Thus, glucagon secreted from α cells stimulates the mobilization of glucose through gluconeogenesis and glycogenolysis, whereas β cell secreted insulin promotes glucose storage. Defects in α and β cell function play a significant role in the ability of individuals with diabetes to maintain glycemic control.
Characterization of the α and β cell enriched transcription factors involved in regulating insulin and glucagon expression has revealed not only their significance to islet function, but also to pancreas organogenesis. For example, Pdx1 is necessary for the growth of the endocrine and exocrine compartments, with pancreatic agenesis observed in pdx1 mutant mice (2, 3). Pdx1 has also been proposed to be a master regulator of β cell activity, in part based on the severe diabetic phenotype observed upon deletion of this factor from β cells in animals, a phenotype at least caused by reduced insulin and glucose transporter type 2 (GLUT2) expression (4).
In contrast to the profound deficiencies in pancreatic endocrine and exocrine cell development observed in the total Pdx1 knockout, the loss of Pax6 (5, 6) or BETA2 [NeuroD1 (7)] only affects endocrine cell production. In fact, changes in endocrine cell development are commonly found in transcription factor knockout models. Thus, loss of Ngn3 leads to a severe reduction in endocrine progenitor cells (8), whereas Nkx6.1-deficient animals display a selective loss of β cells (9). In contrast, there is a switch in cell lineage commitment in Arx (10), Pax4 (11), Pax6 (12), and Nkx2.2 (11) null mutant mice. The notable exceptions are MafA and Brn4 (13), with a deficiency only observed in adult MafA−/− mice, which were glucose intolerant and developed diabetes (14).
The absence of a developmental phenotype in MafA−/− animals was particularly surprising, because this factor is expressed exclusively within the insulin+ cell population produced after embryonic day (E) 13.5 that mature to become islet β cells (15). The Maf basic leucine-zipper-containing transcription factor family consists of two distinct subfamilies, which differ by the presence of an N-terminal transactivation domain in a large Maf (i.e., MafB, MafA, c-Maf, and NRL) that is not found in a small Maf (i.e., MafF, MafG, and MafK). In contrast to MafA, all other closely related members of the large Maf family are involved in processes associated with cell specification, including in the brain [i.e., MafB (16)], kidney [MafB (17, 18)], and immune system [cMaf (19) and MafB (18, 20)].
MafB is expressed in all developing insulin- and glucagon-producing cells, and then in a restricted fashion in adult α cells (21, 22). MafB is also capable of activating insulin- and glucagon-driven transcription in non-hormone-expressing cell types in vitro (21, 23, 24), indicating a possible redundancy with MafA during β cell differentiation. Here we show that MafB is critical for α and β cell differentiation, because the number of insulin+ and glucagon+ cells were substantially reduced in MafB mutant animals throughout embryogenesis. Strikingly, no difference was found between the wild type and MafB mutant in total endocrine cell numbers, which were assessed by pan-endocrine Islet1 (Isl1), NeuroD1, and Pax6 expression. In addition, the level of many α (e.g., Arx, Nkx2.2, Brn4) and β (Nkx2.2, Nkx6.1) cell markers was unchanged in α and β cells (both hormone-expressing and -deficient) at E15.5. However, insulin production was delayed until the onset of MafA expression, and several other gene products required for β cell maturation and function were selectively reduced in hormone− β cells by E18.5 (Pdx1, MafA, Nkx6.1, GLUT2). These results revealed that MafB is required for the generation of the physiologically functional α and β cell population by directly activating hormone gene transcription and key regulators of β cell differentiation and function.
Results and Discussion
MafB−/− Animals Have Reduced Numbers of Insulin+ and Glucagon+ Cells, Whereas the Number of Endocrine Cells Remains Unchanged During Development.
MafB is expressed in all insulin+ and glucagon+ cells during pancreas organogenesis but only in α cells in the adult islet (21, 22). The importance of MafB in endocrine cell formation was examined in homozygous MafB mutant animals, which die at birth because of central apnea [MafB−/− (16)] or renal failure [krENU/krENU (17)]. Glucagon+ cells first appear in the mouse at E9.5, followed by ghrelin+ cells at E10.5 (11), insulin+ cells at E11.5 (25), and somatostatin+ and PP+ at E15.5 and E18.5, respectively (26). MafB is expressed in the distinct first- and second-phase insulin+ and glucagon+ cells made during development, with only second-phase cells producing the α and β cells that mature and populate the islet (27).
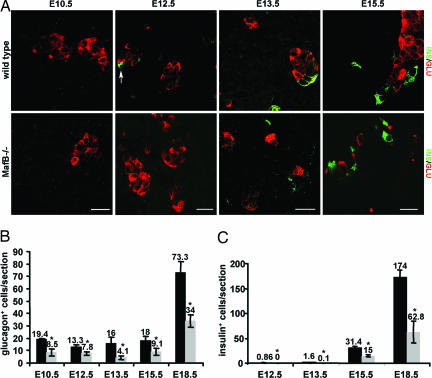
The size and gross appearance of the MafB−/− embryonic pancreata was normal, and insulin- and glucagon-producing cells were detected (Fig. 1A). However, the number of glucagon+ cells was drastically lower at all embryonic stages when compared with the wild type (Fig. 1B). Moreover, insulin+ cells were not detected until E13.5, and their number was reduced to <50% of wild type (Fig. 1B). Glucagon+ cell numbers were also significantly lower at E15.5 in krENU/krENU embryos, in which krENU is a hypomorphic allele of MafB (wild type, 39.1 ± 6.7 glucagon+ cells per section; krENU/krENU, 19.8 ± 6.0), and trended toward fewer insulin+ cells (wild type, 63.3 ± 23.5; krENU/krENU, 40.7 ± 26.8). In contrast, the number of somatostatin- and PP-producing cells was not changed from wild type in either MafB−/− or krENU/krENU pancreata (data not shown). There was also no replacement of α and β cells with the normally minor ghrelin+ cell population (data not shown), an abnormality observed in Pax6 (12), Nkx2.2, and Pax4 null mutants (11).
Fig. 1.
MafB−/− pancreata have fewer insulin+ and glucagon+ cells. (A) Immunofluorescence staining for insulin (green) and glucagon (red) was performed in wild-type and MafB−/− pancreas at E10.5 (glucagon only), E12.5, E13.5, and E15.5. Insulin+ cells are indicated by an arrow at E12.5. Notably, insulin expression was delayed until E13.5 in the MafB mutant, whereas glucagon was observed at all stages. (B) Quantification of insulin+ and glucagon+ cells in wild-type (black bars) and MafB−/− (gray bars) embryos. Because the wild-type and heterozygous MafB embryos showed no difference in hormone cell numbers (data not shown), both were incorporated into the “wild-type” data. The average number of cells per section is shown ± SD, with the asterisk denoting the significance between wild-type and mutant embryos (P < 0.02). (Scale bars: 20 μM.)
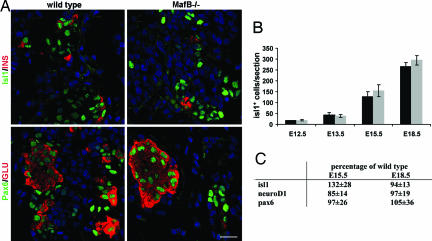
Endocrine cell number is often compromised in animals deficient for islet-enriched transcription factors [e.g., Ngn3 (8) and Nkx6.1 (9)]. To assess whether the reduction of insulin+ and glucagon+ cells was due to a defect in either endocrine cell formation or maintenance, wild-type and MafB−/− pancreata were analyzed for expression of transcription factors produced in the entire endocrine cell population, specifically Islet1 (Isl1) (28), NeuroD1 (7), and Pax6 (29). The number of Isl1+ cells was unchanged at all developmental stages (Fig. 2 A and B). Isl1, Pax6, and NeuroD1 mRNA levels were also similar between wild-type and mutant animals (Fig. 2C). In addition, there was no change in the endocrine cell replication rate as measured by bromodeoxyuridine incorporation in MafB−/− pancreata (data not shown). Collectively, these results demonstrated that endocrine cells are produced in normal numbers in MafB mutant animals, but that a substantial proportion of these cells are incapable of expressing the hormone characteristic of a mature cell.
Fig. 2.
Total endocrine cell numbers are unchanged in MafB−/− pancreata. (A) Immunofluorescence staining of E15.5 wild-type and MafB mutant pancreas sections with α-Isl1 (green), α-insulin (red), α-Pax6 (green), and/or α-glucagon (red). Nuclei were stained with YoPro1 (blue). In the mutant, sections containing a larger number of glucagon and insulin producing cells were selected to illustrate Isl1 and Pax6 coexpression, and not the quantitative decrease in insulin+ or glucagon+ cells found between wild-type and MafB−/− mice. (B) Quantification of Isl1+ cells in wild-type (black bars) and MafB−/− (gray bars) embryos. No significant difference was found in Isl1+ cell numbers at E12.5, E13.5, E15.5, and E18.5. The average number of cells per section is shown ± SD. (C) The real-time PCR mRNA levels of isl1, pax6, and neuroD1 are shown as the percentage of wild type (100%) at E15.5 and E18.5. (Scale bar: 20 μM.)
MafA Is only Produced in the Insulin+ Cells of the MafB Mutant, Whereas Other α and β Cell Differentiation Markers Are Present in Hormone− Endocrine Cells.
Many of the transcription factors required in differentiation and hormone expression are predominantly expressed within one adult islet cell type. For example, MafA, Nkx6.1, and Pdx1 are enriched in β cells, whereas Arx, Brn4, and MafB are principally in α cells, and Nkx2.2 is found in both α and β cells (reviewed in ref. 30). The expression pattern of these cell identity markers was next used to evaluate the differentiation state of the endocrine cells produced in MafB−/− mice.
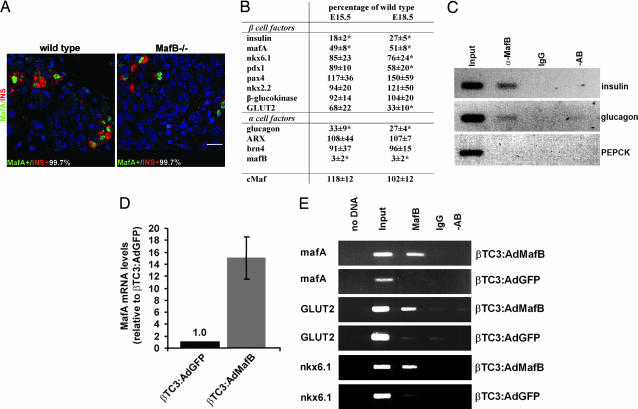
The endocrine cell population was analyzed immunohistochemically for cells expressing MafA, Nkx2.2, Brn4, or both a transcription factor and hormone (MafA/insulin, Nkx2.2/insulin, Brn4/glucagon). The number of cells producing a given transcription factor was set to 100% in this analysis. Nkx2.2 and Brn4 were detected in both hormone− and hormone+ endocrine cells of mutant pancreata at E15.5 [ supporting information (SI) Fig. 6 ]. The percentage of cells expressing both hormone and transcription factor was decreased in MafB mutant pancreatic tissue, whereas cells producing only Nkx2.2+ or Brn4+ increased (SI Fig. 6). The total number of Nkx2.2 and Brn4 producing cells appears to be unchanged despite the presence of fewer insulin+ and glucagon+ cells in MafB mutant embryos. In addition, the mRNA levels for other key α and β cell gene products were unchanged in E15.5 MafB mutant samples (e.g., Isl1, NeuroD1, and Pax6 in Fig. 2C; Pdx1, GLUT2, ARX, glucokinase, and Pax4 in Fig. 3B). These results demonstrated that hormone− endocrine cells express many α and β cell differentiation markers in the MafB mutant.
Fig. 3.
α and β cell identity markers are maintained in hormone− β cells at E15.5. (A) Double immunofluorescence detection of MafA (green) and insulin (red), at E15.5 in wild-type and MafB−/− embryos. Nuclei were stained with YoPro1 (blue). The percentage of total cells expressing MafA and insulin (MafA+/INS+) is shown. (Scale bar: 20 μM.) (B) Relative mRNA level of various α and β cell markers at E15.5 and E18.5 in MafB-deficient pancreata. MafB−/− mRNA levels are shown as the percent of wild type. The asterisk denotes the significance between wild-type and mutant mRNA levels (P < 0.05). (C) ChIP analysis was performed on E18.5 pancreata treated with α-MafB antibody. The precipitated chromatin was analyzed by PCR for insulin (−378/−46), glucagon (−353/+7) and PEPCK (−434/−96) control region sequences. As controls, PCRs were run with input chromatin (1:200 dilution) and DNA obtained after treatment with rabbit IgG or no antibody. (D) MafA mRNA levels in βTC3 cells infected with Ad:GFP (AdGFP:βTC3) or Ad:MafB (AdMafB:βTC3). The real-time PCR data represent the normalized fold difference relative to GFP alone. (E) Chromatin from AdGFP or AdMafB-infected β TC3 cells was precipitated with α-MafB or IgG and analyzed by PCR with mafA (−8120 to −7750), GLUT2 (−1012 to −667), and nkx6.1 (−480 to −141) control region primers. rVISTA and TRANSFAC analysis of upstream GLUT2 and nkx6.1 sequences identified within the primer spanned regions many potential large Maf binding sites very near [GLUT2 (40)] or within [Nkx6.1 (41)] transcriptional regulatory domains.
MafA is first detected during pancreas development in the second wave insulin+ cells that mature and populate the islet (15, 27). The presence of MafA in only insulin+ cells in the MafB mutant distinguished this transcription factor from all others analyzed (Fig. 3A). MafA is presumably produced at normal levels in these cells, because mafA mRNA (Fig. 3B) was reduced by 50%, as found for the total insulin+ cell population. In contrast, mRNA levels were unchanged in the MafB mutant for the related cMaf transcription factor (Fig. 3B), which has been reported to be expressed in low amounts in the embryonic pancreas and a few adult α and β cells (22). Our inability to detect a change in islet development in cMaf −/− animals is also consistent with a much more limited role than MafB in hormone gene transcription or endocrine pancreatic cell formation (data not shown). These results strongly suggest that large Maf factors, principally, if not exclusively MafA and MafB, are critical to insulin transcription during pancreas development.
MafB Binds Within the insulin and glucagon Transcriptional Control Region of Endocrine Cells.
Not only was insulin and glucagon protein expression lost in ≈50% of α and β cells, hormone mRNA levels were reduced even further to 27% of wild type (Fig. 3B). Chromatin immunoprecipitation (ChIP) studies were next performed to determine whether MafB binds within both the insulin and glucagon control regions of wild-type E18.5 pancreata. α-MafB precipitated the large Maf element spanning sequences of both genes, whereas normal IgG or no antibody treatment did not (Fig. 3C). Additionally, α-MafB did not precipitate control region sequences for PEPCK, a gene not expressed in the pancreas. This analysis further supports a direct role for MafB in insulin and glucagon activation in α and β precursor cells.
MafB Stimulates mafA Transcription.
To examine whether the loss of MafA in insulin− endocrine cells results from the direct actions of MafB on mafA transcription, MafB was over-expressed in insulinoma βTC3 cells. Adenoviral-mediated MafB (Ad:MafB) does not affect endogenous insulin, pdx1, or γ-actin expression under these conditions (21), whereas mafA mRNA levels were induced 15-fold (Fig. 3D). Presumably, transcription of insulin and pdx1 is already maximal in β cell lines. ChIP analysis demonstrated that MafB was capable of binding to the β cell-specific control domain of mafA found between base pairs −8118 and −7750 (Fig. 3E) (31), with binding at the base pair −7810/−7782 element detected in gel-shift assays (SI Fig. 7). The combined data support a role for MafB in mafA transcription during β cell development.
Pdx1 and GLUT2 Are only Produced in Insulin+ Cells in E18.5 MafB−/− Embryos.
The maturation of β cells in regards to glucose responsiveness and the promotion of high insulin expression occurs during late gestation in mice (32). Thus, β cells do not become organized into islet structures, obtain high insulin production capacity, or produce significant amounts of membrane-associated GLUT2 until around E18.5 (33). During this period, Pdx1 and Nkx6.1 transcription factor expression becomes principally restricted to developing β cells, with much higher Pdx1 produced in this cell population than surrounding acinar cells (42).
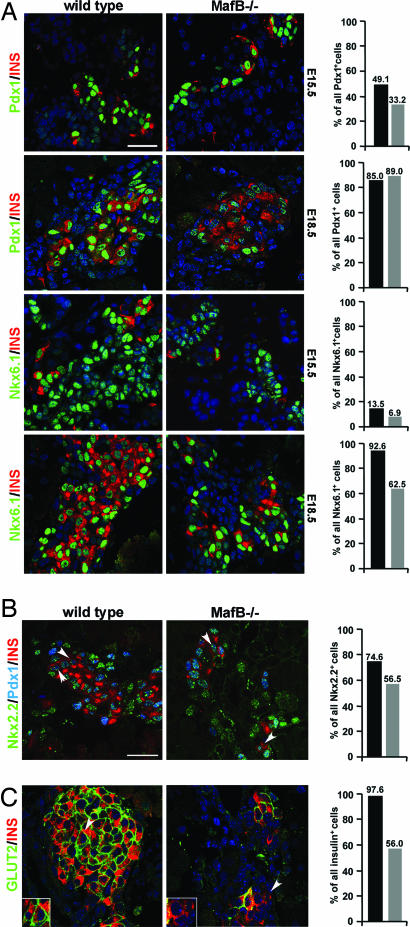
Pdx1 and Nkx6.1 mRNA and protein expression is maintained at wild-type levels in the insulin− endocrine cell population at E15.5 (Figs. 3B and 4A), despite the ≈50% decrease in insulin+ cell number in MafB mutant embryos. However, pdx1 mRNA levels decreased significantly by E18.5 (Fig. 3B), whereas the percentage of mutant Pdx1+/insulin+ cells was the same as wild type (Fig. 4A). In addition, pan endocrine marker expression was unaffected within this developmental time frame (Fig. 2). These results strongly indicated that pdx1 mRNA expression was restricted to insulin+ cells in E18.5 MafB−/− endocrine cells, and that sustaining high Pdx1 expression involves MafB actions in β precursor cells.
Fig. 4.
Pdx1 and GLUT2 are not detected in hormone− β cells at E18.5. (A) Double immunofluorescence staining of wild-type and MafB−/− E15.5 and E18.5 pancreata. Pdx1 (green) and Nkx6.1 (green) are detected in both insulin+ (red) and insulin− endocrine cells in E15.5 MafB−/− embryos. At E18.5, Pdx1 expression is only retained in insulin+ cells, whereas Nkx6.1 expression was lost in some insulin− β cells. The percentage of Pdx1+/insulin+ or Nkx6.1+/insulin+ cells in wild-type (black bars) and MafB mutant embryos (gray bars) within the total Pdx1+ or Nkx6.1+ cell population is shown in the histogram. Only cells producing high levels of Pdx1+ were counted in the analysis, because these represent β precursor cells (42). Nuclei were stained with YoPro1 (blue). (B) Analysis of Nkx2.2 (green), Pdx1 (blue), and insulin (red) coexpression in E18.5 wild-type and MafB−/− pancreata. Nkx2.2 expression is unchanged from wild type in E15.5 and E18.5 MafB−/− embryos (Fig. 3B and SI Fig. 6). The arrows depict the location of Nkx2.2/Pdx1/insulin-expressing cells, with their percentage in wild type (black) and mutant (gray) shown in the graph. The number of Nkx2.2+ only cells increased in MafB mutant pancreata to 37% (wild type, 22%) at the expense of Nkx2.2+/Pdx1+/insulin+ cells, whereas the level of Pdx1+/Nkx2.2+ cells remained constant (E18.5: wild type, 3% of all Nkx2.2+ cells; MafB−/−, 6%). (C) GLUT2 (green) is expressed in all wild-type insulin+ (red) cells at E18.5. Nuclei were stained with YoPro1 (blue). In contrast, MafB mutant pancreata have decreased numbers of GLUT2+/INS+ cells. The histogram depicts the percentage of wild-type (black bars) and MafB−/− (gray bars) GLUT2+/INS+ cells. The arrows indicate the region magnified in Inset. The sections were selected to illustrate insulin and marker gene coexpression. (Scale bars: 20 μM.)
To further examine whether Pdx1 was decreased or specifically lost within a β cell precursor population, immunohistochemical staining for Nkx2.2, Pdx1, and insulin was performed at E18.5. Nkx2.2 is expressed in α and β cell precursors (34) and serves as a positive control in this analysis, because its mRNA and protein levels were unchanged at E15.5 and E18.5 in MafB−/− pancreata (Figs. 3B and 4B, and SI Fig. 6). As predicted Pdx1 was selectively lost in insulin− β cells, the percentage of cells expressing Nkx2.2+/Pdx1+/insulin+ decreased by E18.5 in MafB mutant embryos from 75% to 57% (Fig. 4B), whereas the percentage of cells expressing Nkx2.2 alone increased from 22% to 37% (Fig. 4B). These results demonstrate that Pdx1 is selectively lost from the insulin− endocrine cell population by E18.5, whereas Nkx2.2 expression is maintained in the MafB mutant. We conclude that the loss of Pdx1 reflects the importance of MafB in maintaining expression through activation at the large Maf response element present in the pdx1 regulatory region (35), whereas retention in insulin+ cells represents the actions of MafA.
In contrast to Pdx1, Nkx6.1 expression was still detected in both insulin+ and insulin− β cells in MafB−/− embryos at E18.5 (Fig. 4A). However, Nkx6.1 mRNA levels were reduced in the mutant (Fig. 3B). ChIP analysis performed over the nkx6.1 regulatory region in Ad:MafB-infected βTC3 cells supports that MafB directly regulates transcription (Fig. 3E). Interestingly, the E18.5 MafB−/− embryo also had a higher proportion of Nkx6.1+ only cells, implying that immature β cells accumulate in the absence of MafB.
A hallmark of β cell maturation is the establishment of membrane bound GLUT2. At E18.5, GLUT2 expression was undetected in hormone− endocrine cells and was present in only 56% of the remaining insulin+ cells (Fig. 4C). Consistent with a loss of GLUT2 in insulin+ cells, the E18.5 mutant pancreata contained but 32% of the wild-type GLUT2 mRNA level (Fig. 3B). This effect is unlikely to simply be due to the reduction in Pdx1, a known transcriptional regulator (4), because GLUT2 expression was lost in many insulin and Pdx1 producing cells. The ability to detect MafB binding to upstream GLUT2 regulatory sequences supports a direct role for this factor in activation (Fig. 3E). GLUT2 expression is also decreased in adult MafA-deficient animals (14), further indicating a crucial role for large Maf factors in GLUT2 transcription in β cells. These results demonstrate that MafB activates genes like GLUT2 and pdx1 that are critical to β cell maturation.
Conclusions
Our findings establish MafB as a transcriptional regulator of terminal α and β cell differentiation with previously uncharacterized control properties. A notable phenotype of MafB-deficient animals was the loss of ≈50% of insulin+ and glucagon+ cells while retaining the endocrine cell population and expression of α and β cell identity markers in both hormone− and hormone+ endocrine cells during early islet cell development. Significantly, MafA expression was only detected in insulin+ cells and pdx1 and nkx6.1 levels were specifically reduced in hormone− endocrine cells between E15.5 and E18.5 cells. MafB also profoundly impacted GLUT2 expression by E18.5, resulting in reduced levels in insulin+ cells and complete loss in hormone− cells.
Importantly, the defects in α and β cell development in the MafB null animals are very different from other mouse models with impaired endocrine cell development, which often impact islet cell production by affecting cell lineage commitment, differentiation, and/or the generation of endocrine progenitors. In contrast, MafB appears to be a key regulator of α and β cell maturation, by directly affecting transcription of genes enriched in these cells and crucial to function (Fig. 5). Thus, MafB did not affect the expression of genes broadly expressed in islet cell types (e.g., Pax6, Isl1, and NeuroD1) but only those selectively present in α or β cells (e.g., α: glucagon; β: insulin, MafA, and Pdx1).
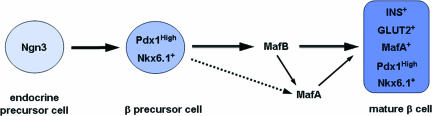
Fig. 5.
Potential role of MafB in β cell development. The model proposes that MafB is required for β cell differentiation and maturation, as supported by the loss of first wave insulin+ cells and the delay in appearance of hormone-producing cells until MafA is made in the MafB mutant. The ≈75% decline in insulin mRNA in MafA+/insulin+ cells (dashed line illustrates the minor role of MafA in insulin production) also indicates that MafB is the principal insulin activator during β cell development. Our results also suggest that MafB is directly required for inducing mafA transcription in β precursor cells and is a primary regulator of pdx1, nkx6.1, and GLUT2 expression in maturing β cells.
Our data also suggests the presence of at least two distinct β cell populations within the developing pancreas. In one, transcription of many key β cell genes is principally dependent on MafB (e.g., insulin, pdx1, mafA, and GLUT2), whereas it is only partially dependent in the other cell population because of the presence of MafA (Fig. 5). The presence of a Maf transcription factor appears to be key to insulin transcription, because the production of insulin+ cells was delayed until the onset of MafA expression in the MafB−/− embryos. MafB may play a similar important role in α cell differentiation, although this was not apparent in our analysis of the comparatively few α cell-enriched products available (i.e., Arx and Brn4). Our findings also strongly indicate that MafB is the principal regulator of insulin and glucagon transcription during development.
Materials and Methods
Animals.
MafB mutant mice were generated by gene targeting (16), with the loxP flanked neomycin selection cassette removed by using ubiquitously expressing sox2 Cre mice (36). KreislerENU/KreislerENU (krENU/krENU) mice were produced by chemical mutagenesis resulting in a point mutation within the MafB DNA binding domain (Asn248 to Ser), which decreased MafB binding affinity by 80% (37). Homozygous MafB mutant animals die at postnatal day 1 because of central apnea [MafB−/− (16)] or renal failure [krENU/krENU (17)]. The day of vaginal plug discovery was designated E0.5.
Immunohistochemistry.
Immunofluorescence and confocal image analyses were performed on paraffin sections as described (23). The primary antibodies used were rabbit α-MafA (1:2,000; Bethyl Laboratories, Montgomery, TX), rabbit α-Pdx1 (1:10,000; Chris Wright, Vanderbilt University Medical School), guinea pig α-insulin (1:2,000; Linco Research, St. Charles, MO), guinea pig α-glucagon (1:2,000; Linco Research), rabbit α-glucagon (1:2,000, Linco Research), rabbit α-Brn4 (1:1,000; Palle Serup, Hagedorn Research Institute, Gentofte, Denmark), rabbit α-Nkx6.1 (1:1,000; Palle Serup), mouse α-Nkx2.2 (1:500; Developmental Studies Hybridoma Bank, Iowa City, IA), mouse α-Isl1 (1:300 Developmental Studies Hybridoma Bank), rabbit α-GLUT2 (1:1,000; Chemicon, Billerica, MA), and rabbit α-Pax6 (1:300; Covance, Princeton, NJ). Secondary antibodies were Cy3- or Cy5-conjugated donkey α-guinea pig, α-mouse, and α-rabbit IgG (1:500; Jackson ImmunoResearch, West Grove, PA). Nuclear counterstaining was performed by using YoPro1 (Molecular Probes, Eugene, OR). Fluorescent images were captured with a Zeiss LSM 510 confocal microscope, using an optical depth of 1 μm.
Adenovirus Infection and RNA Analysis.
Purified recombinant mouse MafB (Ad:MafB) and the GFP control (Ad:GFP) adenoviruses (21) were used to infect β TC3 (2 × 106) cells for 5 h at a multiplicity of infection of 100. Total RNA was prepared from β TC3-infected cells and E15.5 embryonic pancreas anlagen from wild-type and MafB−/− mice by using the RNeasy kit (Qiagen, Valencia, CA). Isolated RNAs were subjected to DNaseI treatment (Versagene RNA DNase kit; Gentra Systems, Minneapolis, MN). Reverse-transcript PCR reagents (TaqMan; Applied Biosystems, Foster City, CA) were used to generate cDNAs from 0.5 μg of DNaseI-treated RNA. PCR mixes were assembled by using SYBR Green Master Mix reagents (Applied Biosystems), and reactions were performed and analyzed by using the Applied Biosystems Prism 7000 sequence detection system and software. The real-time PCR cDNA primer sets for ARX, Brn4, cMaf, γ-actin, glucagon, β-glucokinase, GLUT2, insulin II, MafA, MafB, BETA2, Pax4, Pax6, Nkx2.2, Nkx6.1, and 18s rRNA are available upon request. Relative changes were calculated by the comparative ΔCt method in which γ-actin was used for normalization (38).
ChIP Assay.
β TC3 cells (2 × 107) infected with Ad:GFP or Ad:MafB at a multiplicity of infection of 100 and E18.5 pancreata were formaldehyde cross-linked, and the sonicated protein–DNA complexes were isolated under conditions described previously (23, 39). Sonicated chromatin was incubated 12–14 h at 4°C with α-MafB, and the complexes were isolated with A/G-agarose (Santa Cruz Biotechnology, Santa Cruz, CA). PCR was performed on 1/10 of the purified immunoprecipitated DNA by using Ready-to-Go PCR beads (Amersham Pharmacia Biotech, Rockville, MD) and 15 pmol of the mafA Region 3 (31), insulin (23), PEPCK, glucagon (21), GLUT2 (40), and Nkx6.1 (41) transcriptional control region primers. PCR primer sequence information is available upon request. Amplified products were resolved on a 1.4% agarose gel in Tris-acetate EDTA buffer containing ethidium bromide.
Statistical Analysis.
The entire pancreas anlagen from several animals (n ≥ 3 for each genotype) were used to obtain a representative average of the number of endocrine or hormone+ cells. Immunofluorescence staining was performed on 6-μm sections; positive cells were counted every eighth section throughout the pancreas at E15.5 and E18.5, every fifth section at E12.5 and E13.5, and on all sections at E10.5. The average cell number per section was determined for each individual pancreas (all sections containing pancreatic tissue were examined). The percentage of single-positive (Pdx1+, Nkx2.2+, MafA+, Nkx6.1+, Brn4+) cells to double-positive (Pdx1+/INS+, Nkx2.2+/INS+, Nkx6.1+/INS+, MafA+/INS+, Brn4+/GLU+) or triple-positive (Nkx2.2+/Pdx1+/INS+) cells was measured by counting expressing cells in at least 20 random microscopy fields. Mean differences were tested for statistical significance by using the Student's two tail t test.
Supplementary Material
Acknowledgments
We thank Drs. M. Gannon and C. Wright for critical comments on the manuscript, and Eva Henderson and Amy Schellhaas for technical assistance. This work was supported by National Institutes of Health Grant P01 DK42502 (to R.S.), American Diabetes Association Grant 7-04-RA-116 (to R.S.), and Juvenile Diabetes Research Foundation Advanced Postdoctoral Grant 10-2006-5 (to I.A.). Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (Public Health Service Grant P60 DK20593).
Abbreviation
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700013104/DC1.
References
- 1.Steiner DF, Bell GI, Rubenstein AH, Chan SJ. In: Endocrinology. DeGroot LJ, Jameson JL, editors. Philadelphia: Saunders; 2001. pp. 667–696. [Google Scholar]
- 2.Jonsson J, Carlsson L, Edlund T, Edlund H. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 3.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. Development (Cambridge, UK) 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 4.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 6.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 7.Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Proc Natl Acad Sci USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Development (Cambridge, UK) 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 10.Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Proc Natl Acad Sci USA. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller RS, Jenny M, Collombat P, Mansouri A, Tomasetto C, Madsen OD, Mellitzer G, Gradwohl G, Serup P. Dev Biol. 2005;286:217–224. doi: 10.1016/j.ydbio.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 13.Heller RS, Stoffers DA, Liu A, Schedl A, Crenshaw EB, III, Madsen OD, Serup P. Dev Biol. 2004;268:123–134. doi: 10.1016/j.ydbio.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, et al. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. Proc Natl Acad Sci USA. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchi B, Kelly LM, Viemari JC, Lafon I, Burnet H, Bevengut M, Tillmanns S, Daniel L, Graf T, Hilaire G, et al. Nat Neurosci. 2003;6:1091–1100. doi: 10.1038/nn1129. [DOI] [PubMed] [Google Scholar]
- 17.Sadl V, Jin F, Yu J, Cui S, Holmyard D, Quaggin S, Barsh G, Cordes S. Dev Biol. 2002;249:16–29. doi: 10.1006/dbio.2002.0751. [DOI] [PubMed] [Google Scholar]
- 18.Moriguchi T, Hamada M, Morito N, Terunuma T, Hasegawa K, Zhang C, Yokomizo T, Esaki R, Kuroda E, Yoh K, et al. Mol Cell Biol. 2006;26:5715–5727. doi: 10.1128/MCB.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao S, Liu J, Chesi M, Bergsagel PL, Ho IC, Donnelly RP, Ma X. J Immunol. 2002;169:5715–5725. doi: 10.4049/jimmunol.169.10.5715. [DOI] [PubMed] [Google Scholar]
- 20.Aziz A, Vanhille L, Mohideen P, Kelly LM, Otto C, Bakri Y, Mossadegh N, Sarrazin S, Sieweke MH. Mol Cell Biol. 2006;26:6808–6818. doi: 10.1128/MCB.00245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, Bonner-Weir S, Sharma A. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 25.Herrera PL, Huarte J, Sanvito F, Meda P, Orci L, Vassalli JD. Development (Cambridge, UK) 1991;113:1257–1265. doi: 10.1242/dev.113.4.1257. [DOI] [PubMed] [Google Scholar]
- 26.Teitelman G, Alpert S, Polak JM, Martinez A, Hanahan D. Development (Cambridge, UK) 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 27.Pictet RL, Clark WR, Williams RH, Rutter WJ. Dev Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- 28.Thor S, Ericson J, Brannstrom T, Edlund T. Neuron. 1991;7:881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- 29.Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, Berry A, Gruss P. Dev Biol. 2004;269:479–488. doi: 10.1016/j.ydbio.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 30.Collombat P, Hecksher-Sorensen J, Serup P, Mansouri A. Mech Dev. 2006;123:501–512. doi: 10.1016/j.mod.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R. Mol Cell Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hole RL, Pian-Smith MC, Sharp GW. Am J Physiol. 1988;254:E167–E174. doi: 10.1152/ajpendo.1988.254.2.E167. [DOI] [PubMed] [Google Scholar]
- 33.Pang K, Mukonoweshuro C, Wong GG. Proc Natl Acad Sci USA. 1994;91:9559–9563. doi: 10.1073/pnas.91.20.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Development (Cambridge, UK) 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 35.Samaras SE, Zhao L, Means A, Henderson E, Matsuoka TA, Stein R. J Biol Chem. 2003;278:12263–12270. doi: 10.1074/jbc.M210801200. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi S, Tenzen T, McMahon AP. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- 37.Sadl VS, Sing A, Mar L, Jin F, Cordes SP. Dev Dyn. 2003;227:134–142. doi: 10.1002/dvdy.10279. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonny C, Thompson N, Nicod P, Waeber G. Mol Endocrinol. 1995;9:1413–1426. doi: 10.1210/mend.9.10.8544849. [DOI] [PubMed] [Google Scholar]
- 41.Watada H, Mirmira RG, Leung J, German MS. J Biol Chem. 2000;275:34224–34230. doi: 10.1074/jbc.M004981200. [DOI] [PubMed] [Google Scholar]
- 42.Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD, Serup P. Diabetes. 2000;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- 43.Kataoka K, Noda M, Nishizawa M. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.